Secondary metabolites, which are small-molecule organic compounds produced by living organisms, provide or inspire drugs for many different diseases. These natural products have evolved over millions of years to provide a survival benefit to the producing organism and often display potent biological activity with important therapeutic applications. For instance, defensive compounds in the environment may be cytotoxic to eukaryotic cells, a property exploitable for cancer treatment. Here, we describe the genome of an uncultured symbiotic bacterium that makes such a cytotoxic metabolite. This symbiont is losing genes that do not endow a selective advantage in a hospitable host environment. Secondary metabolism genes, however, are repeated multiple times in the genome, directly demonstrating their selective advantage. This finding shows the strength of selective forces in symbiotic relationships and suggests that uncultured bacteria in such relationships should be targeted for drug discovery efforts.
KEYWORDS: Verrucomicrobia, metagenomics, natural products, polyketides, symbiosis
ABSTRACT
A symbiotic lifestyle frequently results in genome reduction in bacteria; the isolation of small populations promotes genetic drift and the fixation of deletions and deleterious mutations over time. Transitions in lifestyle, including host restriction or adaptation to an intracellular habitat, are thought to precipitate a wave of sequence degradation events and consequent proliferation of pseudogenes. We describe here a verrucomicrobial symbiont of the tunicate Lissoclinum sp. that appears to be undergoing such a transition, with low coding density and many identifiable pseudogenes. However, despite the overall drive toward genome reduction, this symbiont maintains seven copies of a large polyketide synthase (PKS) pathway for the mandelalides (mnd), cytotoxic compounds that likely constitute a chemical defense for the host. There is evidence of ongoing degradation in a small number of these repeats—including variable borders, internal deletions, and single nucleotide polymorphisms (SNPs). However, the gene dosage of most of the pathway is increased at least 5-fold. Correspondingly, this single pathway accounts for 19% of the genome by length and 25.8% of the coding capacity. This increased gene dosage in the face of generalized sequence degradation and genome reduction suggests that mnd genes are under strong purifying selection and are important to the symbiotic relationship.
IMPORTANCE Secondary metabolites, which are small-molecule organic compounds produced by living organisms, provide or inspire drugs for many different diseases. These natural products have evolved over millions of years to provide a survival benefit to the producing organism and often display potent biological activity with important therapeutic applications. For instance, defensive compounds in the environment may be cytotoxic to eukaryotic cells, a property exploitable for cancer treatment. Here, we describe the genome of an uncultured symbiotic bacterium that makes such a cytotoxic metabolite. This symbiont is losing genes that do not endow a selective advantage in a hospitable host environment. Secondary metabolism genes, however, are repeated multiple times in the genome, directly demonstrating their selective advantage. This finding shows the strength of selective forces in symbiotic relationships and suggests that uncultured bacteria in such relationships should be targeted for drug discovery efforts.
Author Video: An author video summary of this article is available.
INTRODUCTION
Microbes frequently associate with higher organisms, and under certain circumstances, such a relationship leads to genome erosion in the microbial partner (1, 2). Host restriction, where an organism is an obligate symbiont with no free-living phase in its life cycle (such as in strict vertical transmission), reduces the need to maintain functions required for independent life. Likewise, adaptation to an intracellular lifestyle further reduces the need to synthesize metabolites available from the host. Sequence degradation and genome reduction occur in the absence of selection pressure, often accompanied by a change in symbiont population structure. Isolated populations of symbionts within hosts undergo frequent population bottlenecks at host-cell division or vertical transmission. In this setting, slightly deleterious mutations can easily become fixed, due to these bottlenecks and the unavailability of horizontal gene transfer (HGT) processes (3). Such sequence degradation leads to weakening of protein function until coding sequences (CDSs) become nonfunctional pseudogenes, which tend to be deleted (1). Transitions to a symbiont lifestyle are therefore accompanied by a proliferation of pseudogenes and apparent lowering of coding density (4) before intergenic sequences are deleted, resulting in vastly reduced genomes.
We have a longstanding interest in symbionts that make bioactive natural products (secondary metabolites) and previously identified a tunicate symbiont that was the source of the patellazoles (5, 6), potent cytotoxins that likely act as chemical defenses for the host. The biosynthetic genes for the patellazoles showed an unusual degree of fragmentation, whereas genes in secondary metabolite pathways tend to be clustered (7). As more symbiont genomes have been sequenced, we have noted that fragmentation of biosynthetic gene clusters (BGCs) appears to be common in symbionts (7).
As part of our efforts to discover novel biosynthetic pathways, we focused on the mandelalides (8–10), which are cytotoxic compounds isolated from the marine tunicate Lissoclinum sp. Given the propensity of this genus to have both intra- and extracellular symbionts and the resemblance of the mandelalides to bacterial compounds made by trans-AT polyketide synthases (PKS), we embarked on a metagenomic sequencing campaign to characterize the mnd pathway and the genome of the producing symbiont. Here, we describe a symbiont in the phylum Verrucomicrobia, the genome for which contains a complete set of biosynthetic genes that likely produce the mandelalides. This genome shows signs of ongoing degradation, with numerous pseudogenes and low coding density. To our surprise, the mandelalides gene cluster had much higher coverage than the rest of the genome, and we found evidence that it is connected to multiple parts of the otherwise well-assembled genome. The cluster is repeated seven times and is likely under strong selective pressures to enhance mandelalide production. We also found evidence that the mnd cluster is not a recent acquisition and that it is undergoing degradation and sequence divergence. The repeat structure may represent a paradigm for the ancestral state of older symbionts with pathways formed from fragmented secondary metabolite genes.
RESULTS
Identification of a bacterial symbiont associated with mandelalide-containing Lissoclinum sp.
In an effort to investigate the biosynthesis of the mandelalides, we recollected the Lissoclinum sp. tunicate that had previously yielded the mandelalides (8), near the original collection site of Algoa Bay, South Africa. The individual animal that we collected yielded mandelalides A to D (Fig. 1A), as well as eight new analog mandelalides, E through L (9, 11). Tunicates in the genus Lissoclinum are colonial, consisting of many tiny individual animals (zooids) enveloped in a protective coat or “tunic.” The mandelalide-containing tunicate was dissected to separate the tunic from the zooids, since we previously found a bacterial symbiont of the related tunicate Lissoclinum patella to be localized to zooids (5). Unlike L. patella (12), the mandelalide-containing animal appeared not to harbor Prochloron didemni or other photosynthetic symbionts in the cloacal contents. Total DNA was extracted separately from each of the tunic and zooid fractions and subjected to shotgun metagenomic sequencing (Illumina 101-bp paired end). Retrobiosynthetic analysis of mandelalide structures revealed several features suggestive of synthesis via a trans-acyltransferase (AT) polyketide synthase (PKS) pathway (13, 14). These included the presence of a cis double bond, a β-methyl moiety, and multiple tetrahydrofuran (THF) and -pyran (THP) rings (Fig. 1B). Therefore, we searched initial assemblies for fragments of trans-AT pathways. One putative pathway was found; however, it was fragmented due to low coverage and there was a general lack of bacterial contigs in the metagenome. Since coverage of this putative pathway was higher in the zooid fraction than in the tunic, additional sequencing was obtained from the zooid extract.
FIG 1 .
(A) Structures of mandelalides A to D. (B) Retrobiosynthetic analysis of the two types of mandelalide carbon skeleton, showing probable building blocks and features suggestive of trans-AT-type polyketide synthases (PKS). (C) Visualization of the metagenomic assembly obtained from the zooid fraction, where each point represents a contig of >3,000 bp in length. Points are colored based on taxonomic group, and their size is proportional to contig length. The contig bearing the mnd pathway is outlined in black. (D) Approximately maximum-likelihood tree based on 16S rRNA gene sequences from “Candidatus Didemnitutus mandela” and 100 other bacteria in the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum obtained from the Ribosomal Database Project (RDP) (83), showing the placement of “Ca. Didemnitutus mandela” in the family Opitutae in the phylum Verrucomicrobia. Bootstrap proportions greater than 70% are expressed to the left of each node as a percentage of 1,000 replicates. (E) Amplicon analysis of ketosynthase (KS) domains in Lissoclinum sp. zooid and tunic fractions. mnd accounts for the vast majority of KS domains in the zooid fraction and for trans-AT KS domains in both fractions.
With the additional sequencing data in hand (Table 1), a new metagenome assembly was constructed from all zooid-derived sequence reads, with tunic-derived reads excluded. Predicted open reading frames (ORFs) within the contigs were used to infer probable taxonomy based on the lowest common ancestor of BLASTP hits in the NCBI nr database (15, 16). In visualizations of the assembly, a discrete cluster in the bacterial phylum Verrucomicrobia was observed, comprised of contigs with low coverage and high GC content (Fig. 1C). This set, termed Ver_v1 here, consisted of 15 contigs that we predicted to represent a bacterial genome that was 94.2% complete and 100% pure, based on analysis of single-copy markers (Table 2) (17). This assembly included a large 108-kbp contig containing a complete trans-AT PKS pathway that we termed mnd. Unexpectedly, this large contig showed much higher coverage than the other contigs in Ver_v1, suggesting either a sequence misassembly or that the mnd pathway is repeated within the symbiont genome.
TABLE 1 .
