ABSTRACT
Human papillomavirus (HPV) genomes are replicated and maintained as extrachromosomal plasmids during persistent infection. The viral E2 proteins are thought to promote stable maintenance replication by tethering the viral DNA to host chromatin. However, this has been very difficult to prove genetically, as the E2 protein is involved in transcriptional regulation and initiation of replication, as well as its assumed role in genome maintenance. This makes mutational analysis of viral trans factors and cis elements in the background of the viral genome problematic and difficult to interpret. To circumvent this problem, we have developed a complementation assay in which the complete wild-type HPV18 genome is transfected into primary human keratinocytes along with subgenomic or mutated replicons that contain the minimal replication origin. The wild-type genome provides the E1 and E2 proteins in trans, allowing us to determine additional cis elements that are required for long-term replication and partitioning of the replicon. We found that, in addition to the core replication origin (and the three E2 binding sites located therein), additional sequences from the transcriptional enhancer portion of the URR (upstream regulatory region) are required in cis for long-term genome replication.
KEYWORDS: DNA replication, HPV, keratinocyte, papillomavirus, partitioning, persistent replication, stable replication, viral replication
IMPORTANCE
Human papillomaviruses infect cutaneous and mucosal epithelial cells of the host, and this results in very-long-lived, persistent infection. The viral genomes are small, circular, double-stranded DNA molecules that replicate extrachromosomally in concert with cellular DNA. This replication strategy requires that the virus has a robust mechanism to partition and retain the viral genomes in dividing cells. This has been difficult to study, because viral transcription, replication, and partitioning are regulated by the same viral proteins and involve overlapping elements in the viral genome. We developed a complementation assay that allows us to separate these functions and define the elements required for long-term replication and stable maintenance replication of the HPV genome. This has important implications, as disruption of viral maintenance replication can eliminate viral genomes from infected cells, thus curing persistent HPV infection.
INTRODUCTION
Papillomaviruses are the causative agent of virtually all cervical cancers, and a viral etiology has been demonstrated for many other human cancers (1). The family Papillomaviridae is made up of over 300 genetically distinct papillomaviruses (NIAID PaVE database; [https://pave.niaid.nih.gov]) (2, 3) that are taxonomically classified into distinct genera named according to the Greek alphabet (4, 5). The papillomavirus life cycle is closely linked to the differentiation process of infected cells. After establishing a persistent infection in the basal layer of the epidermis, the viral double-stranded circular genome is maintained as a low-copy-number, replicating plasmid in the stratum basale (6). Remarkably, viral infections often persist for months and even years (7). This implies that the papillomavirus extrachromosomal genome has to undergo stable maintenance replication in the actively dividing cells within the basal layer.
Persistent, extrachromosomally replicating viruses require replicator elements to replicate and maintain the viral genome. The replicator consists of cis elements that are required to initiate viral DNA synthesis (ori, or origin) as well as cis elements which efficiently distribute the newly replicated genomes to daughter cells (partitioning elements) (8). Gammaherpesviruses, such as Epstein-Barr virus, contain bipartite replicator sequences known as OriPs. Replication is initiated at the DS (dyad symmetry) element within oriP, whereas the adjacent FR (family of repeats) element serves to partition the viral genomes. The FR element contains binding sites for the viral EBNA1 protein (9), and EBNA1 binds to FR and tethers the viral DNA to host mitotic chromosomes to maintain and partition the genomes (10). A similar bipartite replicator sequence has been defined for bovine papillomavirus 1 (BPV1) (11). In the case of BPV1, the viral E2 protein cooperatively binds with the viral E1 helicase to conserved E1 and E2 binding sites in the origin of replication (12, 13). The minimal origin of replication is sufficient to support the transient replication of viral DNA, but long-term stable maintenance replication of the genomes requires an additional cis element known as the minichromosome maintenance element (MME) (11). For BPV1, the MME consists of at least six E2 binding sites (E2BS), in addition to those in the replication origin (11).
As with other viruses, the ability to interact with mitotic chromosomes is required for stable maintenance of the BPV1 genome (14, 15). One chromosomal target is the bromodomain-containing protein Brd4, which interacts with E2 to form a physical tether between the viral genome and host chromosomes. This physical link ensures efficient segregation of the viral genome to the daughter cells (16–18). While the BPV1 “replicator” is fairly well understood, recent research suggests that these insights cannot be directly translated from BPV1 (of the Deltapapillomaviridae genus) to members of the Alphapapillomaviridae (alpha-PVs). The alpha-PV E2 proteins interact with Brd4 to regulate viral transcription, but the role for Brd4 as a molecular tether is less obvious (19–21). In addition, the BPV1 genome contains 17 E2BS, 6 of which are required to form the MME (11), but most Alphapapillomavirus genomes contain only 3 or 4 canonical E2BS (22), hinting toward different requirements for the alpha-PV replicator sequence. Indeed, targeted mutagenesis of human papillomavirus 31 (HPV31, an alpha-PV type) indicated that only three of the four E2BS are required for stable maintenance replication (23). However, analysis of these data is complicated because the viral E2 protein is an essential regulator of both viral replication and transcription. Furthermore, a recent study showed that mitotic partitioning of HPV18 genomes (in the absence of replication) may only require two canonical E2BS (24).
