Abstract
The inwardly rectifying potassium channel Kir 2.3 is specifically targeted and expressed on the basolateral membrane of certain renal epithelial cells. In the present study, the structural basis for polarized targeting was elucidated. Deletion of a unique COOH-terminal domain produced channels that were mistargeted to the apical membrane, consistent with the removal of a basolateral membrane-sorting signal. By characterizing a series of progressively smaller truncation mutants, an essential targeting signal was defined (residues 431–442) within a domain that juxtaposes or overlaps with a type I PDZ binding motif (442). Fusion of the COOH-terminal structure onto CD4 was sufficient to change a random membrane-trafficking and expression pattern into a basolateral membrane one. Using metabolic labeling and pulse–chase and surface immunoprecipitation, we found that CD4-Kir2.3 COOH-terminal chimeras were rapidly and directly targeted to the basolateral membrane, consistent with a sorting signal that is processed in the biosynthetic pathway. Collectively, the data indicate that the basolateral sorting determinant in Kir 2.3 is composed of a unique arrangement of trafficking motifs, containing tandem, conceivably overlapping, biosynthetic targeting and PDZ-based signals. The previously unrecognized domain corresponds to a highly degenerate structure within the Kir channel family, raising the possibility that the extreme COOH terminus of Kir channels may differentially coordinate membrane targeting of different channel isoforms.
Transport proteins in epithelial cells must be specifically targeted and retained at particular membrane domains to affect vectorial transport of fluid, solutes, and electrolytes. Current opinion holds that polarized protein expression is determined by specific sorting and retention mechanisms (1, 2). In renal epithelial cells, most newly synthesized membrane proteins are segregated within trans-Golgi apparatus into distinct transport vesicles destined to the apical or basolateral membranes (2, 3). Directed trafficking is determined by signals contained within the membrane protein structure. Sorting machineries in the trans-Golgi network read, interpret, and act on the signals to shuffle molecules to the proper destination (2). Once delivered to the appropriate domain, many of these integral membrane proteins are effectively anchored at these polarized locales through interactions with the cytoskeleton or other associated proteins, completing the polarization program (4, 5).
Recently, several distinct classes of sorting motifs have been identified. Glycerophosphatidylinsitol linkage seems to be an important component for sorting numerous apical membrane proteins (6). Transmembrane domain structures (7) and N-linked glycosylation are required for apical membrane sorting in others (8). Two different basolateral sorting signals have been defined from work on several unrelated proteins. One class is similar to coated pit localization signals (2, 9, 10). The other class, distinguished only by being unrelated to endocytic signaling motifs, may actually represent several distinct kinds of sorting signals (3, 11–13). To date, most of the targeting determinants, particularly the basolateral ones, have largely been identified and characterized in several unrelated receptor proteins, e.g., low density lipoprotein (LDL) (9, 14), transferrin (15), polymeric immunoglobin receptor (16, 17), and IgG Fc receptors (18). Consequently, the extent to which basolateral membrane transport proteins share common targeting mechanisms largely remains undetermined.
The recent identification of a plausible gene candidate, Kir 2.3, for a basolateral membrane potassium channel (19, 20) provided the opportunity to elucidate the basis for polarized targeting of a native renal epithelial transport protein. Mutational and chimeric analysis revealed a structure that is both necessary and sufficient for basolateral sorting in the biosynthetic pathway. Kir 2.3 channels lacking the domain are mistargeted to the apical membrane. Fusion of the Kir 2.3 sorting domain onto CD4 was sufficient to change a random membrane-trafficking and expression pattern into a basolateral membrane one. Interestingly, the sorting determinant lies at the extreme COOH terminus and juxtaposes or is colinear with a PDZ binding motif (21). Collectively, the data indicate that basolateral membrane expression of the Kir 2.3 channel is coordinated by a sorting signal, perhaps working in conjunction with a nearby PDZ ligand.
Experimental Procedures
Construction of Epitope-Tagged KIR 2.3 Mutants.
Stop codons or alanine substitutions were introduced into the vesicular stomatitis virus (VSV) (Y2F) epitope-tagged-Kir 2.3 cDNA by PCR (20). The Y2F form of the VSV G protein epitope contains a phenylalanine replacement of the tyrosine residue to disrupt a potential sorting signal (10). Like the native 12-aa epitope, VSV (Y2F) is recognized by the P5D4 mAb (Sigma).