Sequence data used in this study
| Method of sequencing and type of value | Tunic | Zooids |
|---|---|---|
| Whole metagenome | ||
| Illumina HiSeq reads (2 × 101 bp), millions | 96.2 | 279.1 |
| Assembly, thousands of contigs | 2,575 | |
| Assembly, Mbp | 1,009 | |
| Assembly, N50, bp | 2,453 | |
| KS PCR, Illumina MiSeq reads (2 × 251 bp), thousands | 545.3 | 57.3 |
| 16S PCR, Illumina MiSeq reads (2 × 251 bp), thousands | 92.6 | 0.53 |
TABLE 2 .
Assembly characteristics
| Characteristic | Value for assembly: |
|
|---|---|---|
| Ver_v1 | Ver_v2 | |
| Size (Mbp) | 2.17 | 2.17 |
| No. of contigs | 15 | 10 |
| N50 (kbp) | 224.8 | 319.6 |
| GC content (%) | 51.93 | 51.93 |
| Completeness (%) | 94.2 | 94.2 |
| Purity (%) | 100 | 100 |
To obtain greater coverage of the symbiont genome and potentially achieve a better assembly, the subset of reads from both zooid and tunic DNA extracts that aligned to Ver_v1 contigs was segregated and reassembled. The lower-coverage contigs from Ver_v1 did indeed coalesce into fewer contigs, although the higher-coverage contig, including the section containing the mnd genes, became fragmented. We hypothesize that in the single-genome setting and with higher coverage, the repeat structure of mnd was more apparent to the assembler (see below), resulting in fragmented contigs that could not be resolved automatically. By alignment of paired-end reads, we confirmed that the original mnd sequence obtained from Ver_v1 was consistent with most of the repeats. We combined the genomic contigs obtained from the new assembly, along with the original mnd contig, in Ver_v2 (Table 2). This genome assembly is 2.17 Mbp in total length and estimated to be 94.2% complete and 100% pure. Phylogenetic analysis of the full-length 16S rRNA gene places the symbiont, which we termed “Candidatus Didemnitutus mandela,” in a clade with the family Opitutaceae in the phylum Verrucomicrobia (Fig. 1D), and this placement is consistent with a phylogenetic tree derived from concatenated protein markers (see Fig. S1 in the supplemental material). In a similar vein as the naming of Opitutus (“protected by the Roman Earth and harvest goddess Ops” [18]), “Didemnitutus” denotes a bacterium protected by a member of the family Didemnidae, and “mandela” is an allusion to the collection site, near the Nelson Mandela Bay municipality in South Africa. The closest relative to the symbiont that has a publicly available genome sequence is Opitutus sp. strain GAS368 (92% 16S rRNA sequence identity). According to the sequence cutoffs proposed by Yarza et al. (19), this level of identity would be consistent with a new genus in the family Opitutaceae.
Approximately maximum-likelihood tree generated by FastTreeMP from 31 concatenated single-copy marker gene protein sequences from the “Ca. Didemnitutus mandela” genome and 1,068 other reference genomes (species outside the PVC superphylum and Elusimicrobia not shown). Bootstrap proportions greater than 70% are expressed to the left of each node as a percentage of 1,000 replicates. Download FIG S1, EPS file, 0.9 MB (991.9KB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Given that the symbiont “Ca. Didemnitutus mandela” 16S rRNA sequence could not be amplified with standard MiSeq universal primers, we were not able to directly quantify the abundance of “Ca. Didemnitutus mandela” relative to other bacteria represented in the metagenome of the tunicate consortium. We obtained a relatively low number of 16S reads from the zooid amplification product, compared to that of the tunic, potentially due to a low copy number of amplifiable 16S genes in the zooids (Table 1). Degenerate ketosynthase (KS) primers targeted to polyketide synthase (PKS) genes were then used to quantify the levels of the mnd BGC and other pathways. This revealed that 96.8% of reads in the zooid fraction originated from mnd, as well as 50.5% of reads from the tunic. A very low number of the non-mnd reads were identified as being from trans-AT-type PKS pathways (Fig. 1E), and therefore, mnd is the only detectable pathway capable of making the mandelalides (see also details of the proposed biosynthetic scheme, below).
Multiple copies of the mnd biosynthetic gene cluster are maintained in the symbiont genome despite streamlining.
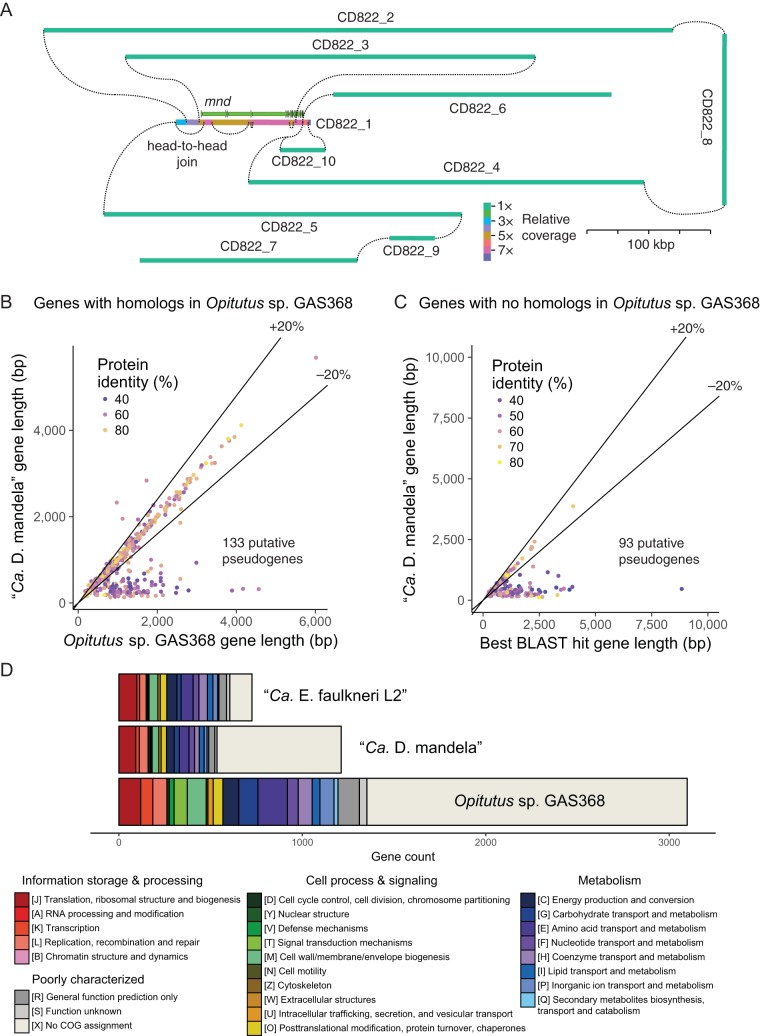
In order to resolve the repeat structure within Ver_v2, we examined the read coverage of all contigs in the assembly. We aligned paired-end reads from both the zooid and tunic metagenomes to Ver_v2 contigs, to detect joins suggested by read pairs that aligned to different contigs. Joins were considered between the ends of contigs and also to middle regions, especially where abrupt changes in read coverage suggested that the assembler had joined two repeats of differing copy number (Fig. 2A). The high-coverage region of the genome includes the mnd pathway except for mndR, which has a relative coverage of 1× and resides on contig CD822_6. Sections of the repeat region range in relative coverage from 3× to 8× because the ends of these regions are variable, with several relatively rare deletions within the mnd cluster. The deleted regions have 5× coverage, and the rest of the mnd pathway has 7× coverage, suggesting either that there are two mnd repeats with three deletions each or that these deletions are evenly distributed among each of the seven repeats, each occurring twice. There is evidence of multiple alternate connections between both ends of the repeat region and 1× contigs, supporting the notion that the mnd pathway is repeated multiple times in the genome and not simply embedded in a different genome. A number of connections could also be made between 1× contigs, suggesting five 1× scaffolds, several of which are joined to mnd at both ends (Fig. 2A). With only two loose ends in the connection map, it appears that the majority of the genome is represented in the assembly. Our estimation of 94.2% completeness from single-copy marker genes, therefore, might be due to a bona fide absence of a small number of markers, potentially due to genome reduction. We have observed much lower apparent completeness values in the complete chromosome of a more extensively eroded genome (15). If the genome consists of a single circular chromosome, our findings mean that there are five repeat regions in the genome, with some adjacent mnd repeats. Consistent with this notion, paired-end alignments showed evidence of head-to-head connections at least two mnd repeats.
FIG 2 .
(A) Scale map of connections between contigs in Ver_v2 suggested by the alignment of paired-end Illumina reads (insert size, ~300 bp). Colors denote different relative coverages. Deletions are suggested by joins between distal regions in the mnd gene cluster. While deleted regions have 5× coverage, the rest of mnd has 7× coverage, indicating that deletions occur in 2/7 repeats. (B) Comparison of gene length in “Ca. Didemnitutus mandela” and Opitutus sp. GAS368. Genes <80% of the length of the Opitutus sp. GAS368 homolog are putative pseudogenes. (C) For genes without a homolog in Opitutus sp. GAS368, length is compared to their closest BLASTP hit. (D) Analysis of COG gene categories in “Ca. Endolissoclinum faulkneri,” “Ca. Didemnitutus mandela,” and Opitutus sp. GAS368, for genes that are not putative pseudogenes.