To be able to define the replicator sequence, viral replication must be uncoupled from transcriptional control of the virus. It is impossible to achieve this in the background of the viral genome, where the E2 protein supports replication but also regulates transcription of the E1 and E2 genes. To circumvent this problem, we developed an in vivo complementation assay. In this assay, primary human keratinocytes are cotransfected with wild-type (wt) HPV18 genomes and HPV18 subgenomic replicons. The wild-type genome expresses viral proteins at physiologically relevant levels, thus enabling genetic manipulation of the cotransfected replicons. This effectively uncouples replication and transcription. Using this assay, we provide evidence that the viral URR is both necessary and sufficient for long-term HPV18 stable maintenance replication. Furthermore, we mapped the minimal HPV18 replicator and found it consists of three E2BS in the replication origin, along with a portion of the upstream regulatory region (URR) that overlaps with the viral transcriptional enhancer.
(Parts of this work [Figures 1 and 2] were adapted from the thesis of Sandra Chapman [https://etda.libraries.psu.edu/files/final_submissions/5555].)
RESULTS
The viral URR is required for long-term maintenance of an oncogenic HPV.
A complementation assay was developed to identify the cis elements required for stable maintenance replication of the HPV18 genome. In this assay, primary human foreskin keratinocytes (HFKs) are transfected with the wild-type HPV18 genome and a test replicon plasmid. Under these conditions, the HPV18 genome provides physiological levels of expression of the viral replication proteins in trans and allows us to assay stable maintenance replication of the cotransfected replicon, effectively unlinking replication and transcription. In addition, expression of the E6 and E7 oncogenes from the wild-type HPV18 genome provides the transfected primary cells with a selective growth advantage over nontransfected cells.
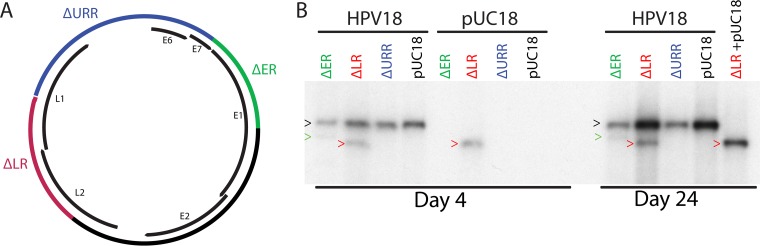
Although we expected that the replicon would require the HPV minimal origin of replication, and perhaps additional E2 binding sites, we initially generated three different subgenomic replicons that contained deletions of large regions of the HPV18 genome (Fig. 1A) (see Materials and Methods for details). The ΔURR replicon contains a deletion spanning the entire URR (including the minimal replication origin). The ΔLR replicon contains a deletion disrupting both late proteins L1 and L2. Finally, a large fragment encoding the early genes was deleted from the ΔER replicon. The ΔURR, ΔLR, and ΔER replicons were transfected into primary HFKs in the presence or absence of a full-length wild-type HPV18 genome to complement the replicons. At different time points posttransfection, low-molecular-weight DNA was extracted from the cells. The extracted DNA was digested with DpnI to remove bacterial input DNA and with EcoRI to linearize the genomes. Since the subgenomic replicons are smaller, they were readily distinguished from the full-length complementing HPV18 genome by Southern blotting (Fig. 1B). In the absence of complementation, the ΔER replicon, which lacks part of the early region encoding the replication proteins, was unable to replicate. However, trans factors provided by the complementing wild-type genome allowed the ΔER replicon to replicate and be stably maintained for at least 24 days. Therefore, the region deleted in the ΔER replicon is not required in cis for long-term replication as long as the replication proteins are provided in trans. On the other hand, the ΔLR replicon did not require complementation with the full-length HPV18 genome to be maintained long term. Therefore, the late region does not contain cis elements that contribute to efficient long-term maintenance. Finally, as expected, the ΔURR replicon lacking the entire URR (and minimal replication origin) was unable to replicate regardless of whether the full-length wild-type genome was present. Similar results were obtained using an HPV31 complementation system, illustrating the general use of the complementation system (data not shown).
FIG 1 .
A complementation assay demonstrated that the URR is required for long-term genome maintenance. (A) Diagram of deletions made in the background of the HPV18 genome. The ΔER replicon (deleted region highlighted in green) lacks part of the E7 and E1 open reading frames. The 3′ end (L1) of the entire URR and part of the E6 gene are deleted from ΔURR (blue). A large region of the L1 and L2 genes is deleted from ΔLR. (B) Different subgenomic replicons (or pUC18 as a control) were cotransfected with the HPV18 genome into primary HFKs. Low-molecular-weight DNA was extracted at different time points, digested with DpnI and a linearizing enzyme, and then analyzed by Southern blotting. Colored arrowheads indicate the positions of the HPV18 genome (black), ΔER (green), and ΔLR (red). A representative blot is shown. Each replicon was tested in similar assays at least three times.
Role of the E2 binding sites in replication can be assessed via the complementation assay.