CD4-Kir 2.3C chimeras were constructed by PCR by using overlapping oligonucleotides. Constructs comprised the entire extracellular and transmembrane domains of the human CD4 protein (GenBank accession no. M12807; ref. 22) and COOH-terminal cytoplasmic domains of Kir 2.3. Channel domains were fused at amino acid 422 of CD4 to create CD4 chimeras of either the entire COOH-terminal tail of Kir 2.3 [CD4 Kir 2.3 (170)] or portions of the Kir 2.3 extreme COOH terminus [CD4 Kir 2.3 (413) or CD4-Kir 2.3 (413–431STOP)]. The control CD4 construct for these experiments (CD4-stop) was obtained by introducing a stop codon at the fusion site, truncating most of the cytoplasmic COOH terminus except for the two amino acids that follow the transmembrane domain of CD4.
All of the constructs were sequenced with chain-terminating inhibitors by using sequenase to verify proper construction. Each construct was subcloned into the expression vector Pcb6 containing the G418 resistance gene (23) for generation of stably transfected Madin–Darby canine kidney (MDCK) cell lines.
Transfection and Stable Cell Line Generation.
MDCK cells were plated at a density of 2 × 105 cells per 35-mm plastic dish and grown in DMEM supplemented with 10% FBS. One day after seeding, the MDCK cells were transfected with 5 μg of recombinant PCB6 plasmid by using Lipofectamine (BRL) according to the manufacturer's specifications. The cells were trypsinized, diluted in DMEM (4:1), and seeded onto 100-mm Petri dishes 48 h after transfection. Stably transfected MDCK cells clones were isolated in a selection medium containing 500 μg/ml G418 (BRL). Control MDCK cell clones were produced by transfecting cells with the Pcb6 plasmid alone. LLC-PK1 cells (ATCC Cl-101 as used in ref. 24) were processed for transfection in an identical manner, except they were grown in MEMα supplemented with 10% FBS.
Vectorial Delivery of the CD4-Kir 2.3 (170) Chimera.
In these studies, confluent MDCK cells were grown on permeable supports. Cells were washed twice and then incubated at 37°C for 30 min in cysteine and methionine-free DMEM (GIBCO/BRL). The cells were then incubated for 10 min at 37°C with 400 μCi/ml [35S]methionine and [35S]cysteine (Express Protein Labeling Mix, NEN). They were then rinsed three times and incubated for 30 min or 1 h in complete DMEM containing a 10-fold excess of cold cysteine and methionine. After the chase period, anti-CD4 (2 μg/ml) and anti-uvomorulin (1 μg/ml) (Sigma) antibodies were added to either the apical or basolateral membrane compartments in PBS, containing 1 mM CaCl2 and 1 mM MgCl2 (PBSCM). After a 2-h incubation at 4°C, cells were extensively rinsed with PBSCM at 4°C and lysed in buffer A (150 mM NaCl/20 mM Tris/5 mM EDTA, pH 7.5) containing 1% Triton X-100, 0.1% BSA, and a protease inhibitor mixture (antipain, leupeptin, and pepstatin, 10 μg/ml each, and 1 mM PMSF) (buffer A). CD4 proteins were recovered on a suspension of prewashed protein-A Sepharose CL4B beads (50 μl) for 2 h at 4°C (Sigma). The beads were washed three times with buffer A, twice with buffer A without Triton X-100, and once with 50 mM Tris, pH 8.0. The beads were then boiled with Laemmli buffer, and the proteins were resolved by SDS/PAGE. The immunoprecipitated CD4 proteins from the apical membranes (CD4ap) or the membranes (CD4bl) were then quantified by using a PhosphorImager (Molecular Dynamics).
The amount of CD4 protein appearing in the basolateral membrane relative to the total plasma membrane (apical + basolateral) was determined in following manner. Two CD4-transfected and two mock-transfected MDCK cell filters were processed simultaneously. For each group, cells were incubated with antibodies on either the apical or the basolateral compartment. Relative basolateral delivery was calculated as %Bl = cCD4bl/(cCD4bl + cCD4ap), where cCD4bl and cCD4ap are the amounts of CD4 proteins immunoprecipitated from the basolateral and apical compartments, respectively. In parallel studies, antibodies were added to both compartments. In these cases, the total CD4 recovery was identical to the sum of labeling (cCD4bl + cCD4ap) from individual experiments. Immunoprecipitation of an intracellular marker, uvomorulin, was also done to check for intracellular CD4 contamination that can occur when cells become damaged before or during the antibody binding period. Gels showing an uvomorulin signal were discarded.
Anti-CD4 antibody (Sim.2, National Institutes of Health AIDS Research and Reference and Reagent Program) was purified from mice ascite fluid according to standard protocols (25).
Immunofluorescence Experiments.