Factoring in the copy number of repeat regions, the intact chromosome of “Ca. Didemnitutus mandela” was calculated to be 2.68 Mbp in size. The coding density of the repeat region is 84.9%, with an average gene size of 1,574 bp, whereas the coding density of the 1× contigs is 64.3%, with an average gene size of 470 bp. The copy number of each mnd gene was calculated from the total number of repeats in which the respective gene is not truncated, and such intact genes should occupy 514,374 bp, accounting for 19.2% of the genome and 25.8% of its entire coding capacity. We analyzed a set of symbiont genomes and genomes of free-living bacteria (Table S1) and found that several facultative and transitional symbionts have a similar or higher fraction of repeats. However, in all other cases, there seemed to be a general proliferation of many repeat loci, with few long repeats, and “Ca. Didemnitutus mandela” is unique in having repetition of genes from an entire pathway. The lower coding density within the 1× regions of the “Ca. Didemnitutus mandela” genome suggested sequence degradation characteristic of symbionts shortly after a change in lifestyle such as restriction to a particular host or a switch to an intracellular habitat (4). Therefore, we examined the gene inventory beyond mnd.
Comparison of repeat structure in symbionts and free-living bacteria. Download TABLE S1, PDF file, 0.1 MB (56.4KB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Out of 2,864 predicted protein-coding genes in “Ca. Didemnitutus mandela,” only 780 were found to have homologs in Opitutus sp. GAS368. A further 162 genes had BLAST hits in the NCBI NR database, and the remainder (n = 1,922) were found to have much shorter average length and slightly lower GC content (Table 3). Of the “Ca. Didemnitutus mandela” genes found to be homologous to genes in Opitutus sp. GAS368, 133 were significantly truncated (>20%), suggesting that they are pseudogenes (20) (Fig. 2B). Of the remaining genes with BLAST hits, 93 of them were truncated more than 20% compared to their best BLAST hit and are counted here as putative pseudogenes (Fig. 2C). The genes that are truncated may reflect functions that are under reduced selection for retention in the symbiotic relationship (Table S2); for example, there are many putative pseudogenes involved in lipopolysaccharide (LPS) biosynthesis. Key enzymes involved in the biosynthesis of the amino acids isoleucine, valine, leucine, proline, and tryptophan are also truncated. All but one of these amino acids cannot be synthesized by eukaryotic organisms (21). Consequently, it is unlikely that “Ca. Didemnitutus mandela” serves a nutritional function for the host organism. Putative pseudogenes were also found in the pathways for some cofactors, including riboflavin and folate.
TABLE 3 .
Characteristics of genes and intergenic sequences in the “Ca. Didemnitutus mandela” genome
| Type of sequence in “Ca. Didemnitutus mandela” | Avg length (bp) |
GC% | n |
|---|---|---|---|
| Gene with homolog in Opitutus sp. GAS368 | 981 | 53.9 | 780 |
| Gene with no homolog in Opitutus sp. GAS368 but BLAST hit in NR | 993 | 55.0 | 162 |
| Gene with no homolog in Opitutus sp. GAS368, no BLAST hit in NR | 259 | 51.1 | 1,922 |
| Intergenic sequence | 295 | 49.0 | 2,794 |
Putative pseudogenes with functional annotations in the “Ca. Didemnitutus mandela” genome. Download TABLE S2, PDF file, 0.1 MB (64.3KB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The number of genes with annotated functions in “Ca. Didemnitutus mandela” is on a par with “Ca. Endolissoclinum faulkneri,” an intracellular symbiont of L. patella and source of the patellazoles, cytotoxic polyketides (5, 6) (Fig. 2D). However, a number of factors suggest that “Ca. Endolissoclinum faulkneri” is in a more advanced state of genome reduction than “Ca. Didemnitutus mandela.” “Ca. Endolissoclinum faulkneri” has a lower coding density than “Ca. Didemnitutus mandela” (Table 4), and the intergenic regions of the former are degraded to the point that there are few recognizable pseudogenes and there is a pronounced AT-skew compared to coding regions (5, 6) not seen in “Ca. Didemnitutus mandela.” “Ca. Didemnitutus mandela” also possesses some key genes that “Ca. Endolissoclinum faulkneri” has lost, including dnaA and ftsZ, which are central to chromosome replication and cellular division, respectively. This suggests that “Ca. Didemnitutus mandela” maintains more control over these processes than “Ca. Endolissoclinum faulkneri.”
TABLE 4 .
Comparison of “Ca. Didemnitutus mandela,” Opitutus sp. GAS368, and “Ca. Endolissoclinum faulkneri” L2 genomes
| Characteristic | Value for genome: |
||
|---|---|---|---|
| “Ca. Didemnitutus mandela” | Opitutus sp. GAS368 | “Ca. Endolissoclinum faulkneri” L2 | |
| Genome length, Mbp | 2.68 | 4.15 | 1.48 |
| % coding (length) | 69.0a | 89.4 | 57.2 |
| GC% | 51.9 | 65.7 | 34.1b |
| No. of nonhypothetical genes | 654 | 2,016 | 688 |
| No. of hypothetical genes | 2,031 | 1,389 | 95 |
| No. of pseudogenes | 226 | 183 | 5 |
| % secondary metabolism (fraction of coding length) | 25.8 | 3.2 | 10.2 |
Repeat region density, 84.9%; 1× contigs, 63.4%.
Coding regions are 40.9% GC, whereas noncoding regions are 24.7% GC.
We did, however, find that several DNA repair pathways have deficiencies in the “Ca. Didemnitutus mandela” genome. The nucleotide excision repair pathway is complete, as is the mismatch repair pathway (in contrast to “Ca. Endolissoclinum faulkneri”). However, the base excision repair pathway is missing two DNA glycosylases (alkA and tag) responsible for removing 3-methyladenine adducts, one of which is still present in “Ca. Endolissoclinum faulkneri.” Additionally, the homologous recombination system is missing several key genes (recB, recF, recN, and recQ). The loss of these genes should preclude both RecBCD-dependent and RecBCD-independent homologous recombination, as well as incorporation of horizontally transferred DNA into the chromosome (22). Taken together, this pattern of maintenance and loss suggests that “Ca. Didemnitutus mandela” is still able to copy its genome with fidelity but is likely vulnerable to strand breaks due to impaired homologous recombination systems. No transposases, integrases, or restriction-modification or phage genes (3) were annotated, and “Ca. Didemnitutus mandela” does not have a nonhomologous end-joining system. Therefore, it would appear that further genome rearrangement is possible only through RecA-independent “illegitimate recombination” (3) during chromosome replication.
Secondary metabolism is by definition more variable among close relatives than are central functions (23), and it is thought that BGCs are often disseminated via HGTs (24). Since HGTs are likely no longer possible in “Ca. Didemnitutus mandela,” and genome rearrangements are probably rare at this stage, the mnd cluster was probably obtained before homologous recombination was lost, with duplication occurring shortly after acquisition. Notably, the cluster repeats have been present long enough to allow divergence through deletions and the accumulation of a small number of single nucleotide polymorphisms (SNPs) (Fig. S2). Consistent with this timeline, we found that the codon adaptation index (CAI) (25) of mnd genes is not significantly different from those of other genes with annotated function (Fig. S3). Interestingly, we found that the CAIs of both pseudogenes and hypothetical genes are significantly different from those of genes with annotated functions, suggesting that these ORFs are degraded with concomitantly reduced codon selection.
SNPs identified within the mnd pathway (mnd genes are green, other protein coding genes are gray, and tRNAs are magenta). Read coverage is plotted above the contig. Download FIG S2, EPS file, 1.1 MB (1.2MB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Box plots of codon adaptation index (CAI) of different groups of genes within the “Ca. Didemnitutus mandela” genome. (B) Individual P values for pairwise comparisons of the groups compared through one-way ANOVA and Tukey HSD. Significant P values (P < 0.05) are highlighted in red. Annotated genes are not significantly different in codon usage from mnd genes. However, both pseudogenes and hypothetical genes are significantly different from annotated genes in this respect. Download FIG S3, EPS file, 0.9 MB (913.7KB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Model for mandelalide biosynthesis by mnd.
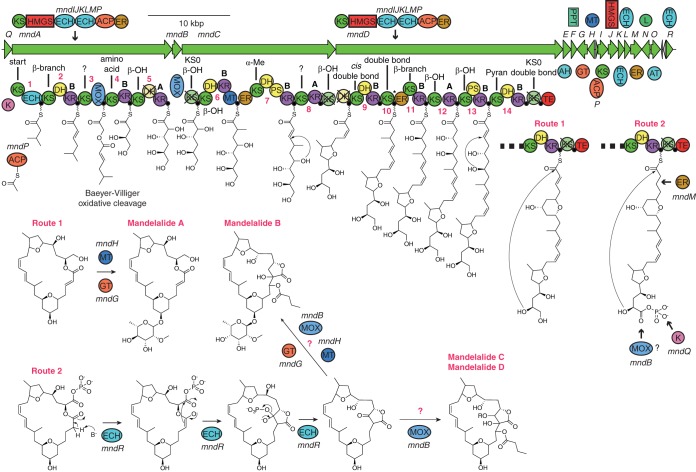
Our model for the biosynthesis of mandelalides is shown in Fig. 3. The pathway consists of three large PKS proteins and 15 accessory proteins. These accessory proteins include a phosphopantetheinyltransferase (PPT, MndF), required to postranslationally modify acyl carrier protein (ACP) domains within the PKS with a phosphopantetheine arm (26, 27), and a trans-acting acyltransferase (AT, MndO), which is responsible for loading malonyl-S-coenzyme A (CoA) extender units onto these phosphopantetheine arms (13, 14). The mnd cluster also contains a suite of proteins predicted to install the β-methyl at carbon 11 (MndIJKLMP) (28); a glycosyltransferase (GT, MndG), which may attach a sugar unit to the polyketide core structure; and a methyltransferase (MT, MndH), which could supply the O-methyl group to the sugar units observed in the known mandelalide structures (8, 9).