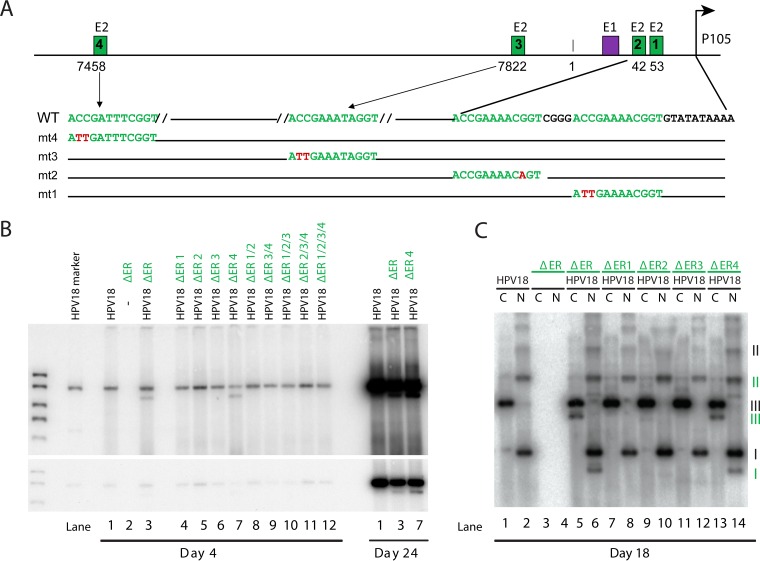
The HPV18 URR contains only four canonical E2BS, and while previous studies have tried to elucidate their precise role in replication of the full HPV genome, this has sometimes been difficult to interpret because of the requirement for the sites to support replication as well as to regulate transcription of the viral replication proteins (23, 25). The ΔER replicon generated in this study is a convenient background to study the role of the E2BS in genome maintenance, because it can be stably replicated long term, it is dependent on the full-length HPV18 genome to provide the replication proteins in trans, and it is distinguishable because of the size difference. Therefore, E2BS were mutated in the ΔER replicon as described previously with minimal nucleotide substitutions (26) that are sufficient to abrogate E2 binding but less likely to alter the function of neighboring elements (Fig. 2A). Replicons with individual and combined E2BS mutations were cotransfected into HFKs with the full-length HPV18 genome, and low-molecular-weight DNA was isolated and analyzed. As shown in Fig. 2B, only replicons containing a mutated distal E2BS (E2BS4) supported short-term replication (4 days posttransfection). This finding is in agreement with findings of previous studies that showed that all three E2BS proximal to the E1 binding site are required for efficient transient replication (27, 28), and so we could not further evaluate the role of these sites in stable maintenance replication. However, the replicon containing a mutated E2BS4 was efficiently maintained for 24 days (Fig. 2B); therefore, we concluded that stable maintenance replication does not require E2BS4. Southern blotting of nonlinearized DNA confirmed that both the wild-type and E2BS4 mutated subgenomic replicons were maintained extrachromosomally (Fig. 2C).
FIG 2 .
E2BS4 is not required for long-term genome maintenance. (A) Diagram of the positions and sequences of E2 binding sites in the HPV18 URR. Shown is the position of the core origin of replication, the constitutive enhancer, E1 and E2 binding sites, and the TATA box and position of the P105 early promoter. Nucleotide substitutions generated in each binding site are shown in red and labeled mt1 to -4, corresponding to E2BS1 to -4. Previous studies have shown that these mutations abrogate E2 binding (26). (B) ΔER replicons containing the E2BS mutated alone or in combination (e.g., ΔER 1/2 has mutations in E2BS1 and -2 [mt1 and mt2, as shown in panel A]) were cotransfected with the HPV18 genome into primary HFKs. pUC18 was used to balance the amount of transfected DNA. Low-molecular-weight DNA was extracted at the times shown, digested with DpnI and a linearizing enzyme, and then analyzed by Southern blotting. Two exposures of a representative blot are shown (n = 2, with different collection times). (C) Low-molecular-weight DNA from a subset of the transfected cells shown in panel B was harvested from cells at 18 days posttransfection and digested with an enzyme that either linearized (C) or did not cut (N) the HPV18 genome (and derived replicons). DNA was analyzed using Southern blotting. The different forms of extrachromosomal DNA are indicated (black roman numerals indicate the wild-type genome; subgenomic replicons are represented by green numerals). I, supercoiled; II, nicked circle; III, linear. n = 2, with different collection times.
Generation of a minimal replicon.
The data presented above indicate that the URR is required for stable maintenance replication. To test whether it is also sufficient for maintenance, we developed a minimal plasmid into which we could insert cis elements and test for their ability to replicate in the presence of the complete HPV18 genome. To avoid excess prokaryotic sequences and to maximize the stable maintenance replication potential of the test replicons, we used a plasmid backbone that contained minimal bacterial sequence and was mostly devoid of CpG dinucleotides, pCpG-neo (Fig. 3A). We previously showed that the NeoR expression cassette derived from this plasmid can be inserted into the late region of the HPV18 genome (replacing the ΔLR deletion described above) to generate a “marker genome” (29). These marker genomes are able to persistently and stably replicate in keratinocytes, demonstrating that the vector components are not detrimental to viral DNA replication and persistence (29).
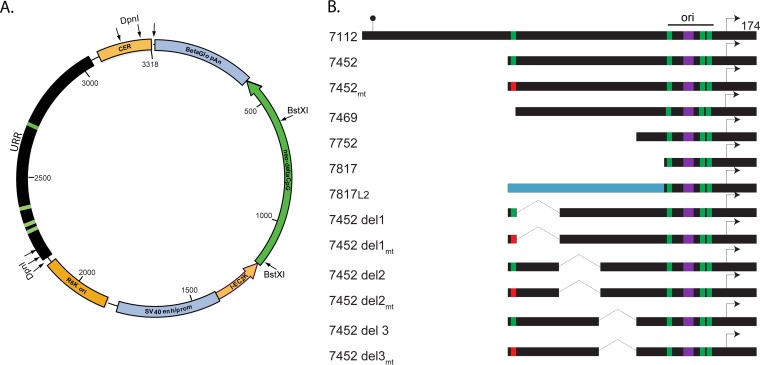
FIG 3 .