Stably transfected cells were grown on permeable supports until they formed a confluent monolayer. They were rinsed twice with PBSCM and fixed with 3% paraformaldehyde for 30 min. Cultures were then washed three times with PBSCM and incubated for 15 min in PBSCM containing 50 mM NH4Cl. After three PBSCM washes, the cultures were permeabilized with 0.1% Triton X-100 in PBSCM for 30 min and incubated for 2 h with the P5D4 antibody (Sigma) (1:1,000) in PBSCM supplemented with 1% BSA. After three washes, the cultures were incubated for 30 min with rhodamine-conjugated goat anti-mouse IgG (1:200) (Pierce) in the same buffer. Filters were washed with PBSCM, mounted in PBS/glycerol, and observed with a Leitz confocal microscope. In some cases, images were quantified for basolateral and apical membrane expression using National Institutes of Health image software according to the methods of Gottardi and Caplan (7).
Yeast Two-Hybrid Studies.
The yeast two-hybrid interaction trap system was used according to established methods (26) to test for interaction between the COOH-terminal sorting signal in Kir 2.3 and the μ1B clathrin adapter subunit. LexA fusion proteins of the Kir 2.3 COOH-terminal targeting domain with (amino acids 417–445) and without (417–442X) the PDZ binding motif were constructed in the yeast expression plasmid, pJK202, containing an HIS3-selectable marker. A cDNA encoding the μ1B subunit (GenBank accession number AA100486) was cloned in-frame with the AD transcriptional activation domain in the pJG4-5 plasmid (TRP4-selectable marker). A positive control was constructed by subcloning an oligonucleotide encoding three tandem repeats of the consensus uIB binding motif (YRRL) (27, 28) in-frame with the Lex A DNA binding module in pJK202. Saccharomyces cerevisiae (EGY 48, Mat a-ura3, his3, trp1, ura3, 3lexAop-leu2) was cotransfected with the reporter, pJG4-5 containing μ1B, and pJK202 containing Kir 2.3C or Kir 2.3C442X. To select for yeast that were triple transfected with pJG4–5/μ1B, the pJK202 constructs, and the reporter plasmid, yeast were plated onto medium that lacks uracil, histidine, and tryptophan. In this system, expression of the AD-fusion protein is under the control of the GAL1-inducible promoter, and interaction of the LexA and AD fusion partners causes transcriptional activation of a LacZ reporter gene. Accordingly, yeast that turned blue on plates containing galactose and 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal) [(uracil, histidine, and tryptophan), 2% galactose, and 1% raffinose as a carbon source] but remained white on X-gal plates containing glucose were considered positive.
Results
The Cytoplasmic COOH Terminus of KIR 2.3 Contains a Basolateral Sorting Signal.
Several conspicuous features of the large cytoplasmic COOH terminus (see below) provided reason to suspect that this domain contained basolateral membrane-sorting information. To test this hypothesis, we constructed a chimeric molecule between the human CD4 protein (22) and Kir 2.3 in which the extracellular and transmembrane domains of CD4 were fused to the entire COOH terminus of Kir 2.3 (CD4-Kir 2.3C, 170–445) (Fig. 1). The extracellular location of the SIM2 epitope in CD4 permits direct evaluation of the targeting mechanism by biosynthetic labeling, pulse–chase, and surface immunoprecipitation. Moreover, the approach allows direct determination of whether the Kir 2.3 COOH terminus is sufficient to coordinate basolateral membrane targeting.
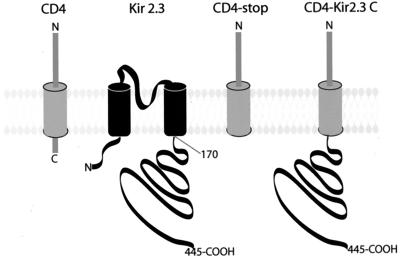
Figure 1.
CD4-Kir 2.3 chimeric proteins. The extracellular and transmembrane domains of CD4 were used to construct a chimeric protein with Kir 2.3. A stop codon was introduced 2 aa after CD4 transmembrane domain to create the control CD4 construct (CD4-Stop). The entire cytoplasmic COOH terminus of Kir 2.3 (amino acids 171–445) was fused at the same position to create the CD4-Kir 2.3 chimera.
In these studies, MDCK cells were grown on permeable supports and were pulse-labeled for 10 min with [35S]methionine. After 0.5-h or 1-h chase periods, CD4-Kir 2.3C protein appearing in the apical and basolateral membranes was bound with extracellular anti-CD4 antibodies, immunoprecipitated, and quantified. As illustrated in Fig. 2C, 77.6 ± 4.3% (n = 3) and 73.8 ± 5 (n = 4) of the CD4-KIR 2.3C was detected in the basolateral membranes of the cells after 30-min and 1-h chase periods, respectively. In contrast, the control CD4-stop construct was expressed equally on the apical and basolateral membranes (48.9 ± 6.1% at t = 30 min and 45.7 ± 7% at t = 1 h, n = 3).