FIG 3 .
Proposed mandelalide biosynthetic pathway. The trans-AT PKS pathway consists of 14 modules with proposed chain shortening mediated by a monooxygenase. Butyrolactone formation (as in mandelalides B to K) is proposed through the action of MndR, a homolog of Dieckmann cyclase MenB. Modules are numbered in red, and predicted substrates (Fig. S4) are shown next to the respective KS domain. A cross indicates domains predicted to be catalytically inactive. Abbreviations: ACP, acyl carrier protein, also denoted by a filled black circle; AH, acylhydrolase; AT, acyltransferase; DH, dehydratase; ECH, enoyl-CoA reductase; ER, enoylreductase; GT, glycosyltransferase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; K, kinase; KR, ketoreductase; KS, ketosynthase; L, phospholipase; MOX, flavin monooxygenase; MT, C-methyltransferase; PPT, phosphopantetheinyltransferase; PS, pyran synthase; TE, thioesterase.
Expansions of clades containing mnd KS domains in an approximately maximum-likelihood tree made from 665 KS domain sequences. Colors in each clade correspond to the chemical structures shown immediately to its right side. Download FIG S4, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Type I PKS proteins, such as MndACD, cause the synthesis of specific structures through the presence or absence of specific enzymatic domains within “modules,” which each add a C2 unit, analogous to an assembly line (29). This process results from repeated Claisen condensation of a ketosynthase (KS)-bound thioester intermediate onto ACP-bound malonate to make a β-keto thioester with the loss of CO2. If the module additionally contains a ketoreductase (KR) domain, then the β-position is reduced to a hydroxyl moiety. Inclusion of a dehydratase (DH) domain in addition to a KR results in an α-β double bond, and the presence of an enoylreductase (ER) domain with DH and KR results in complete reduction of the β-position to give an alkyl chain. Many other exotic variants of module structure are known in trans-AT systems (13, 14), and KS domains in these pathways have diversified to accept specific substrate structures (30–32). Through phylogenetic analysis, substrate specificity was predicted for the majority of mnd KS domains (Fig. S4) and found to be almost completely congruent with our proposed biosynthetic scheme (Fig. 3). In this case, the order of PKS proteins appears to be colinear with the gene order, as the first KS of MndA is closely related to others in starter modules of trans-AT PKS pathways and because MndD contains a thioesterase (TE) domain for cleaving and macrocyclization of the final PKS product.
Similar to other trans-AT pathways, several domains in MndACD were predicted to be nonfunctional due to disrupted catalytic residues or truncated sequences (Fig. S5). However, the three PKS proteins still contain 14 extending modules—three more than would be needed to make the mandelalides (Fig. 1B). We rationalize this discrepancy by proposing that the monooxygenase (MOX) domain in MndA carries out a Baeyer-Villiger-type oxidation, thereby effecting oxidative cleavage of the intermediate. The following KS is related to amino-acid-accepting KSs, even though it does not follow a nonribosomal peptide synthetase (NRPS) module. The predicted intermediate accepted by this KS is a hydroxy acid (glycolic acid), similar to the amino acid glycine except that the glycine nitrogen is replaced by an oxygen. A similar mechanism of chain cleavage is thought to occur in the pederin (33) and diaphorin (34) pathways. This mechanism would also generate the apparent starter unit for mandelalide A, 3,4-dihydroxybutanoic acid. There are a number of other features that are consistent with the final mandelalide structures. In particular, there are two modules containing pyran synthase (PS) domains (35) (modules 7 and 12, Fig. S6), which are the correct distance apart to install both the tetrahydrofuran (THF) and tetrahydropyran (THP) rings in mandelalide A. Module 10 also contains a specialized β-branching ACP (36), which would cause the installation of a β-methyl in the expected location next to the THP (Fig. S5). Additionally, based on previous findings (37), KR domains were analyzed to predict the configuration of installed hydroxyl groups (Fig. S5). In all cases, the predicted configurations match configurations confirmed in mandelalide A by total synthesis (10, 38–42).
(A) Alignment of mnd KS domains, with the CHH catalytic triad highlighted. The domains where this triad is disrupted are predicted to be catalytically inactive. (B) Alignment of mnd KR domains, along with two from the erythromycin biosynthetic gene cluster to allow comparison to previous alignments. The NADPH-binding site (GxxxGxG) is highlighted, as is the LDD motif that is thought to control product stereoconfiguration. The catalytic triad (KSY) is also highlighted. (C) Alignment of mnd DH domains. Highlighted are the conserved motifs HxxxGxxxxP and DxxxQ, which are involved in catalysis. (D) Alignment of mnd ACP domains, with the extender unit attachment point serine highlighted. In addition, residues characteristic of β-branching ACPs in MndD_ACP3 are shown in red. Download FIG S5, PDF file, 0.3 MB (313KB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Approximately maximum-likelihood tree based on DH and PS domains. The PS domains form a distinct clade, as shown previously (35). Download FIG S6, EPS file, 1.9 MB (2MB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Often, trans-AT PKS proteins deviate from strict gene and domain colinearity (13, 14); for example, in the mnd pathway modules 8 and 13 lack DH domains even though they should introduce double bonds. These modules may utilize DH domains in the following module, similar to many other trans-AT pathways, such as those that produce bacillaene, calyculin, and oxazolamycin (13, 14). Module 13 contains a PS domain, which is related to dehydratases but lacks key catalytic residues (35). This pattern is consistent with the structures of the mandelalides, but the reaction requires the installation of an α-β double bond, which could be installed by the DH in module 14. We also found that the KR domain in module 5 has a noncanonical catalytic triad—KSH instead of KSY (43) (Fig. S5). This mutation appears to be rare; the only other example that could be found is the KR domain of the ena5920 protein within the pathway for enacycloxin (44). This KR domain is annotated as functional, and we propose that histidine likely fulfills the same proton source role as tyrosine during generation of the β-keto group of the substrate (Fig. S7).
(A) Canonical mechanism of KR domains with catalytic triad KSY. (B) Proposed mechanism of functional KR domains with catalytic triad KSH. Download FIG S7, EPS file, 2.2 MB (2.2MB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A compelling mystery in the biosynthesis of mandelalides is the mechanism by which a butyrolactone is installed in mandelalides B to K at the point of macrocyclization, which would require both ester and C–C bond formation. A further question is how the pathway produces butyrolactone-containing mandelalides alongside mandelalide A, which lacks this moiety. We propose that the butyrolactone is generated by MndR, a homolog of crotonase superfamily member MenB. For both mandelalide A-type and B-type compounds, we predict that the thioesterase of MndD produces an initial macrocycle. MenB catalyzes a Dieckmann cyclization to produce dihydroxynaphthoyl-CoA in the vitamin K biosynthesis pathway (45, 46), and we predict that MndR could analogously form the lactone C–C bond in a Dieckmann reaction (Fig. 3). In order to allow for the action of MndR, we propose that the terminal hydroxyl is oxidized to a carboxylic acid, which is then phosphorylated by kinase MndQ, so as to activate the carbonyl to nucleophilic attack. Additionally, the α-β double bond could be removed by trans-acting ER MndM, to allow the formation of an enolate which can attack the phosphoester.
The mandelalide-containing Lissoclinum sp. is a novel species of tunicate.
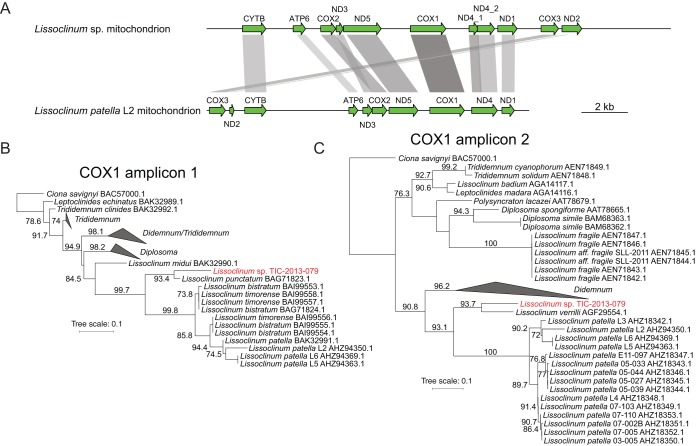
A BLAST search using L. patella cytochrome c oxidase 1 (COX1) protein sequences as queries identified a contig in the metagenomics assembly that appeared to represent the majority of the host mitochondrial genome (Fig. 4). This contig is 20.7 kbp in length and contains all the protein-coding genes previously identified in the L. patella mitochondrial genome (47). Comparison of the coding sequences in Lissoclinum sp. and L. patella L2 (Fig. 4A) revealed several genes that appeared to be shorter in L2. The NADH dehydrogenase subunit 4 (ND4) gene appears to be disrupted by a frameshift in Lissoclinum sp., but this disruption could be an artifact of sequencing errors due to low sequence complexity and prevalent homopolymers within the contig. Assuming a circular chromosome, the gene order in Lissoclinum sp. is very similar to the gene order in the L. patella L2 mitochondrial genome, except for a swap in the positions of ATP synthase Fo subunit 6 (ATP6) and cytochrome c oxidase subunit 2 (COX2). Both the mitochondrial genomes of Lissoclinum sp. and L. patella L2 are very low in GC content (12.4% and 21.2%, respectively). The resulting low complexity of the sequences makes it difficult to detect rRNA genes. Previously, we suggested that the rRNA genes are in the space between the CYTB and ATP6 genes in L. patella L2 (47). A corresponding space without detectable CDSs is present in the Lissoclinum sp. mitochondrial genome, after the ND2 gene, potentially signifying a second rearrangement (Fig. 4A). It has been suggested previously that gene order in tunicate mitochondrial genomes could be used as a phylogenetic signal (48). The gene rearrangements observed therefore suggest that Lissoclinum sp. is phylogenetically distinct from Lissoclinum patella.