Map of the pCpGneo plasmid and URR-derived inserts. (A) Diagram of pCpGneo plasmid. The prokaryotic elements (CER, R6Kori, and I-EC2K) are shown in orange, and eukaryotic elements [simian virus 40 (SV40) enhancer/promoter and beta-globin poly(A)] are shown in blue. The neomycin resistance gene is shown in green. The URR is shown in black, with E2 binding sites indicated as green bars. The position of the DpnI and BstXI restriction sites used in Southern blotting are indicated. (B) Diagram of inserts derived from the HPV18 URR. Plasmids were named according to the first nucleotide (numbering according to the HPV18 reference genome on PaVE). All inserts end at viral position 174. The lollipop indicates the position of the L1 stop codon, while the arrow indicates the major early viral promoter (p105). Green boxes represent wild-type E2BS; red boxes are mutated E2BS. The E1 binding site is shown in purple. The replication origin (ori) is indicated. The blue bar represents a fragment of L2 (HPV18 bases 5867 to 6231) that was used as stuffer DNA.
The HPV18 URR (HPV18 nucleotides [nt] 7112 to 174) was cloned into pCpG-neo, and initial studies showed that the cotransfected HPV18 genome could support transient replication and stable extrachromosomal maintenance replication of this plasmid in keratinocytes. Therefore, a series of plasmids containing deletions in the URR (but maintaining the HPV18 replication origin) were generated (Fig. 3B). In the URR replicon series, 5′ truncations were generated beginning at nucleotide positions 7452 (with and without a mutation in E2BS4), 7469, 7752, and 7817. The smallest insert (nt 7817 to 174) contains the replication origin, consisting of an E1 binding site and three E2 binding sites. To ensure that results were not biased by topological constraints due to the small size of the plasmid, a fragment of L2 (HPV18 bases 5154 to 4790) was used as stuffer DNA and was inserted upstream of nucleotide 7817. This resulted in an insert equivalent in size to the replicons truncated at nt 7452. We showed above (Fig. 1) that this region of L2 is not required for HPV18 stable maintenance replication. Finally, three deletions (del1, -2, and -3) were generated in the enhancer region between E2BS3 and -4. To further explore the requirement for E2BS4, the mutation shown in Fig. 2A was also generated in the 7452 and del1, -2, and -3 replicons.
Early experiments showed that the minimal pCpG plasmids had a tendency to multimerize in the GT115 Escherichia coli host, and so a ColE1 CER element (cer resolution of multimer site) (30) was inserted into the plasmids, as shown in Fig. 3A. The CER element only partially reduced the propensity for multimerization, and so supercoiled, monomeric plasmids were gel purified for subsequent experiments.
The pCG-neo URR series of replicons can replicate transiently in keratinocytes.
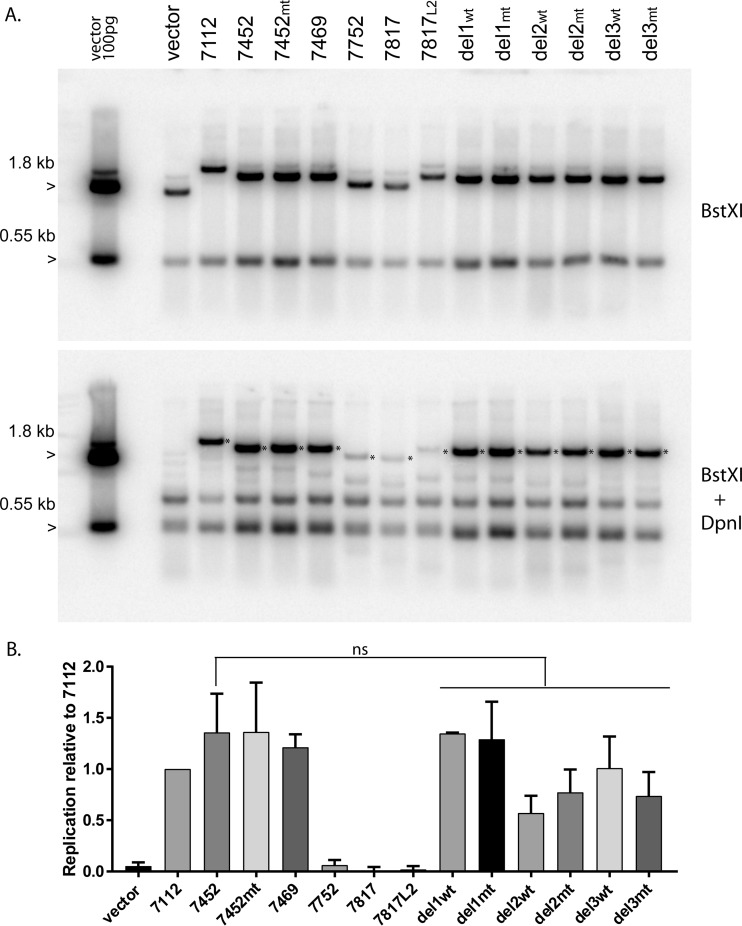
To ensure that the series of pCpGneo URR replicons could replicate transiently, they were cotransfected into primary keratinocytes with the wild-type HPV18 genome, and cellular DNA was harvested at 48 to 72 h. Cellular DNA was prepared and digested with BstXI (cleaves the plasmids into two fragments), or with BstXI and DpnI to eliminate unreplicated input plasmid DNA. As shown in Fig. 4A, and as expected, the empty pCpGneo vector was unable to replicate transiently. However, all URR replicons had replicated by 48 h after transfection. By this time point, there was already a difference in the levels of replication of the minimal origin-containing plasmids (URR 7752, 7817, and 7817L2) from those containing upstream fragments (URR 7452, 7469, and 7452 del1, -2, and -3), indicating that the upstream fragments could promote replication. This was not due to plasmid size, since URR 7817 and 7817L2 replicated at equivalent levels, and it was not due to the presence of E2BS4, since there was no discernible difference in replicons with a mutation in this site (Fig. 4B).