Figure 2.
The COOH tail of Kir 2.3 is necessary and sufficient for basolateral membrane expression. Amounts of CD4-stop or CD4-Kir 2.3 appearing in the apical (Ap) or basolateral (bl) membrane were evaluated by surface immunoprecipitation in stably transfected MDCK cells after metabolic labeling (35S-methionine, 10 min) and a 30-min to 1-h chase period. (A) Audioradiogram of a representative surface immunoprecipitation on experiment with cells transfected with CD4-stop (Left) or CD4-Kir 2.3C (Right). In contrast to the random targeting of CD4-stop, the CD4-Kir 2.3C is preferentially targeted to the basolateral membrane. Lanes marked “C” are surface immunoprecipitations (apical + basolateral surface) from mock-transfected cells. (B) Summary. Relative basolateral delivery was calculated as %Bl = cCD4bl/(cCD4bl + cCD4ap), where cCD4bl and cCD4ap are the amounts of CD4 proteins measured in the transfected cells.
These studies reveal that the Kir 2.3 COOH terminus is sufficient to direct basolateral membrane targeting in MDCK cells. The routing mechanism seems to be direct rather than random and transcytosic. Indeed, the chimera is rapidly and preferentially delivered to the basolateral membrane. Basolateral membrane delivery of the CD4-KIR 2.3C construct is not absolute (e.g., 77% basolateral membrane delivery), perhaps reflecting a lower sorting efficiency that occurs in the context of a CD4 monomer compared with the native channel, a tetramer. Nevertheless, these data provide strong support for a sorting signal that is located within the COOH-terminal domain of Kir 2.3, a sorting signal that is sufficient to coordinate a critical basolateral membrane-trafficking step in the biosynthetic pathway, presumably within the trans-Golgi network.
Putative Polarization Signals in the Extreme COOH-Terminal Domain of Kir 2.3.
Alignment of the COOH-terminal sequence of Kir 2.3 with a related apical membrane inward-rectifying K channel, Kir 1.1a (ROMK) (29), revealed that the extreme COOH-terminal tail of Kir 2.3 is most different, being 55 residues longer. Interestingly, this unique domain contains several conspicuous motifs, suggesting a structural basis for basolateral membrane targeting. The domain contains a potential tyrosine-based sorting signal, similar to the distal signal in the LDL receptor (9), and a PDZ binding motif (21).
Role of the COOH-Terminal Tyrosine-Containing Motif in Basolateral Membrane Localization.
A COOH-terminal domain, composed of tyrosine 372 and a downstream cluster of glutamic acid residues (382), is similar to the tyrosine-based basolateral membrane-sorting signal (distal determinant) in the LDL receptor (3, 9, 14). To address the importance of this potential sorting signal in Kir 2.3 basolateral targeting, the tyrosine 372 was mutated to an alanine as was previously described for the LDL receptor (9). In contrast to the apical missorting phenotype in the LDL receptor, this point mutation (Kir 2.3–Y372A) did not modify basolateral expression of the Kir 2.3 channel protein (not shown).
The Extreme COOH-Terminal Tail of Kir 2.3 Contains a Basolateral Membrane Localization Signal.
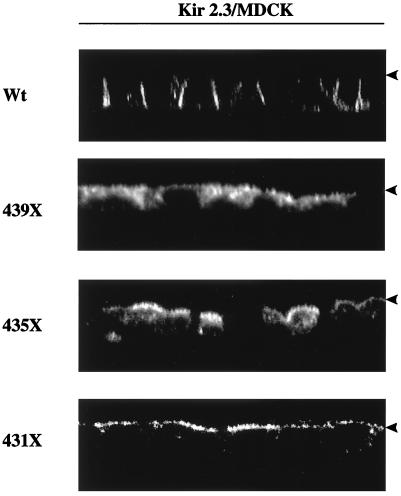
To test whether the basolateral membrane expression depended on other information contained within the unique COOH-terminal tail, deletion analysis was performed. Truncation of the last 89 (Kir 2.3, 356X, where X is a stop codon), 75 (370X), or 45 (400X) amino acids produced channels that were predominantly expressed in a perinuclear compartment, presumably the Golgi (not shown). In contrast, deletion of the last 15 (431X), 11 (435X), or seven (439X) amino acid residues resulted in a dramatic change in plasma-membrane targeting. The 439X mutant exhibited a mixed apical, basolateral, and intracellular localization (Fig. 3). Channels bearing the two larger deletions mutants, 431X and 435X, were exclusively expressed in the apical membrane (Fig. 3). The shift from basolateral to apical expression is reminiscent of other plasma membrane-sorting signals. Removal of a dominant signal often reveals a cryptic contralateral membrane-sorting signal (2, 3). In any regard, these data indicate that the last 10–15 aa of Kir 2.3 form a signal that is necessary for basolateral membrane localization.
