Abstract
The levels of 8-oxo-2-deoxyguanosine (oxo8dG) in DNA isolated from tissues of rodents (male F344 rats, male B6D2F1 mice, male C57BL/6 mice, and female C57BL/6 mice) of various ages were measured using sodium iodide to prevent oxidative damage to DNA during DNA isolation. Oxo8dG was measured in nuclear DNA (nDNA) isolated from liver, heart, brain, kidney, skeletal muscle, and spleen and in mitochondrial DNA (mtDNA) isolated from liver. We observed a significant increase in oxo8dG levels in nDNA with age in all tissues and strains of rodents studied. The age-related increase in oxo8dG in nDNA from old mice was shown not to the result of the tissue's reduced ability to remove the oxo8dG lesion. Rather, the increase in oxo8dG levels appears to arise from an age-related increase in the sensitivity of these tissues to oxidative stress. We also observed an age-related increase in oxo8dG in mtDNA isolated from the livers of the rats and mice. Dietary restriction, which is known to retard aging and increase the lifespan of rodents, was shown to significantly reduce the age-related accumulation of oxo8dG levels in nDNA in all tissues of male B6D23F1 mice and in most tissues of male F344 rats. Our study also showed that dietary restriction prevented the age-related increase in oxo8dG levels in mtDNA isolated from the livers of both rats and mice.
The oxidative stress hypothesis of aging (or the free radical hypothesis as it was first proposed) is currently one of the most popular explanations for how aging occurs at the biochemical level. The basic tenet of the oxidative stress hypothesis is that the age-related loss of physiological function and aging are because of the progressive and irreversible accumulation of oxidative damage (1). Over the past decade, the oxidative stress hypothesis of aging has gained wide acceptance because numerous studies have shown a strong correlation between increasing age and the accumulation of oxidative damage to cellular macromolecules (2, 3) and because the increased survival observed with dietary restriction has been correlated to reduced oxidative damage (3, 4). It also appears that certain types of pathological lesions that arise with age are associated with increased levels of oxidative damage to cellular macromolecules (5, 6).
In 1990, Ames' laboratory reported the first data on the effect of aging on DNA oxidation (7). They observed a significant (≈2-fold) increase in 8-oxo-2-deoxyguanosine (oxo8dG) levels in nuclear DNA (nDNA) isolated from liver, kidney, and intestine of male rats between 2 and 24 months of age. Later, Ames et al. (5) reported that the levels of oxo8dG in mitochondrial DNA (mtDNA) isolated from male rat liver increased 2- to 3-fold with age. Since 1990, a number of research groups have observed an age-related increase in the level of oxo8dG in both nDNA and mtDNA in a variety of tissues of rats and mice (8).
On the other hand, many investigators have been unable to detect a significant increase in DNA oxidation in rodent tissues with increasing age (9). The most likely explanation for the contradictory results is artifactual DNA oxidation, which arises during the isolation and analysis of the DNA samples. Since the report by Claycamp (10) that phenol can induce DNA oxidation during DNA extraction, there has been a great deal of debate about the actual in vivo levels of oxo8dG in DNA. Recently, two groups showed that DNA isolated using sodium iodide (NaI) had very low levels of oxo8dG compared with the commonly used phenol method (11–13), and our laboratory showed that the generation of oxo8dG during DNA isolation was eliminated using the NaI isolation method (14). All of the studies reported to date on the effect of aging on DNA oxidation have isolated DNA from tissues of rodents using either phenol or other organic solvents, which give relatively high levels of oxo8dG because of oxidative damage to DNA during the isolation process. In a recent review, Lindahl and Wood (15) concluded, “previous reports on the presence of high quantities of oxo8dG in organ DNA as an apparent consequence of physiological aging should be interpreted with caution.”
The purpose of this study was to determine the effect of age and dietary restriction on the levels of oxo8dG in DNA isolated from rodent tissues using the NaI method to eliminate/minimize the artifactual generation of oxo8dG during DNA isolation. We measured the levels of oxo8dG in DNA isolated from a wide variety of tissues in both rats and various mouse strains to determine the universality of the age-related changes in DNA oxidation, i.e., were the changes tissue- or strain-specific. We observed an age-related increase in oxo8dG levels in nDNA isolated from all tissues studied from both rats and mice and an increase in oxo8dG levels in mtDNA isolated from the livers of both rats and mice. Dietary restriction reduced the levels of oxo8dG in nDNA from most tissues studied as well as in liver mtDNA of both rats and mice.
Materials and Methods
Animals and Diet.
Male Fischer 344 (F344) rats and male B6D2F1 (C57BL/6 × DBA/2) mice fed either ad libitum or food-restricted diets were obtained from the National Institute on Aging. The F344 rats were 6, 18, and 24 months of age, and the B6D2F1 were 6 and 25 months of age when used in the experiments described below. The F344 rats and B6D2F1 mice were housed individually under barrier conditions. Starting at 16 weeks of age, the food-restricted rats or mice were given 60% of the diet consumed by a companion group of rats or mice fed ad libitum as described previously (16). The mean survival for rats and mice maintained by the National Institute on Aging is 26 and 35 months for male F344 rats fed ad libitum and a food-restricted diet, respectively, and 31 and 47 months for male B6D2F1 mice fed ad libitum and a food-restricted diet, respectively (17).