FIG 4 .
(A) Scale map of Lissoclinum sp. mitochondrion genome and comparison to the genome of the Lissoclinum patella L2 mitochondrion. (B and C) Approximately maximum-likelihood trees based on two nonoverlapping amplicons in the mitochondrial cytochrome c oxidase I (COX1) gene. Bootstrap proportions greater than 70% are expressed to the left of each node as a percentage of 1,000 replicates. Sequence identities of the Lissoclinum sp. COX1 gene to its close relatives are shown in Fig. S8.
Sections of the COX1 gene have been used for molecular barcoding in tunicates, but unfortunately, two nonoverlapping regions of the gene have been employed (47). In two phylogenetic trees based on COX1 protein sequence (Fig. 4B and C), the mandelalide-containing Lissoclinum sp. appears to be a divergent Lissoclinum, distinct from a major group that includes L. patella, Lissoclinum bistratum, and Lissoclinum timorense. The closest relatives to Lissoclinum sp. are Lissoclinum punctatum and Lissoclinum verrilli. The Lissoclinum sp. COX1 protein sequence has 72 to 75% identity to L. patella specimens, 76 to 78% identity to L. bistratum and L. timorense, and 82% identity to its closest relative, L. punctatum (Fig. S8). The divergence of Lissoclinum sp. and L. punctatum is on par with the evolutionary distance between different subpopulations of L. patella that we believe represent multiple cryptic species with a common ancestor that existed 6 to 31 million years ago (6). Therefore, the mandelalide-containing Lissoclinum sp. may be a novel species of tunicate in the family Didemnidae.
Matrices showing pairwise amino acid identities for the full-length Lissoclinum sp. COX1 gene and other COX1 sequences from the NCBI database. Download FIG S8, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Culture-independent sequencing has revealed evidence of the phylum Verrucomicrobia in a variety of terrestrial and marine environments, although relatively few species have been isolated and/or sequenced (49). However, both free-living and symbiotic verrucomicrobial species are known. For instance, Akkermansia muciniphila is a prevalent member of the human gut microbiota that degrades mucins (50). Intracellular and genome-reduced Verrucomicrobia members are also known, such as “Candidatus Xiphinematobacter” in nematodes (51) and “Candidatus Nucleococcus,” which lives inside the nucleus of protists in the termite gut (52). In contrast to “Ca. Didemnitutus mandela,” these and related symbionts have yet to be associated with what appears to be highly defensive functions by virtue of toxin biosynthesis. Secondary metabolite pathways have been noted in the genomes of Verrucomicrobia (53, 54), but to the best of our knowledge “Ca. Didemnitutus mandela” is the only species in this phylum that has been linked to secondary metabolites that have been isolated, structurally characterized, and shown to be potently cytotoxic. Our findings here reiterate the sentiment that uncultured bacterial lineages may be a prolific source of bioactive natural products for drug discovery.
The mandelalides were previously found to be potent cytotoxins (8, 9) and may therefore serve as chemical defenses for the tunicate host, similar to what has been noted for the patellazoles in L. patella (5). The structures of mandelalides B to K are unique among cyclic polyketides, which invariably are cyclized through formation of an ester bond (29). The macrocycles of mandelalides B to K are formed through both an ester and C–C bonds that constitute a butyrolactone not found in mandelalides A and L. To the best of our knowledge, the only comparable macrocyclization occurs in the biosynthesis of lankacidin, in which an amine oxidase produces an imine for attack by an acidic carbon between two carbonyl groups, thus forming a C–C bond (55). In both cases, the resulting C–C macrocycle may be more pharmacokinetically stable by virtue of its resistance to circulating esterases in vivo, which break down ester-containing drugs and thus limit efficacy and duration of action (56). Accordingly, further study of the biochemical mechanisms of these macrocyclizations is likely to aid the design of pharmacokinetically stable polyketide drugs. In the case of the mnd pathway, confirmation of the mechanism of butyrolactone production would require heterologous expression of mnd genes and characterization of their biochemical activities in vitro.
The duplication of a very long gene cluster such as mnd in “Ca. Didemnitutus mandela” has not been observed in nature, to the best of our knowledge, especially in a genome-reduced symbiont. As gene amplification is a rapid and common process (57), it is reasonable to suppose that such duplications of secondary metabolite pathways do occur in nature given the right selective environment. Indeed, the duplication or amplification of pathways has been observed in industrial actinomycete strains that have been heavily mutagenized and selected for higher-level production (58, 59). A similar effect has been achieved in actinomycetes through purposeful pathway amplification (60, 61). This suggests that mnd is under strong selection, in a manner similar to the trpEG genes in the aphid endosymbiont Buchnera aphidicola, which provide tryptophan to the host (62). B. aphidicola has been an endosymbiont for ~150 million years (63) and now has a very small genome (~640 kbp). Remarkably, despite extreme genome reduction, some strains harbor a plasmid with multiple copies of trpEG, although there are often pseudogenes among the copies (64). In the early stages of the symbiosis, the plasmid location of these genes may have increased gene dosage and tryptophan production. However, at some later point, the Buchnera chromosome became polyploid, and there is evidence of back-transfer of these genes to the chromosome in some lineages (65). These back-transfers and trpEG copy number variants are the only recombination events known to have occurred since the divergence of extant Buchnera strains, which lack RecA but maintain RecBCD (65).
The genome of “Ca. Didemnitutus mandela” shows signs of degradation and reduction consistent with host restriction, although this change in lifestyle is likely to have happened relatively recently. With only one sequenced strain of “Ca. Didemnitutus mandela” and the paucity of known close relatives to either the symbiont or host, it is difficult to date the symbiosis, except through loose comparisons to unrelated symbiotic systems. Mandelalide-producing tunicates have been found in only one place on Earth, and so, more complete investigation of the evolution of “Ca. Didemnitutus mandela” is currently very challenging. The erosion of the “Ca. Didemnitutus mandela” genome is not as severe as in B. aphidicola (~150 million years) (63) or “Ca. Endolissoclinum faulkneri,” which has been an intracellular symbiont for at least ~6 to 31 million years (6). However, the “Ca. Didemnitutus mandela” genome contains fewer recognizable pseudogenes relative to a very recent symbiont such as Sodalis glossinidius, which diverged from a close free-living relative ~30,000 years ago (66) and has 972 pseudogenes in its genome (67). The current repeat structure of mnd in the chromosome is also likely not recent, as we predict “Ca. Didemnitutus mandela” to be recombination deficient, and at this point, the gene order is likely fixed. Accordingly, we found the codon usage in mnd to be consistent with the rest of the genome. We found that only a small number of mutations had accumulated in mnd since duplication, perhaps because DNA repair pathways remain largely intact. Nevertheless, with the cessation of recombination and the segregation of small populations within individual hosts, degradation continues through a process known as “Muller’s ratchet” (68). Mutations and deletions that are not outright lethal tend to become irreversible through population bottlenecks and the inaccessibility of HGT or recombination events. Due to the deletion and AT mutation bias of bacteria, along with weakened selection caused by small effective populations (1), nonfunctional pseudogenes and intergenic sequences will be lost quickly and the sequence of essential genes will drift. This process will accelerate when DNA repair pathways become compromised. The copies of mnd are likely to continue diverging from each other as their sequences degrade, before individual gene copies become nonfunctional pseudogenes and are deleted. The result of such a process would appear similar to the genome of “Ca. Endolissoclinum faulkneri,” where genes from a single pathway are fragmented across many loci in the chromosome. In nonsymbiotic bacteria, there is a strong tendency for the genes of secondary metabolite pathways to remain clustered on a contiguous region of a chromosome or plasmid (7), and it is thought that this colocalization is advantageous in coregulating genes and operons (69). It has been suggested that clustering aids HGTs in the “selfish operon” hypothesis (70). However, such events may be quite rare (24) and therefore have little influence on selective pressures to maintain clustering. We have observed that secondary metabolite BGCs in symbionts tend to be fragmented more often than expected (7). A potential explanation for this fragmentation is a reduced need for fine regulation of a product that is always needed (e.g., for defense), in an environment where production has little survival cost since nutritional needs are met by the host. Our results here suggest that biosynthetic pathway fragmentation in symbionts could also arise through strong selection for high production at the onset of symbiosis, causing pathway duplication prior to genome degradation.