Figure 3.
Basolateral targeting determinant is located within the extreme COOH terminus of Kir 2.3. Reconstructed z-plane confocal images of MDCK cell monolayers stably transfected with various Kir 2.3 constructs. In contrast to the basolateral membrane expression of the wild-type (Wt) channel, mutant Kir 2.3 channels lacking the last 10–15 aa (435X, 431X) are expressed on the apical membrane (arrowhead), consistent with the removal of a basolateral membrane-targeting signal.
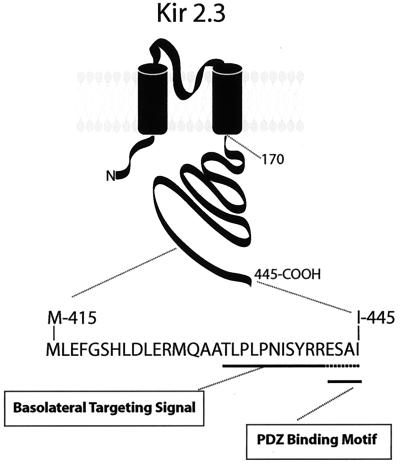
Interestingly, the COOH-terminal sorting domain contains a consensus motif for type I PDZ binding, S/TXV/I/L (442-ESAI-445) (21). In fact, we recently found that Kir 2.3 interacts with a PDZ protein, Lin-7, on the basolateral membrane to stabilize expression of the channel on the plasmalemma (O.O., H. Liu, J.M., and P.A.W., unpublished results). Consistent with a retention function, removal of the PDZ ligand caused channels to accumulate into an endocytic compartment (O.O., H. Liu, J.M., and P.A.W., unpublished results). Accordingly, to delimit the basolateral membrane signal in the context of the archetypal PDZ binding motif, an internal deletion mutant (Δ431–441) was introduced into Kir 2.3, leaving the last four residues intact. As illustrated in Fig. 4, Kir 2.3 Δ431–441 was largely delivered to the apical membrane of MDCK cells, suggesting that the basolateral sorting motif requires residues 431–441, upstream from the PDZ binding site. In this case, the COOH-terminal basolateral sorting determinant juxtaposes or may even overlap with the smaller PDZ binding motif.
Figure 4.
Basolateral sorting information is located upstream from the PDZ binding motif. Confocal image of an MDCK cell monolayer stably transfected with a mutant channel (Kir 2.3 Δ431–441) harboring an internal deletion that leaves the PDZ binding site intact. As shown in x-y (above) or z-plane images, the PDZ binding motif is not sufficient to define a basolateral membrane-targeting domain; Kir 2.3 Δ431–441 channels are largely expressed on the apical membrane, similar to 435X and 431X.
The studies above reveal that the extreme COOH terminus is necessary for basolateral membrane expression. To determine whether this domain is sufficient to coordinate basolateral membrane localization, two CD4 chimeras of the extreme COOH terminus were constructed (CD4-Kir 2.3, 413–445 and CD4 Kir 2.3, 413–431), stably expressed in MDCK cells, and examined for polarized expression. Unfortunately, we could not consistently detect enough of either protein by biosynthetic labeling and surface immunoprecipitation to produce reliable quantitative data for making conclusions about the mechanism of polarized processing. Nevertheless, steady-state expression of both constructs could be detected by immunofluorescence and confocal microscopy. As shown in Fig. 5, the CD4 Kir 2.3, 413–445 was preferentially expressed on the basolateral membrane. In contrast, chimeras lacking the last 14 COOH-terminal residues, CD4 Kir 2.3, 413–431, are largely confined to the apical membrane. Collectively, these studies reveal that the extreme COOH-terminal domain of Kir 2.3 is both necessary and sufficient to coordinate basolateral membrane expression.
Figure 5.
Extreme COOH-terminal domain is sufficient to coordinate basolateral membrane localization. Reconstructed z-plane confocal images of MDCK cells stably transfected with two different CD4Kir 2.3 chimeras. One contains residues 413–445; the other is truncated after residue 431, removing the sorting signal defined above. The final 32 aa are sufficient for basolateral membrane expression. As defined above, the final 14 aa are necessary to direct basolateral membrane targeting.
Basolateral Membrane-Sorting Mechanism of the Kir 2.3 Channel.