The male and female C57BL/6 mice used in this study were obtained from the aging colonies of mice maintained by the San Antonio Nathan Shock Aging Center. The male mice were 6 and 26 months of age when used in the experiments described below, whereas the female mice were 6, 14, and 26 months of age. These mice were group housed, four mice per cage, and fed ad libitum. The mean survival of the male and female C57BL/6 mice in these colonies was 29 and 33 months, respectively.
The rodents were humanely euthanized between 9:00 and 11:00 a.m. to minimize diurnal variation. Tissues were immediately collected and frozen in liquid nitrogen. The tissues were stored at −80°C and analyzed within 30 days of collection. In one experiment, mice were exposed to 2 Gy of whole-body γ-irradiation using a 137Cs GammaCell-40 Irradiator (Atomic Energy, Ottawa) that produced 136 cGy/min. Mice were euthanized at various times (0, 7.5, 15, and 30 min) after γ-irradiation to follow the removal of oxo8dG from the DNA. The tissues were collected and immediately frozen in liquid nitrogen. All procedures for handling the rats and mice were approved by the Institutional Animal Care and Use Committee of the University of Texas Health Science Center at San Antonio and the Subcommittee for Animal Studies at the Audie L. Murphy Memorial Veterans Hospital.
Isolation and Analysis of DNA.
Nuclear DNA was isolated using a DNA Extractor WB kit (Wako Chemicals, Richmond, VA) as modified by Hamilton et al. (14). The liver (50–100 mg), brain (100–150 mg), kidney (150–200 mg), and spleen (40–50 mg) were homogenized in a Dounce homogenizer in ice-cold lysis solution, and the heart was homogenized in a ground-glass homogenizer in ice-cold lysis solution. The skeletal muscle (entire muscle from the two hind limbs, ≈500 mg) was ground into a powder in liquid nitrogen and then homogenized in ice-cold lysis solution using a Dounce homogenizer. Nuclei were collected by centrifuging the homogenate at 10,000 × g for 20 s, and the nuclear pellets were resuspended in the enzyme reaction solution and proteinase K (10 μg/ml) provided with the kit. RNase mixture (Ambion, Austin, TX) was then added to a final concentration of 20 μg/ml. Mitochondrial DNA was isolated using mtDNA Extractor CT kit (Wako Chemicals). Briefly, mitochondria were isolated from livers pooled from five mice or one whole rat liver. The homogenates were centrifuged at 1,000 × g for 1 min. The supernatants were collected and centrifuged at 10,000 × g for 10 min. The pellets, which contained mitochondria, were treated with the solutions in the mtDNA extractor kit.
In total, 50–75 μg of nDNA and mtDNA were hydrolyzed as described by Kasai et al. (18). The 8-oxo-2-deoxyguanosine (oxo8dG) and 2-deoxyguanosine were resolved by high pressure liquid chromatography and quantified by electrochemical detection as described previously (14). The data are expressed as the ratio of nmol of oxo8dG to 105 nmol of 2-deoxyguanosine.
Antioxidant Enzyme Activities.
The activities of Mn and CuZn superoxide dismutase (SOD), catalase, and glutathione peroxidase were measured in tissue extracts obtained from young and old female C57BL/6 mice. The tissues (100 mg) were homogenized in 10 vol of ice-cold homogenization buffer (10 mM KH2PO4, pH 7.4/20 μM EDTA/30 mM KCl) using a ground-glass homogenizer. The homogenates were centrifuged at 13,600 × g for 10 min, and the supernatant was used to measure the activities of the antioxidant enzymes using activity gels to measure simultaneously Mn-SOD and CuZn-SOD activity as described by Beauchamp and Fridovich (19), or glutathione peroxidase and catalase activities as described by Sun et al. (20), using cumene hydroperoxide (0.008%) and hydrogen peroxide (0.003%), respectively. The activities of the enzymes were determined using concentrations of the tissue extracts in which the assays were linear and expressed as relative units per mg of protein.
Results
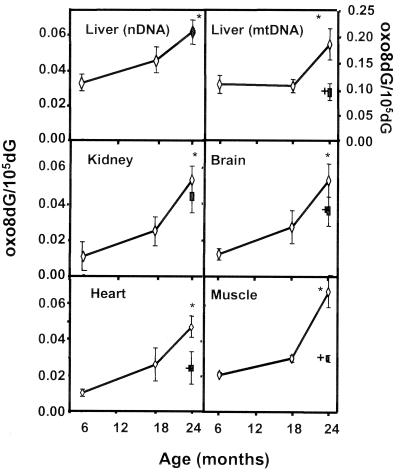
Using the NaI method to isolate DNA, we first measured the levels of oxo8dG in nDNA from male F344 rats of various ages because the initial studies by Ames' laboratory (7) used this strain of rats and because the majority of the studies that have compared DNA oxidation in young and old rodents have used male F344 rats. As can be seen in Fig. 1 and Table 1, we observed a significant age-related increase in oxo8dG levels in nDNA in all tissues studied, e.g., liver, skeletal muscle, brain, kidney, and heart. The age-related increase in oxo8dG from 6 to 24 months of age ranged from 88% for liver, to 225% for skeletal muscle, to 340% for kidney and brain, to 370% for heart. This increase in DNA oxidation occurred primarily in the later half of life (Fig. 1). We also measured the levels of oxo8dG in mtDNA isolated from the livers of the rats. The level of oxo8dG in mtDNA from male F344 liver increased 70% between 6 and 24 months of age.