MATERIALS AND METHODS
Tunicate collection, preservation, and DNA extraction.
A specimen of Lissoclinum sp. was collected at 33°59′55″S, 25°42′43″E on 7 July 2013 from White Sands Reef in Algoa Bay, Eastern Cape Province, South Africa, by scuba at an approximate depth of 18 m. A voucher specimen is maintained with the designation TIC-2013-079 at the South African Institute for Aquatic Biology (SAIAB), Grahamstown, South Africa. Part of the animal was preserved in RNAlater at −80°C. The remainder was used for natural product isolation studies reported elsewhere (9). The preserved tissue was later dissected to separate zooids from the tunic, and DNA was extracted as previously described (5).
Illumina sequencing and metagenome assembly.
Illumina TruSeq libraries were prepared with ~300-bp inserts from DNA obtained from zooids and tunic of Lissoclinum sp. Libraries were sequenced using an Illumina HiSeq 2000 sequencer in multiple 101-bp paired-end runs. Sequence yields are shown in Table 1. Contaminating adaptor sequences were removed with Trimmomatic (71), and the trimmed reads were assembled with metaSPAdes (72).
Construction of the draft “Ca. Didemnitutus mandela” genome.
Contigs in the metagenomic assembly were classified taxonomically from their predicted ORFs as previously described (15, 16). All contigs classified as belonging to the phylum Verrucomicrobia were separated. Trimmomatic-filtered reads from both zooid and tunic fractions were aligned to these contigs with Bowtie 2 (73) (using the --very-sensitive option), and the aligned reads were assembled separately with SPAdes (74), using the --careful parameter. To identify potential connections between contigs and repeats, reads were realigned to contigs or derived sequences using Bowtie 2 (73) and the cytoscapeviz.pl script, part of the Multimetagenome package (75), was run on the alignment. Connections were visualized in Cytoscape (76). The Ver_v2 assembly was annotated with Prokka (77).
Single-copy marker gene analysis.
A set of 139 single-copy marker genes was identified using HMM profiles and cutoffs determined by Rinke et al. (17). The number of different marker genes, expressed as a percentage of 139, was used to estimate bacterial genome completeness. The number of different marker genes unique in a bin, expressed as a percentage of 139, was used to estimate genome purity.
Amplicon sequencing.
An ~430-bp section of 16S rRNA genes was amplified from DNA extracts using primers S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 (78), and an ~700-bp section of ketosynthase domains was amplified using primers KS-F and KS-R (79). In both cases, additional custom 5′ ends were added to primers, specific to each sample, including MiSeq adaptor sequences and a sample-identifying barcode sequence. Pooled amplicons were sequenced on an Illumina MiSeq instrument in a 251-bp paired-end run. For each sample, 16S and KS amplicons were dereplicated by identifying the respective primer sequences in the reads. The forward KS reads were used as queries in BLASTN searches against the mnd pathway, and reads with ≥97% identity and alignment of >90% of the read length were counted as mnd reads. To determine which KS reads were likely part of trans-AT PKS pathways, the forward reads were used as queries in a BLASTX search against the NCBI NR database, using the accelerated BLAST implementation DIAMOND (80). A list of proteins from trans-AT PKS pathways containing KS domains was compiled, and the accession numbers from this list were used to identify KS reads where one or more of the first 500 BLASTX hits were in the trans-AT list. These reads were counted as trans-AT KS reads.
Construction of phylogenetic trees.
Sequences used to make phylogenetic trees were aligned with either ClustalX (81) (small data sets) or Clustal Omega (82) (large data sets), except for the 16S rRNA tree, which used aligned sequences downloaded from the Ribosomal Database Project server (83). Alignments were inspected manually and trimmed before trees were constructed with FastTreeMP (84). The parameters “-slow -spr 5 -mlacc 3 -gamma -gtr -nt” and “-slow -spr 10 -mlacc 3 -bionj -gamma” were used to produce the nucleotide and protein trees, respectively. Trees were visualized on the Interactive Tree of Life server (85). To make the concatenated protein marker tree (see Fig. S1 in the supplemental material), marker protein sequences from the “Ca. Didemnitutus mandela” and Opitutus sp. GAS368 genomes were extracted with AMPHORA 2 (86). AMPHORA 2 was then used to make protein alignments with its reference database, and the corresponding tree was generated by concatenating the alignments, using only those genomes where all of the 31 markers shared by “Ca. Didemnitutus mandela” and Opitutus sp. GAS368 were present.
Repeat analysis.
Representative genomes of bacteria with different lifestyles were assembled from examples listed in the work of Lo et al. (4), using only complete genomes, which should give a more accurate quantification of repeats versus draft genomes (Table S1). Additionally, Opitutus sp. GAS368 and “Ca. Endolissoclinum faulkneri” were included in the analysis. For each genome, repeat regions of >50 bp were identified by using Nucmer (87) to align the genome to itself. Duplicate and self-hits were removed before repeat regions were extracted for quantification of total length and number of loci, etc.
Homolog analysis and identification of pseudogenes.
Predicted genes in the Ver_v2 assembly were used as queries in a BLASTP search against the NCBI NR database, using the accelerated BLAST implementation DIAMOND (80). The accession numbers of all annotated proteins in the Opitutus sp. GAS368 genome were obtained from NCBI, and the BLASTP table was searched for hits from this genome. If a protein from Opitutus sp. GAS368 was found in the first 100 hits, then the query protein was counted as having a homolog in this genome. R (88) was used to plot the comparison of homolog lengths (Fig. 2B). For proteins that did not have homologs in Opitutus sp. GAS368, the best BLASTP hit was instead used for comparison (Fig. 2C). To compare the Opitutus sp. GAS368 genome to that of “Ca. Didemnitutus mandela” (Table 4), the nucleotide sequence was reannotated in the same manner, with Prokka (77). To identify pseudogenes, predicted protein sequences were used as queries in a BLASTP search against the NCBI NR database, and then the annotated Opitutus sp. GAS368 proteins were removed from the results. Protein lengths were then compared to the respective best hit, and genes truncated by >20% compared to their best BLASTP hit were counted as putative pseudogenes. The secondary metabolite fraction of the Opitutus sp. GAS368 genome was calculated after searching for biosynthetic pathways with antiSMASH (89).
Functional gene analysis.
The functional analysis shown in Fig. 2D was carried out as previously described (4, 90). Briefly, the protein sequences of nonpseudogenes from “Ca. Didemnitutus mandela,” “Ca. Endolissoclinum faulkneri,” and Opitutus sp. GAS368 were classified with the KEGG Automatic Annotation Server (KAAS) (91). The resulting KEGG classifications were converted to Cluster of Orthologous Group (COG) categories using mappings supplied through the KEGG database. Bar plots were created using R (88). The presence of specific functions was also assessed through analysis of BLASTP results in MEGAN (92) and with reference to the EcoCyc database (93).
SNP detection.
Trimmed Illumina reads were aligned to the contig containing mnd (CD822_1), using Bowtie 2 (73) with the --very-sensitive parameter. The alignment was then loaded into Geneious (94) for SNP detection.
CAI calculation.
Codon adaptation index (CAI) values were calculated according to the formula of Sharp and Li (25), using the mnd genes to calculate relative synonymous codon usage (RCSU) and w values (Fig. S3). The CAI values were separated into different gene categories and plotted as box plots in R, using the ggplot2 package (95). To test for statistically significant differences between groups, one-way analysis of variance (ANOVA) was carried out in R, using the aov function, followed by Tukey's honest significant difference (HSD) test for significance.
Mitochondrion genome annotation and comparison to the Lissoclinum patella L2 mitochondrion.
An initial annotation was produced using Prokka (77) and manually inspected before additional genes were added, where appropriate, based on protein sequence similarity to other tunicate mitochondrial genes. Attempts were made to detect RNA genes with Rfam (96); however, none were found. Each gene was aligned to its counterpart in the Lissoclinum patella L2 mitochondrion genome using Clustal W (81), and the respective coordinates of the aligned regions were used to produce the diagram in Fig. 4A. The R library genoPlotR (http://genoplotr.r-forge.r-project.org) was used to plot both mitochondrial genomes to scale and the alignments.
Accession number(s).
Raw Illumina reads were deposited in the Sequence Read Archive (SRA) under accession numbers SRR5712450 to SRR5712457. The draft assembly Ver_v2 of the “Ca. Didemnitutus mandela” genome was deposited in GenBank under accession number NJAL00000000. The mitochondrial genome of Lissoclinum sp. was deposited in GenBank under accession number MF573328.
ACKNOWLEDGMENTS
We thank the South African Government for permission to make the tunicate collection under collection permit RES2013/43 issued by the South African Department of Environmental Affairs. We thank Chih-Horng Kuo (Academia Sinica, Taiwan), Michael Thomas (UW—Madison), and Scott Rajski (UW—Madison) for many helpful comments that improved the manuscript. This research was performed in part using the computer resources and assistance of the UW—Madison Center for High Throughput Computing (CHTC) in the Department of Computer Sciences.
K.L.M. collected and prepared tunicate samples for analysis; J.L., I.J.M., and J.C.K. designed and performed the research; J.L., K.L.M., and J.C.K. analyzed data; J.C.K. wrote the paper.