Some basolateral membrane proteins require an epithelial-specific subunit of the AP-1 clathrin adaptor complex, μIb (24), for appropriate basolateral membrane trafficking. To test whether this was the case for Kir 2.3, the channel was stably transfected in a renal epithelial cell line, LLCPK1, which lacks the μIb subunit (27). In contrast to apical mistargeting of the LDL receptor (24) or the beta subunit of the H/K ATPase (30), Kir 2.3 was stably expressed on the basolateral membrane of LLCPK1 cells (Fig. 6A). This result is consistent with observations in the yeast two-hybrid system. In contrast to the interaction of μIb subunit with a tyrosine-based sorting signal (3x YRRL), no interaction was observed between the μIb subunit and the COOH-terminal sorting signal of Kir 2.3. Moreover, removal of the PDZ ligand in Kir 2.3 (442x) did not reveal a cryptic μIb interaction site. These data support the notion that basolateral membrane sorting of the Kir 2.3 channel occurs by a μIb-independent mechanism.
Figure 6.
Basolateral membrane sorting of Kir 2.3 occurs by a mechanism that is independent of the μIb clathrin adaptin subunit. (A) Confocal image of Kir 2.3 stably transfected in LLC-PK1 cells, demonstrating basolateral expression in a cell line that is deficient in the μIb clathrin adaptor subunit. (Upper) X–Y plane is shown. (Lower) Z-plane reconstruction is shown. (B) Yeast two-hybrid interaction study. Yeast were cotransfected with vectors expressing (i) a recombinant LexA protein fused to the Kir 2.3 sorting signal (final 27 aa), a triple repeat of a tyrosine-based sorting signal (YRRL), or the Kir 2.3 sorting signal without the PDZ ligand (442x); (ii) a recombinant AD protein fused to the μIb subunit; and (iii) a LacZ reporter gene under Lex A operator control. Yeast were plated on 5-bromo-4-chloro-3-indolyl β-d-galactoside plates containing galactose (+) or glucose (−) to induce or repress the expression of the AD protein. Yeast that turned blue were scored positively.
Discussion
In the present study, the structural basis for polarized expression of the inwardly rectifying potassium channel, Kir 2.3, was elucidated. Deletion of a unique COOH-terminal domain produced channels that were mistargeted to the apical membrane, consistent with the removal of a basolateral membrane-sorting signal. By characterizing a series of progressively smaller truncations, an essential targeting signal was defined (residues 431–442) within a domain that juxtaposes or is colinear with a PDZ binding motif. Fusion of the extreme Kir 2.3 COOH-terminal signal onto a randomly sorted protein, CD4-stop, conferred a pattern of basolateral membrane localization. Collectively, our studies reveal a sorting signal that is both necessary and sufficient to direct basolateral membrane expression of the Kir 2.3 channel.
Identification of the sorting signal provides important clues about the nature of the sorting machinery. For instance, a plausible mechanism explaining one class of basolateral sorting signals was recently advanced (24, 27). Some basolateral membrane proteins apparently require an epithelial-specific subunit of the AP-1 clathrin adaptor complex, μIb (24), for appropriate trafficking. Such proteins are mistargeted to the apical membrane in LLCPK1 cells (30), a renal epithelial cell line lacking μIb (27). Because tyrosine-based endocytic/basolateral membrane-sorting signals (31) have the capacity to directly interact with uIb subunits (27), it seems likely that the AP-1b complex is recruited to the trans-Golgi network, or conceivably recycling endosomes, by specific membrane protein cargo containing these interaction motifs, effectively marking them for basolateral membrane expression. Although it remains to be experimentally verified, other shared endocytotic, basolateral sorting signals may also engage the AP-1b sorting machinery. For instance, dileucine motifs and dihydrophobic signals are predicted to interact with AP-1b via interactions with another adaptin, the beta chain (32). Similarly, acidic cluster-sorting motifs (13) may interact with AP-1b complex using connector proteins such as PAC-1 (33).
The basolateral sorting signal in Kir 2.3 does not share any resemblance to recognition motifs involved in clathrin adaptor complex association. Although the domain does contain a tyrosine, the residue, Tyr-439, does not fall within a YXXØ (Y is tyrosine, X is any amino acid, and Ø is an amino acid with a bulky hydrophilic side chain) or NPXY (N is asparagine and P is proline) sorting motif (31). Furthermore, our data directly support the idea that basolateral trafficking of the Kir 2.3 channel occurs via an AP-1b independent mechanism. In contrast to apical mistrafficking of the LDL receptor and transferrin in LLPKC1 cells (24), we found that the Kir 2.3 channel is expressed on the basolateral membrane of these μIb-deficient cells. Furthermore, the Kir 2.3 sorting signal did not interact with this AP-1 subunit in the yeast two-hybrid system.