Figure 1.
DNA oxidation in tissues of male F344 rats. nDNA was isolated from the liver, brain, kidney, heart, and skeletal muscle, and mtDNA was isolated from liver tissue of 6-, 18-, and 24-month-old rats fed ad libitum (○) and 24-month-old rats fed a caloric-restricted diet (▪). Each value is the mean ± SEM for data collected from six rats, and the data were analyzed using a one-way ANOVA with a Bonferonni test to show significance. *, The oxo8dG/105dG ratio in nDNA and mtDNA was significantly higher (P < 0.05) for the 24-month-old rats fed ad libitum compared with 6- or 18-month-old rats fed ad libitum. †, The oxo8dG/105dG ratio in nDNA and mtDNA was significantly higher (P < 0.05) for the 24-month-old caloric-restricted rats compared with the 24-month-old rats fed ad libitum.
Table 1.
Effect of age and dietary restriction on the levels of DNA oxidation in tissues from rats and mice
| oxo8dG/105dG ratio
|
||||
|---|---|---|---|---|
| 6 months ad libitum | 14–18 months ad libitum | 24–26 months* ad libitum | 24–26 months calorie restricted | |
| Male F344 rats | ||||
| Liver | 0.033 ± 0.005 (6) | 0.046 ± 0.007 (6) | 0.062 ± 0.001 (6) | 0.062 ± 0.007 (6) |
| Liver (mtDNA) | 0.108 ± 0.017 (6) | 0.105 ± 0.012 (6) | 0.185 ± 0.029 (6) | 0.093 ± 0.016† (6) |
| Brain | 0.012 ± 0.003 (6) | 0.027 ± 0.009 (6) | 0.053 ± 0.009 (6) | 0.036 ± 0.008† (6) |
| Kidney | 0.011 ± 0.008 (6) | 0.025 ± 0.008 (6) | 0.053 ± 0.008 (6) | 0.044 ± 0.009 (6) |
| Heart | 0.010 ± 0.002 (6) | 0.026 ± 0.009 (6) | 0.047 ± 0.006 (6) | 0.024 ± 0.009† (6) |
| Muscle | 0.021 ± 0.001 (6) | 0.030 ± 0.002 (6) | 0.068 ± 0.009 (6) | 0.030 ± 0.002† (6) |
| Male B6D2F1 mice | ||||
| Liver | 0.035 ± 0.003 (18) | ND | 0.048 ± 0.004 (18) | 0.039 ± 0.004† (18) |
| Liver (mtDNA) | 0.133 ± 0.023 (3) | ND | 0.202 ± 0.026 (3) | 0.118 ± 0.024† (3) |
| Brain | 0.012 ± 0.003 (18) | ND | 0.032 ± 0.005 (18) | 0.025 ± 0.003† (18) |
| Kidney | 0.027 ± 0.002 (18) | ND | 0.033 ± 0.003 (18) | 0.029 ± 0.003† (18) |
| Heart | 0.012 ± 0.004 (18) | ND | 0.039 ± 0.007 (18) | 0.026 ± 0.005† (18) |
| Muscle | 0.025 ± 0.002 (7) | ND | 0.038 ± 0.007 (7) | ND |
| Female C57BL/6 mice | ||||
| Liver | 0.029 ± 0.002 (8) | 0.029 ± 0.003 (8) | 0.040 ± 0.003 (8) | ND |
| Liver (mtDNA) | 0.198 ± 0.066 (3) | ND | 0.447 ± 0.041 (8) | ND |
| Brain | 0.012 ± 0.001 (8) | 0.017 ± 0.003 (8) | 0.041 ± 0.003 (8) | ND |
| Kidney | 0.014 ± 0.002 (8) | 0.019 ± 0.004 (8) | 0.029 ± 0.003 (8) | ND |
| Heart | 0.021 ± 0.001 (8) | 0.025 ± 0.003 (8) | 0.051 ± 0.003 (8) | ND |
| Muscle | 0.025 ± 0.004 (7) | ND | 0.037 ± 0.005 (7) | ND |
| Spleen | 0.019 ± 0.002 (5) | 0.019 ± 0.002 (5) | 0.025 ± 0.001 (5) | ND |
| Male C57BL/6 mice | ||||
| Liver | 0.032 ± 0.003 (5) | ND | 0.047 ± 0.003 (5) | ND |
| Brain | 0.012 ± 0.003 (5) | ND | 0.042 ± 0.002 (5) | ND |
| Kidney | 0.022 ± 0.005 (5) | ND | 0.035 ± 0.001 (5) | ND |
| Heart | 0.023 ± 0.009 (5) | ND | 0.041 ± 0.003 (5) | ND |
nDNA and mtDNA (noted in parentheses) were isolated from various tissues of rats and mice. The numbers in parentheses represent the number of animals in each group, except for the mtDNA isolated from the mice, which represents three groups of liver pooled from five mice. All values represent the mean ± SEM, and the data were analyzed using a one-way ANOVA with a Bonferroni test to show significance.
The oxo8dG/105dG ratio in nDNA and mtDNA for the tissue was significantly higher (P < 0.05) for all the old rats or mice fed ad libitum compared to the younger rats or mice.