FIG 4 .
Transient replication of the pCGneo-URR plasmids. (A) Gel-purified monomeric supercoils of the pCpGΔneoURR replicons were cotransfected together with the wild-type HPV18 genome into primary keratinocytes, and total cellular DNA was harvested after 48 h. DNA was digested with either BstXI, or BstX1 and DpnI (to cut unreplicated input DNA), followed by analysis by Southern blotting with a pCpGneo vector probe. One hundred picograms of BstXI-digested vector was run as a marker. DpnI-resistant DNA is indicated by the asterisk in the lower panel. A representative image of two independent experiments is shown. (B) Quantitation of transient-replication levels. DpnI-resistant DNA (indicated by the asterisk in panel A) was quantified by phosphorimaging analysis in two independent experiments. Average replication levels are shown relative to that of the pCpGΔneoURR 7112 replicon. Error bars are the standard errors of the means. A one-way analysis of variance test among 7452 and the 7452 deletions (del1, del2, and del3) showed that any observed differences were not significant (ns), as did paired t tests between 7452 and each individual 7452 deletion (del1, del2, and del3). α = 0.05.
A quantitative colony formation assay showed that the viral replication origin and the viral enhancer are required for efficient stable maintenance replication.
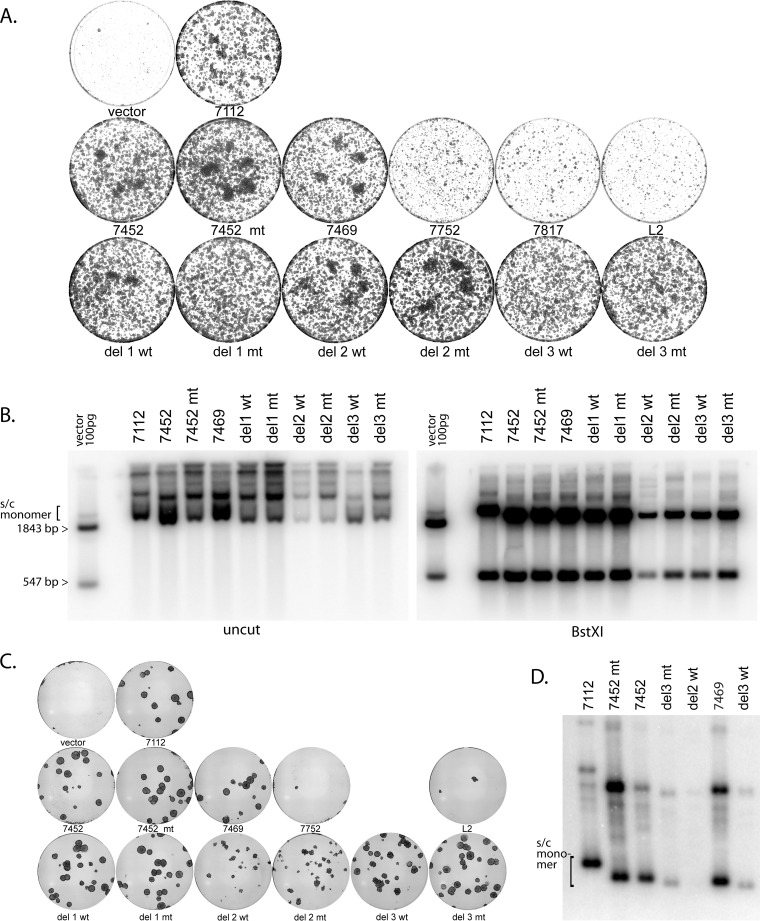
An advantage of the pCpGneo series of replicon plasmids is that they provide cells with antibiotic resistance. Cells cotransfected with the wild-type HPV18 genome and the pCpGneo URR (7112) plasmid gave rise to robust colonies after selection in G418. We showed previously that the formation of robust, expanding colonies reflects stable maintenance replication of recombinant HPV18 marker genomes (29). Therefore, primary keratinocytes were cotransfected with the wild-type HPV18 genome and the series of pCpGneo replicons shown in Fig. 3B, and G418 selective pressure was maintained for the duration of the experiment. The results of the colony formation assay are shown in Fig. 5A, and Southern blotting results showing the extrachromosomal status of the replicons are shown in Fig. 5B.
FIG 5 .