The sorting determinant in Kir 2.3 also is unrelated to other basolateral sorting signals. Indeed, it is not similar to acidic cluster in furin (13); the H/RXXV motif in the polymeric Ig receptor (17) and the cation-dependent mannose-6-phosphate receptor (34); or the transferrin motif (15). However, it is noteworthy that the sorting determinant in Kir 2.3 contains a protein–protein interaction signal called a PDZ binding motif. Generally found at the extreme COOH terminus, these small motifs (3–4 residues) engage PDZ-containing proteins to facilitate multiprotein complex formation and to localize expression on particular membrane domains (35). Consistent with this view, we recently found that a PDZ complex, composed of Lin-7 and CASK, interacts with Kir 2.3 to retain the channel at the basolateral membrane (O.O., H. Liu, J.M., and P.A.W., unpublished results).
An interaction that is coordinated solely by the classic, 4-aa, type I PDZ binding motif in Kir 2.3 cannot account for the entire basolateral membrane-trafficking program, however. Certainly, our mutagenesis studies indicate that the domain proximal to the archetypal PDZ ligand is critical for basolateral membrane sorting. Moreover, the 4-aa PDZ binding motif seems to play a different role than the broader basolateral sorting domain. At least, removal of the archetypal PDZ binding site (442X) produces a completely different sorting phenotype than removal of the upstream domain (Kir Δ431–441). In contrast to the endosomal localization of mutant channels, lacking the last 4 aa (442X) (O.O., H. Liu, J.M., and P.A.W., unpublished results), Kir 2.3 431X or Kir 2.3 Δ431–441 channels are largely mistargeted to the apical membrane, consistent with the removal of a basolateral membrane-sorting determinant.
Whether the proximal domain (431) acts as a discrete sorting signal or acts as a part of a larger structure, involving the PDZ binding motif, remains uncertain. In light of a recent study illustrating that residues proximal to the archetypal PDZ binding motif can influence the affinity and specificity of more complicated PDZ interactions (36), this issue is particularly relevant. Unfortunately, our analysis of the particular determinants of PDZ-dependent Lin 7–Kir 2.3 interaction did not shed light on this issue. We found that Lin 7 binding is not influenced by alanine replacement of individual residues in the proximal sorting domain of Kir 2.3, but binding is attenuated to about one-fourth of control levels when the entire domain is replaced with alanines (431–441A) (O.O., H. Liu, J.M., and P.A.W., unpublished results). In any case, the data collectively indicate that the proximal domain (431), operating independent or jointly with the archetypal PDZ ligand, affects an early basolateral sorting process, presumably in the biosynthetic pathway.
In summary, we discovered that the final 15 residues in the COOH terminus of Kir 2.3 form a basolateral membrane-sorting signal. The previously unrecognized domain corresponds to a highly degenerate structure within the Kir channel family, raising the possibility that the extreme COOH terminus of Kir channels may differentially coordinate membrane targeting of different channel isoforms. Importantly, the structure in the Kir 2.3 channel is composed of a unique arrangement of basolateral sorting signals containing tandem, conceivably overlapping, biosynthetic targeting and PDZ-based signals (Fig. 7).
Figure 7.
Basolateral membrane-sorting domain of Kir 2.3. The final 15 residues in the COOH terminus of Kir 2.3 form a basolateral membrane-sorting domain composed of a unique arrangement of basolateral sorting signals containing tandem, conceivably overlapping, biosynthetic targeting and PDZ-based signals.
Acknowledgments
We thank Hui Liu for expert technical help with the LLCPK1 cells and yeast two-hybrid studies. This study was supported in part by funding from the American Heart Association (Established Investigator) and National Institutes of Health Grant DK 54231 (to P.A.W.) and a North Atlantic Treaty Organization travel award (to J.M. and P.A.W.).