The oxo8dG/105dG ratio in nDNA and mtDNA for the tissue was significantly higher (P < 0.05) for the old caloric-restricted rats or mice compared to the old rats or mice fed ad libitum.
We also measured the levels of oxo8dG in tissues from young and old male B6D2F1 mice and male C57BL/6 mice. The data in Table 1 show that an age-related increase in oxo8dG levels was observed in nDNA isolated from all tissues studied. The age-related increase in oxo8dG levels for male B6D2F1 mice ranged from 21% for kidney, to 37% for liver, to 52% for skeletal muscle, to 167% for brain, to 225% for heart. For the male C57BL/6 mice, the increase in oxo8dG levels ranged from 47% for liver, to 60% for kidney, to 78% for heart, to 250% for brain. We also observed a 50% increase in oxo8dG levels in mtDNA isolated from the livers of old B6D2F1 mice compared with young B6D2F1 mice.
The levels of oxo8dG in nDNA isolated from female C56BL/6 mice of various ages were also observed to increase significantly with age (Table 1). The age-related increase in oxo8dG levels ranged from 32% for spleen, to 38% for liver, to 48% for skeletal muscle, to 107% for kidney, to 143% for heart, to 241% for brain. The levels of oxo8dG in mtDNA from liver also increased significantly (126%) with age. The data in Table 1 also show that the age-related increase in oxo8dG levels in tissues of the female C57BL/6 mice occurred primarily in the later half of life, which is identical to what we observed for male F344 rats.
The effect of dietary restriction on DNA oxidation was also studied in tissues from male F344 rats and male B6D2F1 mice. The levels of oxo8dG in nDNA from the brain, heart, and skeletal muscle of food-restricted, 24-month-old rats were significantly lower than those of 24-month-old rats fed ad libitum. However, dietary restriction had no effect on the levels of oxo8dG found in nDNA from the liver and kidney of the rats. In contrast, dietary restriction completely prevented the age-related increase in oxo8dG levels in liver mtDNA in the male F344 rats. In male B6D2F1 mice, dietary restriction significantly reduced the levels of oxo8dG in nDNA from all tissues studied, e.g., liver, kidney, brain, heart, and kidney. In addition, dietary restriction prevented the age-related increase in oxo8dG levels in liver mtDNA of the B6D2F1 mice.
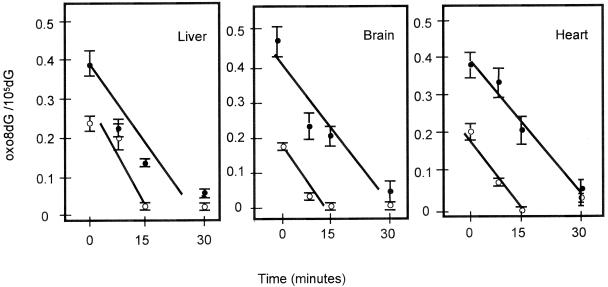
To gain an understanding of the mechanism responsible for the age-related increase in oxo8dG in tissues of rodents, we measured the ability of tissues from young and old mice to remove oxo8dG. We exposed young and old female C57BL/6 mice to a low dose of γ-irradiation (2 Gy), which significantly induced DNA oxidation but was not lethal (21). The ability of tissues from the irradiated mice to repair the oxidative damage was measured by the disappearance of oxo8dG. The data in Fig. 2 show that γ-irradiation induced the levels of oxo8dG in the liver, brain, and heart ≈10-fold compared with the endogenous levels of oxo8dG found in untreated mice (Table 1). The oxo8dG levels rapidly decreased after irradiation in the three tissues of both young and old mice such that the oxo8dG levels returned to untreated levels within 15 min for the young animals and within 30 min for the old animals. The data in Fig. 2 show that the rate of removal of oxo8dG from nDNA in liver, brain, and heart is similar for young and old mice. We also measured the ability of nuclear extracts from young and old female mice to remove oxo8dG as described by Street et al. (22). We observed no decrease in the ability to remove oxo8dG in the liver nuclear extracts from old mice compared with young mice, e.g., the activity was 0.56 ± 0.04 versus 0.64 ± 0.05 fmol of oligonucleotide cleaved/μg protein (mean ± SEM for four mice per group) for young and old mice, respectively.
Figure 2.
Effect of age on the removal of oxo8dG from nDNA following acute whole-body γ-irradiation. Young (○) and old (●) female C57BL/6 mice were exposed to 2 Gy of γ-radiation, and nDNA was isolated from liver, brain, and heart immediately after irradiation and again at 7.5, 15, and 30 min after irradiation. Each value represents the level of oxo8dG induced by γ-irradiation, i.e., the oxo8dG levels in the untreated tissues (given in Table 2) have been subtracted from the levels of oxo8dG obtained after irradiation. Each value is expressed as a mean ± SEM of data from six mice. The rate of removal of oxo8dG from the nDNA was calculated by using a MICROSOFT EXCEL graphing program that generated the best-fitting straight line beginning at the point of maximal damage (time 0) continuing through the time points until the oxo8dG/105dG levels reached baseline. The EXCEL program then generated the slope of the line, and these lines are shown in the figure. The rates of oxo8dG removal were 0.113 ± 0.049 vs. 0.109 ± 0.025 oxo8dG/105dG per min for livers from young and old mice, respectively; 0.134 ± 0.033 vs. 0.086 ± 0.009 oxo8dG/105dG per min for brains from young and old mice, respectively; and 0.118 ± 0.032 vs. 0.100 ± 0.029 oxo8dG/105dG per min for hearts from young and old mice, respectively. The rates of oxo8dG removal for young and old mice were compared statistically using a Student's t test. No statistically significant difference was observed between young and old mice for the three tissues. However, the levels of oxo8dG were significantly higher (P < 0.001 level) for the old mice than the young mice at all time points except the 30-min time point for all three tissues and the 7.5-min time point for liver.