Quantitative colony formation assay results, showing that the viral replication origin and the viral enhancer are required for efficient long-term maintenance. (A) Cells were cotransfected with the HPV18 genome and the series of replicons shown in Fig. 3 and then selected with G418 at 400 µg/ml for 2 days and then 200 µg/ml until visible colonies formed (8 to 9 days posttransfection). Colonies were stained with methylene blue and imaged. The plasmid URR fragment nomenclature is as shown in Fig. 3. (B) Cell DNA was prepared from pooled colonies from a duplicate set of plates and was analyzed using Southern blotting. The samples on the left gel were cleaved with NdeI, which does not cut the pCpG neo URR plasmids; for the right gel, they were cleaved with BstXI. See Fig. 3A for the positions of restriction enzyme sites. The exact experiment shown is representative of two independent experiments. Multiple other replicates (with subsets of plasmids and/or different timings) gave comparable results. (C) Cells were transfected and selected as described for panel A until large colonies were apparent (15 days posttransfection). Colonies were stained and imaged (A) or pooled prior to DNA extraction (D). (D) Cellular DNA was prepared from those colonies that survived selection, and equal amounts were analyzed by Southern blotting. DNA was cut with a restriction enzyme that did not cleave the replicons. The position of supercoiled monomers is indicated.
In the replicon plasmid series, the first three replicons (7452, 7452 mt, and 7469) gave rise to at least as many G418-resistant colonies as the full-length URR (7112) and contained similar or greater levels of extrachromosomally replicating DNA (Fig. 5A and B). Robust replication of 7452 mt confirmed that E2BS4 was not required for maintenance of the HPV18 URR-derived plasmids. The shorter URR-derived replicons (7752 and 7817) showed a drastic reduction in the number and size of G418-resistant colonies, which was not much more than that obtained with the pCpGneo empty vector. There were not enough viable cells to conduct Southern blotting. The URR 7817L2 replicon was also unable to give rise to G418-resistant colonies. Therefore, the inability of the origin-containing replicons 7752 and 7817 to form colonies was not due to their minimal size, and it seems likely that URR sequences between nucleotides 7469 and 7752 are required for stable maintenance replication that is robust enough to generate colonies.
The viral transcriptional enhancer contains cis elements involved in stable plasmid maintenance replication and colony formation.
The central region of the URR between nucleotides 7469 and 7817 (flanked by E2BS3 and -4) has been previously shown to contain the major transcriptional enhancer (31). The viral enhancer consists of three distinct regions: two relatively divergent regions (regions 1 and 3) flanking a relatively conserved core region (region 2). To identify plasmid maintenance elements within this region, the three deletions described above were generated in the URR 7452 wt and URR 7452 mt backgrounds. Replicon plasmids URR del1, del2, and del3 have nucleotides 7470 to 7567, 7568 to 7642, and 7643 to 7750 deleted, respectively, corresponding to the three regions of the enhancer. As shown in Fig. 5A, when cotransfected with the wild-type HPV18 genome, these plasmids were initially able to efficiently give rise to colonies. However, when the colonies shown in Fig. 5A were analyzed by Southern blotting, there was a substantial decrease in extrachromosomal genome copy number of the URR del 2 wt/mt and URR del 3 wt/mt plasmids compared to the URR 7452 wt and URR 7452 mt parental backgrounds. We concluded that these plasmids can replicate and partition enough to give rise to G418-resistant colonies, but the reduced copy number could signify an inability to efficiently and stably maintain and sustain these colonies in the long term.
By plating fewer cells in the colony-forming assay, the colonies were able to expand to a much larger size over a longer period of time (15 days). Under these conditions, differences in the colony-forming abilities of URR del1, del2, and del3 became apparent. As shown in Fig. 5C, the colonies that formed with the URR del2 replicon appeared to be transient and did not continue to expand with time. Therefore, the enhancer portion of the URR is required in addition to the replication origin for optimal stable maintenance replication. Southern blotting of DNA derived from the colonies at this later time point showed that URR del3 replicons can be maintained long term but with a great drop in copy number. However, URR del2 replicons were not maintained.
Replicons containing the minimal replication origin and upstream enhancer sequences replicate as stable extrachromosomal elements for multiple passes.
One consideration is that the formation of colonies in the assay shown in Fig. 5 could be subject to transcriptional enhancement of the neomycin gene, though this should only occur if the replicon is already able to replicate and be maintained for the long term. To alleviate this concern, the complementation assay was carried out without selection.