The CHTC is supported by UW—Madison, the Advanced Computing Initiative, the Wisconsin Alumni Research Foundation, the Wisconsin Institutes for Discovery, and the National Science Foundation and is an active member of the Open Science Grid, which is supported by the National Science Foundation and the U.S. Department of Energy’s Office of Science. This work was supported by the American Foundation for Pharmaceutical Education (to I.J.M.), as well as the School of Pharmacy, Graduate School, and the Institute for Clinical & Translational Research at the University of Wisconsin—Madison.
REFERENCES
- 1.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 2.Bennett GM, Moran NA. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci U S A 112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darmon E, Leach DRF. 2014. Bacterial genome instability. Microbiol Mol Biol Rev 78:1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo WS, Huang YY, Kuo CH. 2016. Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiol Rev 40:855–874. doi: 10.1093/femsre/fuw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. 2012. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci U S A 109:20655–20660. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwan JC, Schmidt EW. 2013. Bacterial endosymbiosis in a chordate host: long-term co-evolution and conservation of secondary metabolism. PLoS One 8:e80822. doi: 10.1371/journal.pone.0080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller IJ, Chevrette MG, Kwan JC. 2017. Interpreting microbial biosynthesis in the genomic age: biological and practical considerations. Mar Drugs 15:165. doi: 10.3390/md15060165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sikorska J, Hau AM, Anklin C, Parker-Nance S, Davies-Coleman MT, Ishmael JE, McPhail KL. 2012. Mandelalides A–D, cytotoxic macrolides from a new Lissoclinum species of South African tunicate. J Org Chem 77:6066–6075. doi: 10.1021/jo3008622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nazari M, Serrill JD, Sikorska J, Ye T, Ishmael JE, McPhail KL. 2016. Discovery of mandelalide E and determinants of cytotoxicity for the mandelalide series. Org Lett 18:1374–1377. doi: 10.1021/acs.orglett.6b00308. [DOI] [PubMed] [Google Scholar]
- 10.Lei H, Yan J, Yu J, Liu Y, Wang Z, Xu Z, Ye T. 2014. Total synthesis and stereochemical reassignment of mandelalide A. Angew Chem Int Ed Engl 53:6533–6537. doi: 10.1002/anie.201403542. [DOI] [PubMed] [Google Scholar]
- 11.Nazari M, Serrill JD, Wan X, Nguyen MH, Anklin C, Gallegos DA, Smith AB III, Ishmael JE, McPhail KL. 2017. New mandelalides expand a macrolide series of mitochondrial inhibitors. J Med Chem 60:7850–7862. doi: 10.1021/acs.jmedchem.7b00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR, Phillippy AM, Schatz MC, Piel J, Haygood MG, Ravel J, Schmidt EW. 2011. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc Natl Acad Sci U S A 108:E1423–E1432. doi: 10.1073/pnas.1111712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piel J. 2010. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep 27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 14.Helfrich EJN, Piel J. 2016. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep 33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- 15.Miller IJ, Weyna TR, Fong SS, Lim-Fong GE, Kwan JC. 2016. Single sample resolution of rare microbial dark matter in a marine invertebrate metagenome. Sci Rep 6:34362. doi: 10.1038/srep34362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller IJ, Vanee N, Fong SS, Lim-Fong GE, Kwan JC. 2016. Lack of overt genome reduction in the bryostatin-producing bryozoan symbiont, “Candidatus Endobugula sertula.” Appl Environ Microbiol 82:6573–6583. doi: 10.1128/AEM.01800-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 18.Chin KJ, Liesack W, Janssen PH. 2001. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division “Verrucomicrobia” isolated from rice paddy soil. Int J Syst Evol Microbiol 51:1965–1968. doi: 10.1099/00207713-51-6-1965. [DOI] [PubMed] [Google Scholar]
- 19.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 20.Lerat E, Ochman H. 2005. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res 33:3125–3132. doi: 10.1093/nar/gki631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guedes RLM, Prosdocimi F, Fernandes GR, Moura LK, Ribeiro HAL, Ortega JM. 2011. Amino acids biosynthesis and nitrogen assimilation pathways: a great genomic deletion during eukaryotes evolution. BMC Genomics 12(Suppl 4):S2. doi: 10.1186/1471-2164-12-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev 58:401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams DH, Stone MJ, Hauck PR, Rahman SK. 1989. Why are secondary metabolites (natural products) biosynthesized? J Nat Prod 52:1189–1208. doi: 10.1021/np50066a001. [DOI] [PubMed] [Google Scholar]
- 24.McDonald BR, Currie CR. 2017. Lateral gene transfer dynamics in the ancient bacterial genus Streptomyces. mBio 8:e00644-17. doi: 10.1128/mBio.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp PM, Li WH. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol 3:923–936. doi: 10.1016/S1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 27.Walsh CT, Gehring AM, Weinreb PH, Quadri LE, Flugel RS. 1997. Post-translational modification of polyketide and nonribosomal peptide synthases. Curr Opin Chem Biol 1:309–315. doi: 10.1016/S1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 28.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. 2006. Convergence of isoprene and polyketide biosynthetic machinery: isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc Natl Acad Sci U S A 103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hertweck C. 2009. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl 48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. 2008. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol 26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- 31.Jenner M, Frank S, Kampa A, Kohlhaas C, Pöplau P, Briggs GS, Piel J, Oldham NJ. 2013. Substrate specificity in ketosynthase domains from trans-AT polyketide synthases. Angew Chem Int Ed Engl 52:1143–1147. doi: 10.1002/anie.201207690. [DOI] [PubMed] [Google Scholar]
- 32.Kohlhaas C, Jenner M, Kampa A, Briggs GS, Afonso JP, Piel J, Oldham NJ. 2013. Amino acid-accepting ketosynthase domain from a trans-AT polyketide synthase exhibits high selectivity for predicted intermediate. Chem Sci 4:3212–3217. doi: 10.1039/c3sc50540e. [DOI] [Google Scholar]
- 33.Piel J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci U S A 99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M, Oldham NJ, van Echten-Deckert G, Okamura K, Yamamoto K, Inoue H, Ohkuma M, Hongoh Y, Miyagishima SY, Hattori M, Piel J, Fukatsu T. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol 23:1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Pöplau P, Frank S, Morinaka BI, Piel J. 2013. An enzymatic domain for the formation of cyclic ethers in complex polyketides. Angew Chem Int Ed Engl 52:13215–13218. doi: 10.1002/anie.201307406. [DOI] [PubMed] [Google Scholar]
- 36.Haines AS, Dong X, Song Z, Farmer R, Williams C, Hothersall J, Płoskoń E, Wattana-Amorn P, Stephens ER, Yamada E, Gurney R, Takebayashi Y, Masschelein J, Cox RJ, Lavigne R, Willis CL, Simpson TJ, Crosby J, Winn PJ, Thomas CM, Crump MP. 2013. A conserved motif flags acyl carrier proteins for β-branching in polyketide synthesis. Nat Chem Biol 9:685–692. doi: 10.1038/nchembio.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keatinge-Clay AT. 2007. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol 14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen MH, Imanishi M, Kurogi T, Smith AB III. 2016. Total synthesis of (−)-mandelalide A exploiting anion relay chemistry (ARC): identification of a type II ARC/CuCN cross-coupling protocol. J Am Chem Soc 138:3675–3678. doi: 10.1021/jacs.6b01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veerasamy N, Ghosh A, Li J, Watanabe K, Serrill JD, Ishmael JE, McPhail KL, Carter RG. 2016. Enantioselective total synthesis of mandelalide A and isomandelalide A: discovery of a cytotoxic ring-expanded isomer. J Am Chem Soc 138:770–773. doi: 10.1021/jacs.5b12318. [DOI] [PubMed] [Google Scholar]
- 40.Brütsch TM, Bucher P, Altmann KH. 2016. Total synthesis and biological assessment of mandelalide A. Chemistry 22:1292–1300. doi: 10.1002/chem.201504230. [DOI] [PubMed] [Google Scholar]
- 41.Willwacher J, Heggen B, Wirtz C, Thiel W, Fürstner A. 2015. Total synthesis, stereochemical revision, and biological reassessment of mandelalide A: chemical mimicry of intrafamily relationships. Chemistry 21:10416–10430. doi: 10.1002/chem.201501491. [DOI] [PubMed] [Google Scholar]
- 42.Willwacher J, Fürstner A. 2014. Catalysis-based total synthesis of putative mandelalide A. Angew Chem Int Ed Engl 53:4217–4221. doi: 10.1002/anie.201400605. [DOI] [PubMed] [Google Scholar]
- 43.White SW, Zheng J, Zhang YM, Rock CO. 2005. The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 44.Mahenthiralingam E, Song L, Sass A, White J, Wilmot C, Marchbank A, Boaisha O, Paine J, Knight D, Challis GL. 2011. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria genomic island. Chem Biol 18:665–677. doi: 10.1016/j.chembiol.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H, Machutta CA, Tonge PJ. 2010. Fatty acid biosynthesis and oxidation, p 231–275. In Mander L, Liu H-W (ed), Comprehensive natural products II. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 46.Sun Y, Song H, Li J, Jiang M, Li Y, Zhou J, Guo Z. 2012. Active site binding and catalytic role of bicarbonate in 1,4-dihydroxy-2-naphthoyl coenzyme A synthases from vitamin K biosynthetic pathways. Biochemistry 51:4580–4589. doi: 10.1021/bi300486j. [DOI] [PubMed] [Google Scholar]