One unexpected observation from the γ-irradiation experiment was the dramatic difference in the levels of oxo8dG induced by γ-irradiation in the tissues of the old mice compared with tissues from young mice. For example, the levels of oxo8dG induced immediately after γ-irradiation in nDNA were higher (60%, 85%, and 170% for liver, heart, and brain, respectively) for old mice compared with young mice. The higher levels of oxo8dG in nDNA from the tissues of the old mice persisted until 30 min after γ-irradiation when the levels of oxo8dG returned to untreated levels. To determine whether the greater induction of oxo8dG in nDNA from old rats after γ-irradiation was caused by a reduced antioxidant defense system, we measured the activities of the four major antioxidant enzymes in the liver, heart, and brain of young and old mice. The data in Table 2 show that the activities of Mn-SOD, CuZn-SOD, catalase, and glutathione peroxidase were similar in tissues from of young and old mice.
Table 2.
The effect of age on the activities of the major antioxidant enzymes
| Tissue | Antioxidant enzyme | Enzyme activity relative units/mg protein
|
|
|---|---|---|---|
| Young | Old | ||
| Liver | Catalase | 4.65 ± 0.17 | 4.56 ± 0.09 |
| Glutathione peroxidase | 7.16 ± 0.60 | 8.36 ± 0.28 | |
| Mn superoxide dismutase | 11.52 ± 1.32 | 12.12 ± 0.31 | |
| CuZn superoxide dismutase | 192.53 ± 20.23 | 174.28 ± 16.19 | |
| Heart | Catalase | 0.42 ± 0.07 | 0.51 ± 0.02 |
| Glutathione peroxidase | 0.40 ± 0.06 | 0.61 ± 0.02 | |
| Mn superoxide dismutase | 63.08 ± 1.95 | 68.04 ± 3.69 | |
| CuZn superoxide dismutase | 66.92 ± 0.33 | 73.20 ± 3.91 | |
| Brain | Catalase | 0.29 ± 0.03 | 0.34 ± 0.06 |
| Glutathione peroxidase | 0.86 ± 0.12 | 0.90 ± 0.07 | |
| Mn superoxide dismutase | 13.76 ± 1.02 | 18.01 ± 1.34 | |
| CuZn superoxide dismutase | 43.31 ± 1.69 | 46.53 ± 4.75 | |
The activities of the various antioxidant enzymes were measured in tissue extracts isolated from young (6–7 months) and old (26–28 months) female C56Bl/6 mice as described in Materials and Methods. Each value represents the mean ± SEM of data from four mice.
Discussion
Since the initial report by Fraga et al. in 1990 (7), there has been a great deal of controversy over whether DNA oxidation (i.e., oxo8dG levels) increases with age. A significant increase in oxo8dG levels in nDNA has been observed with age in a number of studies (7, 23, 24). However, an equal number of studies have reported no significant change in oxo8dG levels with age (9, 25–27). Using the NaI method, which eliminates oxidative damage to DNA during isolation (14), we conducted a comprehensive study on the effect of age on DNA oxidation. A significant increase in oxo8dG levels in nDNA was observed in every tissue and strain of rodent studied. Thus, our data demonstrate that DNA oxidation increases with age and that this increase appears to be universal in all tissues of rodents. However, the increase in oxo8dG levels varies considerably from tissue to tissue and from rodent strain to rodent strain. For example, the age-related increase in DNA oxidation was greater in all tissues of F344 rats than in the same tissues in the mouse strains studied. This greater age-related increase in oxo8dG levels in F344 rats is correlated with a shorter lifespan for the F344 rats compared with the mouse strains studied, e.g., mean survival of 26 months for F344 rats compared with 29 to 33 months for the mice (see Materials and Methods). In addition, the age-related increase in oxo8dG levels in mice was generally the greatest in brain and heart tissue compared with other tissues, e.g., liver. Previous studies have also reported that the age-related increase in oxo8dG was greater in brain and heart of mice compared with liver (28, 29). The most likely explanation for the greater age-related increase in oxidative damage in nDNA from brain and heart is the higher metabolic activity of these tissues compared with other tissues and, thus, the greater potential exposure of the nDNA to oxidative stress. In addition, because these are postmitotic tissues, cell turnover would be minimal in brain and heart compared with other tissues.
Previous studies have reported an age-related increase in oxo8dG levels in mtDNA isolated from liver (5, 8), brain (30), and heart (31) of rats and mice. However, other studies have reported a decrease in oxo8dG levels in mtDNA with age (32, 33). In our experiments, we used large amounts of liver tissue to increase the yield of mtDNA because Beckman and Ames (34) showed that measurements of oxo8dG with small amounts of mtDNA gave apparent elevated levels of oxo8dG in the DNA sample. We observed that the levels of oxo8dG in mtDNA isolated from the livers of old rodents was 50% to 124% higher than that observed in mtDNA isolated from young rodents. Thus, our data show that oxidative damage increases with age for both nDNA and mtDNA.