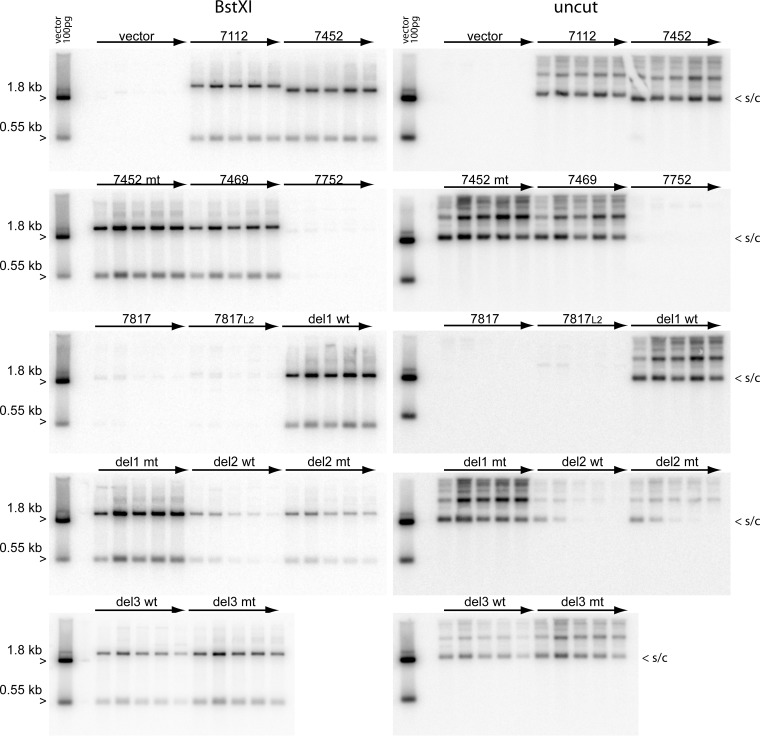
In parallel experiments, primary keratinocytes were cotransfected with the pCpGneo URR replicons and the wild-type HPV18 genome. Cells were passed, without G418 selection, for five passages, and cellular DNA was collected at each pass. Southern blotting was carried out to determine the copy number and extrachromosomal status of the replicons at each pass. As shown in Fig. 6, and as expected from the 48-h analysis results (Fig. 5), there was very little discernible replication of the minimal origin-containing plasmids (URR 7752, 7817, and 7817L2) after long-term cell division. However, the longer replicons URR 7112, 7452, and 7469 replicated stably and were maintained at a constant copy number for the five passes analyzed. There was no difference in the stability or copy number of the 7452 replicons containing a wild-type or mutated E2BS4, confirming that this site was not required for long-term, stable maintenance replication.
FIG 6 .
Replicons containing the minimal replication origin and upstream enhancer sequences replicate extrachromosomally in keratinocytes for multiple passes. Gel-purified monomeric supercoils of the pCpGΔneo URR replicons were cotransfected together with the wild-type HPV 18 genome into primary keratinocytes and cultured for five passages. Each pass corresponded to approximately 4 population doublings, though this is an underestimate because of the low plating efficiency of primary keratinocytes. Total cellular DNA was extracted at each pass and analyzed by Southern blotting with a pCpGneo vector probe. The panel on the left contain samples digested with BstXI, which cleaves the replicon into two fragments. The samples in the panel on the right were digested with NdeI, which does not cut the replicons, allowing visualization of extrachromosomal supercoiled DNA. A representative image of two independent transfection experiments is shown, though subsets of the replicons were also analyzed in multiple other experiments with similar findings.
In the transient-replication experiments, there were no differences in levels of the 7452 series of replicons with deletions in the central URR. However, in the longer-term experiments, it became apparent that replicons with del2 and del3 were impaired in stable maintenance replication. The del3 replicons had decreased copy numbers compared to the wild-type and del1 7452 replicons; however, they were maintained at lower copy number for five passes (the longest time point analyzed). The del2 replicons also had a lower copy number at pass one and pass two, but the extrachromosomal genomes were quickly lost and were not detectable after three passes. E2BS4 did not affect the replication or stability of del2 and del3 7452 replicons. Therefore, the URR between 7568 and 7642 is essential for partitioning, and the region between 7643 and 7750 enhances replicon copy number.
Quantitation of long-term maintenance of pCpG URR replicons in keratinocytes.
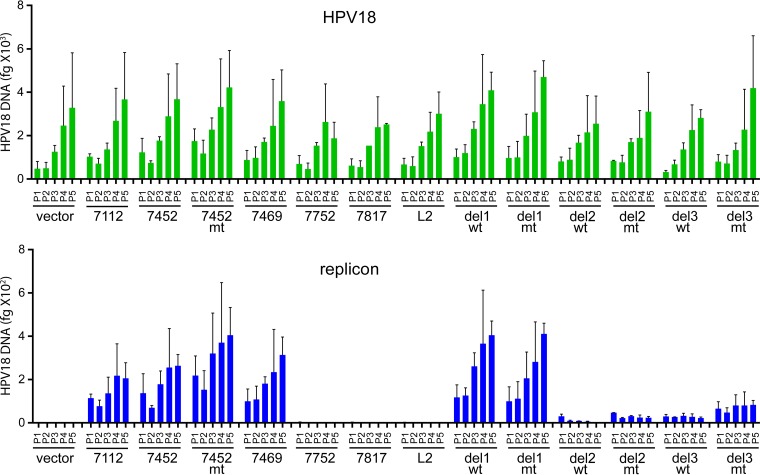
To accurately measure the replication and stability of the pCpGneo URR replicons, a quantitative PCR (qPCR) assay was developed to measure the copy number of the replicon and cotransfected HPV18 genome at each pass. The DNA samples shown in Fig. 6 were digested with EcoRI (to linearize the genomes and replicons) and DpnI to ensure that any observed signal was due to replicated DNA and not input DNA. qPCR primers were designed to span DpnI sites in both the viral genome and replicon. As shown in Fig. 7, the HPV18 genome levels were very consistent across each sample and increased with each pass. This increase was due to the selective effect of the HPV18 oncogenes on transfected cells compared to untransfected cells. The level of each replicon closely mirrored that observed by Southern blotting. The replicons containing only the replication origin were present at very low copy number, only slightly above the background of the empty vector. The amount of the longer replicons was about 10-fold less than that of the wild-type genome, but they were stably replicated and maintained and also increased with cell passage. As observed by Southern analysis, del2 and del3 7452 replicons were maintained at a greatly decreased copy number. However, del 2 mt and del 3 wt and mt replicons were stably maintained at these low levels. The copy number of the del 2 wt replicon was the least stable, as it decreased at every pass. This confirmed that the URR between 7568 and 7642 is essential for efficient partitioning, while the region between 7643 and 7750 enhances replicon copy number.