Abbreviations
- LDL
low density lipoprotein
- MDCK
Madin–Darby canine kidney
- PBSCM
PBS containing 1 mM CaCl2 and 1 mM MgCl2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Drubin D G, Nelson W J. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 2.Caplan M J. Am J Physiol. 1997;272:F425–F429. doi: 10.1152/ajprenal.1997.272.4.F425. [DOI] [PubMed] [Google Scholar]
- 3.Matter K, Mellman I. Curr Biol. 1994;6:545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 4.Mays R W, Nelson W J, Marrs J A. Cold Spring Harbor Symp Quant Biol. 1995;60:763–773. doi: 10.1101/sqb.1995.060.01.082. [DOI] [PubMed] [Google Scholar]
- 5.Yeaman C, Grindstaff K K, Nelson W J. Physiol Rev. 1999;79:73–98. doi: 10.1152/physrev.1999.79.1.73. [DOI] [PubMed] [Google Scholar]
- 6.Lisanti M P, Sargiacomo M, Graeve L, Saltiel A R, Rodriguez-Boulan E J. Proc Natl Acad Sci USA. 1988;85:9557–9561. doi: 10.1073/pnas.85.24.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottardi C J, Caplan M J. J Cell Biol. 1993;121:283–293. doi: 10.1083/jcb.121.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheiffele P, Peranen J, Simons K. Nature (London) 1995;378:96–98. doi: 10.1038/378096a0. [DOI] [PubMed] [Google Scholar]
- 9.Matter K, Hunziker W, Mellman I. Cell. 1992;71:741–753. doi: 10.1016/0092-8674(92)90551-m. [DOI] [PubMed] [Google Scholar]
- 10.Thomas D C, Brewer C B, Roth M G. J Biol Chem. 1993;268:3313–3320. [PubMed] [Google Scholar]
- 11.Le Gall A H, Powell S K, Yeaman C A, Rodriguez-Boulan E. J Biol Chem. 1997;272:4559–4567. doi: 10.1074/jbc.272.7.4559. [DOI] [PubMed] [Google Scholar]
- 12.Reich V, Mostov K, Aroeti B. J Cell Sci. 1996;109:2133–2139. doi: 10.1242/jcs.109.8.2133. [DOI] [PubMed] [Google Scholar]
- 13.Simmen T, Nobile M, Bonifacino J S, Hunziker W. Mol Cell Biol. 1999;19:3136–3144. doi: 10.1128/mcb.19.4.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunziker W, Harter K, Matter K, Mellman I. Cell. 1991;66:907–920. doi: 10.1016/0092-8674(91)90437-4. [DOI] [PubMed] [Google Scholar]
- 15.Odorizzi G, Trowbridge I S. J Cell Biol. 1997;137:1255–1264. doi: 10.1083/jcb.137.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casanova J E, Apodaca G, Mostov K E. Cell. 1991;66:65–75. doi: 10.1016/0092-8674(91)90139-p. [DOI] [PubMed] [Google Scholar]
- 17.Aroeti B, Mostov K E. EMBO J. 1994;13:2297–2304. doi: 10.1002/j.1460-2075.1994.tb06513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunziker W, Fumey C. EMBO J. 1994;13:2963–2967. doi: 10.1002/j.1460-2075.1994.tb06594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welling P A. Am J Physiol. 1997;273:F825–F836. doi: 10.1152/ajprenal.1997.273.5.F825. [DOI] [PubMed] [Google Scholar]
- 20.Le Maout S, Brejon M, Merot J, Welling P A. Proc Natil Acad Sci USA. 1997;94:13329–13334. doi: 10.1073/pnas.94.24.13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fanning A S, Anderson J M. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 22.Maddon P J, Littman D R, Godfrey M, Maddon D E, Chess L, Axel R. Cell. 1985;42:93–104. doi: 10.1016/s0092-8674(85)80105-7. [DOI] [PubMed] [Google Scholar]
- 23.Brewer C B, Roth M G. J Cell Biol. 1991;114:413–421. doi: 10.1083/jcb.114.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folsch H, Ohno H, Bonifacino J S, Mellman I. Cell. 1999;99:189–198. doi: 10.1016/s0092-8674(00)81650-5. [DOI] [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 26.Gyuris J, Golemis E A, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 27.Ohno H, Tomemori T, Nakatsu F, Okazaki Y, Aguilar R C, Foelsch H, Mellman I, Saito T, Shirasawa T, Bonifacino J S. FEBS Lett. 1999;449:215–220. doi: 10.1016/s0014-5793(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 28.Ohno H, Aguilar R C, Yeh D, Taura D, Saito T, Bonifacino J S. J Biol Chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- 29.Ho K, Nichols C G, Lederer W J, Lytton J, Vassilev P M, Kanazirska M V, Hebert S C. Nature (London) 1993;362:31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- 30.Roush D L, Gottardi C J, Naim H Y, Roth M G, Caplan M J. J Biol Chem. 1998;273:26862–26869. doi: 10.1074/jbc.273.41.26862. [DOI] [PubMed] [Google Scholar]
- 31.Bonifacino J S, Dell'Angelica E C. J Cell Biol. 1999;145:923–926. doi: 10.1083/jcb.145.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapoport I, Chen Y C, Cupers P, Shoelson S E, Kirchhausen T. EMBO J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan L, Molloy S S, Thomas L, Liu G, Xiang Y, Rybak S L, Thomas G. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 34.Distel B, Bauer U, Le Borgne R, Hoflack B. J Biol Chem. 1998;273:186–193. doi: 10.1074/jbc.273.1.186. [DOI] [PubMed] [Google Scholar]
- 35.Gomperts S N. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 36.Niethammer M, Valtschanoff J G, Kapoor T M, Allison D W, Weinberg T M, Craig A M, Sheng M. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]