Why does oxidative damage to DNA increase with age? A simple prediction is that the increase is because of a decline in the ability of cells to repair DNA damage. Over the past two decades, reports have shown an age-related decline in nucleotide excision repair (35, 36). However, there is very little information on the effect of age on base excision repair, the DNA repair pathway primarily responsible for repairing oxidative base damage. Our data show that the age-related increase in oxo8dG was not because of a deficit in the ability of the cells from old animals to remove oxo8dG. However, tissues of the old animals appear to be more sensitive to the generation of oxidative damage to DNA when exposed to an oxidative stress, e.g., γ-irradiation. This observation is important because it suggests that the most effective strategy to prevent the age-related increase in the accumulation of oxidative damage to DNA would be the use of compounds that scavenge or prevent the generation of reactive oxygen species, e.g., antioxidants or metal chelators. Other investigators have also observed that cells or tissues from old animals are more sensitive to oxidative stress in vitro than cells or tissues from young animals. For example, Zahn et al. (37) reported that DNA damage (strand breaks) induced after incubating tissues (liver, heart, brain, kidney, lung, skeletal muscle, and intestine) with nitroquinolin-N-oxide in vitro was 2-fold greater for tissues from old mice compared with tissues from young mice. Guo et al. (36) found that more DNA damage (cyclobutane pyrimidine dimers) was induced by UV-irradiation in the nDNA of primary cultures of hepatocytes isolated from old rats compared with hepatocytes isolated from young rats. Rikans and Cai (38) also showed that diquat induced more oxidative damage to lipid and protein in primary cultures of hepatocytes isolated from old rats compared with hepatocytes isolated from young rats. Our report shows that the in vivo sensitivity of tissues of whole animals to oxidative stress increases with age. Although our data do not explain why the tissues of the old mice are more sensitive to the induction of oxo8dG by γ-irradiation, the increase does not appear to be due to age-related changes in the activities of the major antioxidant enzymes (Table 2). Rikans et al. (39) also concluded that the increased sensitivity of hepatocytes from old rats to diquat was not because of changes in enzymatic mechanisms that protect against oxidative damage; rather, they found that hepatocytes from old rats were more sensitive to diquat because of higher levels of ferritin iron in the livers of the old rats. An age-related increase in iron content has been reported in the basal ganglia in human brain tissue (40) as well as in the kidney, liver, and brain tissues of rats (41) and brain tissue of mice (42). Therefore, it is possible that the tissues of the old mice in our study were more sensitive to the induction of oxo8dG in DNA by γ-irradiation because of enhanced levels of iron that potentate the oxidative stress induced by γ-irradiation.
To assess the importance of the increase in DNA oxidation in the aging process, we asked if dietary restriction altered the age-related accumulation of oxo8dG. Over the past two decades, it has been well established that chronic restriction of food (calories) delays the onset of most age-related diseases, alters most physiological processes that change with age, and extends lifespan (3, 43). These data provide convincing evidence that dietary restriction increases survival of rodents by retarding aging. Therefore, if the age-related increase in DNA oxidation is important in aging, the increase should be retarded/reduced by dietary restriction. A few reports indicate that dietary restriction reduces the level of DNA oxidation in tissues of rodents (25, 44, 45); however, all of these studies have used phenol to isolate DNA. We observed a significant decrease in oxo8dG levels in nDNA in all tissues of old mice fed a caloric-restricted diet compared with old mice fed ad libitum. We also observed a decrease in oxo8dG in nDNA from brain, heart, and skeletal muscle in caloric-restricted rats; however, dietary restriction did not decrease the oxo8dG levels in nDNA from liver and kidney of rats. Thus, dietary restriction did not always reduce oxidative damage to nDNA in rodent tissues, and even in those tissues in which dietary restriction significantly reduced oxidative damage to nDNA, the decrease was often relatively mild (e.g., dietary restriction only reduced oxo8dG levels 14–19% in liver and kidney of mice). In contrast, dietary restriction had a dramatic effect on the level of oxo8dG in mtDNA. For example, dietary restriction completely prevented the age-related increase in oxo8dG levels in liver mtDNA in rats and mice even though dietary restriction had no effect on the age-related increase in oxo8dG levels in rat liver nDNA and reduced oxo8dG levels only 19% in mouse liver nDNA. Thus, our data suggest that dietary restriction has a much greater effect on the age-related accumulation of oxidative damage in mtDNA than nDNA. However, additional experiments with mtDNA from other tissues are needed to determine whether this effect of dietary restriction on oxidative damage to mtDNA is seen in tissues other than liver. The observation that dietary restriction has a greater effect on the age-related accumulation of oxo8dG in brain and heart than kidney and liver suggests that oxidative damage to some tissues might be more important than others with regard to the aging phenotype. However, there is not a direct relationship between the effect of dietary restriction on the age-related accumulation of oxo8dG and pathology. For example, dietary restriction has been shown to have a major effect on the age-related increase in pathological lesions in the kidneys of male F344 rats (46). However, we observed no significant effect of dietary restriction on the levels of oxo8dG in the kidneys of old, male F344 rats.