- 47.Kwan JC, Tianero MDB, Donia MS, Wyche TP, Bugni TS, Schmidt EW. 2014. Host control of symbiont natural product chemistry in cryptic populations of the tunicate Lissoclinum patella. PLoS One 9:e95850. doi: 10.1371/journal.pone.0095850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gissi C, Pesole G, Mastrototaro F, Iannelli F, Guida V, Griggio F. 2010. Hypervariability of ascidian mitochondrial gene order: exposing the myth of deuterostome organelle genome stability. Mol Biol Evol 27:211–215. doi: 10.1093/molbev/msp234. [DOI] [PubMed] [Google Scholar]
- 49.Hedlund BP. 2010. Phylum XXIII. Verrucomicrobia phyl. nov., p 795–841. In Kreig NR, Ludwig W, Whitman W, Hedlund BP, Paster BJ, Staley JT, Ward N, Brown D, Parte A (ed), Bergey’s manual of systematic bacteriology, 2nd ed. Springer, New York, NY. [Google Scholar]
- 50.Belzer C, de Vos WM. 2012. Microbes inside—from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown AMV, Howe DK, Wasala SK, Peetz AB, Zasada IA, Denver DR. 2015. Comparative genomics of a plant-parasitic nematode endosymbiont suggest a role in nutritional symbiosis. Genome Biol Evol 7:2727–2746. doi: 10.1093/gbe/evv176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato T, Kuwahara H, Fujita K, Noda S, Kihara K, Yamada A, Ohkuma M, Hongoh Y. 2014. Intranuclear verrucomicrobial symbionts and evidence of lateral gene transfer to the host protist in the termite gut. ISME J 8:1008–1019. doi: 10.1038/ismej.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vollmers J, Frentrup M, Rast P, Jogler C, Kaster AK. 2017. Untangling genomes of novel planctomycetal and verrucomicrobial species from Monterey Bay kelp forest metagenomes by refined binning. Front Microbiol 8:472. doi: 10.3389/fmicb.2017.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arakawa K, Sugino F, Kodama K, Ishii T, Kinashi H. 2005. Cyclization mechanism for the synthesis of macrocyclic antibiotic lankacidin in Streptomyces rochei. Chem Biol 12:249–256. doi: 10.1016/j.chembiol.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 56.Fukami T, Yokoi T. 2012. The emerging role of human esterases. Drug Metab Pharmacokinet 27:466–477. doi: 10.2133/dmpk.DMPK-12-RV-042. [DOI] [PubMed] [Google Scholar]
- 57.Andersson DI, Hughes D. 2009. Gene amplification and adaptive evolution in bacteria. Annu Rev Genet 43:167–195. doi: 10.1146/annurev-genet-102108-134805. [DOI] [PubMed] [Google Scholar]
- 58.Yanai K, Murakami T, Bibb M. 2006. Amplification of the entire kanamycin biosynthetic gene cluster during empirical strain improvement of Streptomyces kanamyceticus. Proc Natl Acad Sci U S A 103:9661–9666. doi: 10.1073/pnas.0603251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Widenbrant EM, Tsai HH, Chen CW, Kao CM. 2008. Spontaneous amplification of the actinorhodin gene cluster in Streptomyces coelicolor involving native insertion sequence IS466. J Bacteriol 190:4754–4758. doi: 10.1128/JB.00131-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Y, Xia L, Ding X, Luo Y, Huang F, Jiang Y. 2011. Duplication of partial spinosyn biosynthetic gene cluster in Saccharopolyspora spinosa enhances spinosyn production. FEMS Microbiol Lett 325:22–29. doi: 10.1111/j.1574-6968.2011.02405.x. [DOI] [PubMed] [Google Scholar]
- 61.Murakami T, Burian J, Yanai K, Bibb MJ, Thompson CJ. 2011. A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc Natl Acad Sci U S A 108:16020–16025. doi: 10.1073/pnas.1108124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moran NA, Plague GR, Sandström JP, Wilcox JL. 2003. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc Natl Acad Sci U S A 100(Suppl 2):14543–14548. doi: 10.1073/pnas.2135345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamas I, Klasson L, Canbäck B, Näslund AK, Eriksson AS, Wernegreen JJ, Sandström JP, Moran NA, Andersson SGE. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 64.Van Ham RCHJ, Martínez-Torres D, Moya A, Latorre A. 1999. Plasmid-encoded anthranilate synthase (TrpEG) in Buchnera aphidicola from aphids of the family Pemphigidae. Appl Environ Microbiol 65:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Latorre A, Gil R, Silva FJ, Moya A. 2005. Chromosomal stasis versus plasmid plasticity in aphid endosymbiont Buchnera aphidicola. Heredity 95:339–347. doi: 10.1038/sj.hdy.6800716. [DOI] [PubMed] [Google Scholar]
- 66.Clayton AL, Oakeson KF, Gutin M, Pontes A, Dunn DM, von Niederhausern AC, Weiss RB, Fisher M, Dale C. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet 8:e1002990. doi: 10.1371/journal.pgen.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toh H, Weiss BL, Perkin SAH, Yamashita A, Oshima K, Hattori M, Aksoy S. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res 16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moran NA. 1996. Accelerated evolution and Muller’s ratchet in endosymbiotic bacteria. Proc Natl Acad Sci U S A 93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ream DC, Bankapur AR, Friedberg I. 2015. An event-driven approach for studying gene block evolution in bacteria. Bioinformatics 31:2075–2083. doi: 10.1093/bioinformatics/btv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawrence JG, Roth JR. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 76.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 78.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng Z, Qi J, Tsuge T, Oba Y, Kobayashi T, Suzuki Y, Sakagami Y, Ojika M. 2005. Construction of a bacterial artificial chromosome library for a myxobacterium of the genus Cystobacter and characterization of an antibiotic biosynthetic gene cluster. Biosci Biotechnol Biochem 69:1372–1380. doi: 10.1271/bbb.69.1372. [DOI] [PubMed] [Google Scholar]
- 80.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 81.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 82.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu M, Scott AJ. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 87.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.R Core Team 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org. [Google Scholar]
- 89.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH. 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lo WS, Chen LL, Chung WC, Gasparich GE, Kuo CH. 2013. Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genomics 14:22. doi: 10.1186/1471-2164-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Schröder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 96.Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Approximately maximum-likelihood tree generated by FastTreeMP from 31 concatenated single-copy marker gene protein sequences from the “Ca. Didemnitutus mandela” genome and 1,068 other reference genomes (species outside the PVC superphylum and Elusimicrobia not shown). Bootstrap proportions greater than 70% are expressed to the left of each node as a percentage of 1,000 replicates. Download FIG S1, EPS file, 0.9 MB (991.9KB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of repeat structure in symbionts and free-living bacteria. Download TABLE S1, PDF file, 0.1 MB (56.4KB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative pseudogenes with functional annotations in the “Ca. Didemnitutus mandela” genome. Download TABLE S2, PDF file, 0.1 MB (64.3KB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SNPs identified within the mnd pathway (mnd genes are green, other protein coding genes are gray, and tRNAs are magenta). Read coverage is plotted above the contig. Download FIG S2, EPS file, 1.1 MB (1.2MB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Box plots of codon adaptation index (CAI) of different groups of genes within the “Ca. Didemnitutus mandela” genome. (B) Individual P values for pairwise comparisons of the groups compared through one-way ANOVA and Tukey HSD. Significant P values (P < 0.05) are highlighted in red. Annotated genes are not significantly different in codon usage from mnd genes. However, both pseudogenes and hypothetical genes are significantly different from annotated genes in this respect. Download FIG S3, EPS file, 0.9 MB (913.7KB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expansions of clades containing mnd KS domains in an approximately maximum-likelihood tree made from 665 KS domain sequences. Colors in each clade correspond to the chemical structures shown immediately to its right side. Download FIG S4, PDF file, 1.6 MB (1.6MB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Alignment of mnd KS domains, with the CHH catalytic triad highlighted. The domains where this triad is disrupted are predicted to be catalytically inactive. (B) Alignment of mnd KR domains, along with two from the erythromycin biosynthetic gene cluster to allow comparison to previous alignments. The NADPH-binding site (GxxxGxG) is highlighted, as is the LDD motif that is thought to control product stereoconfiguration. The catalytic triad (KSY) is also highlighted. (C) Alignment of mnd DH domains. Highlighted are the conserved motifs HxxxGxxxxP and DxxxQ, which are involved in catalysis. (D) Alignment of mnd ACP domains, with the extender unit attachment point serine highlighted. In addition, residues characteristic of β-branching ACPs in MndD_ACP3 are shown in red. Download FIG S5, PDF file, 0.3 MB (313KB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Approximately maximum-likelihood tree based on DH and PS domains. The PS domains form a distinct clade, as shown previously (35). Download FIG S6, EPS file, 1.9 MB (2MB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Canonical mechanism of KR domains with catalytic triad KSY. (B) Proposed mechanism of functional KR domains with catalytic triad KSH. Download FIG S7, EPS file, 2.2 MB (2.2MB, eps) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Matrices showing pairwise amino acid identities for the full-length Lissoclinum sp. COX1 gene and other COX1 sequences from the NCBI database. Download FIG S8, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2017 Lopera et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.