In summary, our data clearly demonstrate that oxidative damage to DNA increases significantly with age in rodents. Based on the values of oxo8dG, we measured in nDNA from female C57BL/6 mice and, assuming a GC content of 40% for nDNA (47), we calculated the steady-state levels of oxo8dG in brain nDNA. The oxo8dG lesions increase from 3 per 108 bases for a young mouse to 8 per 108 bases for an old mouse. In other words, the nuclear genome of the brain of a young mouse would contain a steady-state level of 180 oxo8dG lesions compared with 640 oxo8dG lesions for an old mouse. Based on the half-life of oxo8dG in brain, which we calculated to be 12.5 min for young and old mice (Fig. 2), we estimated that the de novo formation of oxo8dG in brain tissue of a young mouse would be ≈27,000 oxo8dG lesions in a 24-h period, compared with 96,000 oxo8dG lesions in a 24-h period for a brain cell from an old mouse. Because oxo8dG makes up ≈5% of all oxidative lesions (12), it would appear that a brain cell of a young mouse would be exposed to ≈500,000 oxidative lesions per day compared with almost 2 million oxidative lesions per day for a brain cell from an old mouse, assuming the generation and removal of all of the oxidative lesions occur at similar rates as oxo8dG. Thus, although the steady-state levels of oxo8dG are relatively small in the genome of rodent cells, the de novo formation of oxo8dG lesions is considerable, suggesting that oxidative damages to DNA that arise from normal cellular metabolism could be highly relevant in aging and diseases associated with age.
Acknowledgments
This work was supported by a Merit Review grant from the Department of Veteran Affairs, National Institutes of Health Grant AG13319, Program Project Grant PO1AG14674, and Nathan Shock Center of Excellence in Basic Biology of Aging Grant PO3AG13319 at the University of Texas Health Science Center at San Antonio.
Abbreviations
- oxo8dG
8-oxo-2-deoxyguanosine
- nDNA
nuclear DNA
- mtDNA
mitochondrial DNA
- SOD
superoxide dismutase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
To whom reprint requests should be addressed. E-mail: richardsona@uthscsa.edu.
References
- 1.Stadtman E R. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 2.Warner H R. Free Radical Biol Med. 1994;17:249–258. doi: 10.1016/0891-5849(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Sohal R, Weindruch R. Science. 1996;273:59–67. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu B P. J Nutr Sci Vitaminol. 1993;39:575–583. doi: 10.3177/jnsv.39.supplement_s75. [DOI] [PubMed] [Google Scholar]
- 5.Ames B N, Shigenaga M K, Hagen T M. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen D R, Siddique T, Patterson D, Figlewicz D A, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan J P, Deng H-X, et al. Nature (London) 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 7.Fraga C G, Shigenaga M K, Park J, Degan P, Ames B N. Proc Natl Acad Sci USA. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson E K, Hogue B A, Souza-Pinto N C, Croteau D L, Anson R M, Bohr V A, Hansford R G. Free Radical Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 9.Anson R M, Senturker S, Dizdaroglu M, Bohr V A. Free Radical Biol Med. 1999;27:456–462. doi: 10.1016/s0891-5849(99)00091-x. [DOI] [PubMed] [Google Scholar]
- 10.Claycamp H G. Carcinogenesis. 1992;13:1289–1292. doi: 10.1093/carcin/13.7.1289. [DOI] [PubMed] [Google Scholar]
- 11.Nakae D, Mizumoto Y, Kobayashi E, Noguchi O, Konishi Y. Cancer Lett. 1995;97:233–239. doi: 10.1016/0304-3835(95)03980-b. [DOI] [PubMed] [Google Scholar]
- 12.Helbock H, Beckman K, Shigenaga M, Walter P B, Woodall A, Yeo H C, Ames B N. Proc Natl Acad Sci USA. 1998;95:288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakae D, Akai H, Kishida H, Kusuoka O, Tsutsumi M, Konishi Y. Lab Invest. 2000;80:249–261. doi: 10.1038/labinvest.3780028. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton M L, Guo Z M, Fuller C D, Van Remmen H, Ward W F, Austad S N, Troyer D A, Thompson I, Richardson A. Nucleic Acids Res. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl T, Wood R D. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 16.Lipman R D, Smith D E, Bronson R T, Blumberg J. Aging. 1995;7:136–139. doi: 10.1007/BF03324303. [DOI] [PubMed] [Google Scholar]
- 17.Sprott R L. Exp Gerontol. 1997;32:205–214. doi: 10.1016/s0531-5565(96)00065-4. [DOI] [PubMed] [Google Scholar]
- 18.Kasai H, Crain P F, Kuchino Y, Nishimura S, Ootsuyana A, Tanooka H. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 19.Beauchamp C, Fridovich I. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Elwell J H, Oberley L W. Free Radical Res Commun. 1988;5:67–75. doi: 10.3109/10715768809066913. [DOI] [PubMed] [Google Scholar]
- 21.Vijayalaxmi, Meltz M L, Reiter R J, Herman T S, Srinivas U K. Mutat Res. 1999;425:21–27. doi: 10.1016/s0027-5107(98)00246-2. [DOI] [PubMed] [Google Scholar]
- 22.Street K A, Hall K L, Murphy P, Walter C A. J Anti-Aging Med. 1999;2:275–285. [Google Scholar]
- 23.Wang Y-J, Ho Y-S, Lo M-J, Lin J. Chem Biol Interact. 1995;94:135–145. doi: 10.1016/0009-2797(94)03327-5. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko T, Tahara S M, Matsuo M. Mutat Res. 1996;316:277–285. doi: 10.1016/s0921-8734(96)90010-7. [DOI] [PubMed] [Google Scholar]
- 25.Chung M H, Kasai H, Nishimura S, Yu B P. Free Radical Biol Med. 1992;12:523–525. doi: 10.1016/0891-5849(92)90105-p. [DOI] [PubMed] [Google Scholar]
- 26.Sai K, Takagi A, Umemura T, Hasegawa R, Kurokawa Y. J Environ Pathol Toxicol Oncol. 1992;11:139–143. [PubMed] [Google Scholar]
- 27.Hirano T, Yamaguchi R, Asami S, Iwamoto N, Kasai H. J Gerontol. 1996;51:B303–B307. doi: 10.1093/gerona/51a.5.b303. [DOI] [PubMed] [Google Scholar]
- 28.Izzotti A, Cartiglia C, Taningher M, De Flora S, Balansky R. Mutat Res. 1999;446:215–223. doi: 10.1016/s1383-5718(99)00189-8. [DOI] [PubMed] [Google Scholar]
- 29.Sohal R S, Agarwal S, Candas M, Forster M J, Lal H. Mech Ageing Dev. 1994;76:215–224. doi: 10.1016/0047-6374(94)91595-4. [DOI] [PubMed] [Google Scholar]
- 30.Mecocci P, MacGarvey U, Kaufman A E, Koontz D, Shoffner J M, Wallace D C, Beal M F. Ann Neurol. 1993;34:609–616. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 31.Muscari C, Giaccari A, Stefanelli C, Viticchi C, Giordano E, Guarnieri C, Caldarera C M. Aging. 1996;8:429–433. doi: 10.1007/BF03339606. [DOI] [PubMed] [Google Scholar]
- 32.Anson R M, Hudson E, Bohr V A. FASEB J. 2000;14:355–360. doi: 10.1096/fasebj.14.2.355. [DOI] [PubMed] [Google Scholar]
- 33.Agarwal S, Sohal R S. Proc Natl Acad Sci USA. 1994;91:12332–12335. doi: 10.1073/pnas.91.25.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckman K B, Ames B N. Methods Enzymol. 1996;264:442–453. doi: 10.1016/s0076-6879(96)64040-3. [DOI] [PubMed] [Google Scholar]
- 35.Goukassian D, Gad F, Yaar M, Eller M S, Nehal U S, Gilchrest B A. FASEB J. 2000;14:1325–1334. doi: 10.1096/fj.14.10.1325. [DOI] [PubMed] [Google Scholar]
- 36.Guo Z M, Heydari A R, Richardson A. Exp Cell Res. 1998;245:228–238. doi: 10.1006/excr.1998.4269. [DOI] [PubMed] [Google Scholar]
- 37.Zahn R K, Jaud S, Schroder H C, Zahn-Daimler G. Mech Ageing Dev. 1996;89:79–94. doi: 10.1016/0047-6374(96)01738-1. [DOI] [PubMed] [Google Scholar]
- 38.Rikans L E, Cai Y. J Pharmacol Exp Ther. 1992;262:271–278. [PubMed] [Google Scholar]
- 39.Rikans L E, Ardinska V, Hornbrook K R. Arch Biochem Biophys. 1997;344:85–93. doi: 10.1006/abbi.1997.0172. [DOI] [PubMed] [Google Scholar]
- 40.Martin W R, Ye F Q, Allen P S. Movement Disorders. 1998;13:281–286. doi: 10.1002/mds.870130214. [DOI] [PubMed] [Google Scholar]
- 41.Cook C I, Yu B P. Mech Ageing Dev. 1998;102:1–13. doi: 10.1016/s0047-6374(98)00005-0. [DOI] [PubMed] [Google Scholar]
- 42.Sohal R S, Wennberg-Kirch E, Jaiswal K, Kwong L K, Forster M J. Free Radical Biol Med. 1999;27:287–293. doi: 10.1016/s0891-5849(99)00052-0. [DOI] [PubMed] [Google Scholar]
- 43.Weindruch R, Walford R L. The Retardation of Aging and Disease by Dietary Restriction. Springfield, IL: Thomas; 1988. [Google Scholar]
- 44.Kaneko T, Tahara S, Matsuo M. Free Radical Biol Med. 1997;23:76–81. doi: 10.1016/s0891-5849(96)00622-3. [DOI] [PubMed] [Google Scholar]
- 45.Sohal R S, Agarwal S, Candas M, Forster M J, Lal H. Mech Ageing Dev. 1994;76:215–224. doi: 10.1016/0047-6374(94)91595-4. [DOI] [PubMed] [Google Scholar]
- 46.Masoro E J, Iwasaki K, Gleiser C A, McMahan C A, Seo E J, Yu B P. Am J Clin Nutr. 1989;49:1217–1227. doi: 10.1093/ajcn/49.6.1217. [DOI] [PubMed] [Google Scholar]
- 47.Fasman G. Handbook of Biochemistry and Molecular Biology. Boca Raton, FL: CRC; 2000. [Google Scholar]