FIG 7 .
Quantitation of long-term maintenance of the pCpG URR replicons in keratinocytes. The cellular DNA samples shown in Fig. 6 were digested with EcoRI and DpnI and analyzed using qPCR with primers specific for the for HPV18 genome, the pCpGneo URR replicons, and β-actin. The bar graph at the top shows the amounts of HPV18 DNA measured by qPCR in 15 ng of cellular DNA at each of five passes posttransfection, and the bar graph on the bottom shows the amounts of the pCpG-neoURR replicons in the same samples. Each sample was normalized to the amount of actin in each sample. The graphs show the mean values in two identical, independent transfection experiments, and the error bars represent standard deviations. Similar results were found in multiple, similar (though not identical) replicates.
DISCUSSION
The complementation assay developed in this study enabled the direct analysis of the role of the E2BS and other cis elements in genome maintenance by efficiently uncoupling replication and transcription. The HPV18 genome complemented the replication and maintenance of replicons in primary keratinocytes. The viral genomes provided the E1 and E2 replication proteins in trans, and expression of the E6 and E7 oncogenes provided the primary cells with a selectable growth advantage. Initially we tested the ability of large, subgenomic fragments of HPV18 to replicate long term when complemented by the HPV18 complete genome. A replicon lacking a substantial part of the L1 and L2 open reading frames was maintained very efficiently, even without complementation. This led to the development of HPV18-derived marker genomes that encoded selectable markers (neomycin resistance, zeocin resistance, and green fluorescent protein) inserted into the late region (29). Not surprisingly, replicons with the URR deleted were unable to replicate, even when complemented. However, replicons with the ER fragment deleted could replicate long term, but this required the complementing viral genome. Therefore, the ΔER replicon provided a good genetic background in which to study the role of the E2 binding sites.
The importance of individual E2 binding sites during stable viral maintenance replication has been studied previously. However, by binding to E2BS, the viral E2 protein regulates replication and transcription, and therefore outcomes based on mutational disruptions of the viral E2BS in the background of the whole viral genome are hard to interpret. To circumvent this, Lace and colleagues introduced mutations into the E1 and E2 open reading frames of separate HPV16 genomes (32). When cotransfected, both mutated genomes complemented each other, thus allowing for viral transcription and replication. By using this assay, those authors found that short-term HPV16 replication required three promoter-proximal E2BS (32). However, since the expression of either of the essential replication proteins was lost over time in replication-deficient genomes, this system could not be used in long-term maintenance replication studies. A recent study by Ustav and colleagues indicated that partitioning of viral genomes (in the absence of replication) requires only two E2BS. However, since the test plasmids did not replicate, those authors were unable to test very-long-term maintenance (24). In our assay, we found that in the background of the ΔER replicon, the three origin-proximal E2BS were required for transient replication, but E2BS4 was not required for either transient or stable maintenance replication (Fig. 2).
To investigate the importance of distinct regions within the URR, we cloned fragments of the URR into a minimal bacterial plasmid. This plasmid was based on vectors designed for gene therapy. In addition to containing very minimal bacterial sequence, the plasmid is largely devoid of CpG dinucleotides. It also encodes aminoglycoside 3′-phosphotransferase, which confers resistance to kanamycin in E. coli and to G418 in mammalian cells. This plasmid backbone was selected to minimize cellular innate immune responses (30, 33), thereby maximizing the ability of the plasmids to replicate long term in primary cells. Replicon maintenance was assayed by quantifying replicon DNA at various times posttransfection or the ability of the replicon to support colony formation in the presence of antibiotic selection. Using these assays, we showed that stable maintenance does not require the 5′-URR (upstream of E2BS4). This is similar to a previous observation made for the background of the HPV31 genome (25). Furthermore, confirming the observation made in the background of ΔER, E2BS4 was not needed for stable maintenance replication and in fact seemed to reduce plasmid maintenance.
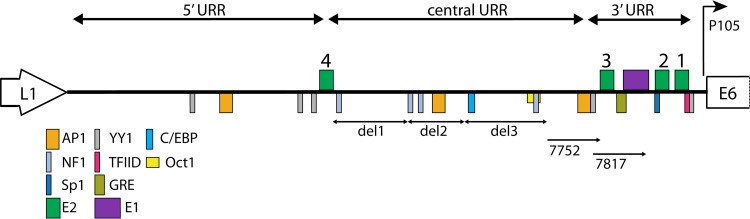
Our mapping studies suggest a key role for the region of the viral URR known as the tissue-specific enhancer. Specifically, the core region (fragment 2) of this viral enhancer plays a significant role in optimal replicon maintenance. The region of the enhancer closest to the origin was also important for the replicon copy number; when this region was deleted, the resulting replicon was maintained at very low copy number. Figure 8 shows a map of the HPV18 URR and the location of binding sites for cellular transcription factors. However, further mutational analysis was unable to identify specific parts of the enhancer as essential (unpublished data). This inability to find a specific sequence is reminiscent of the frustrating quest to identify mammalian replication origins (34). Current thinking is that epigenetic, as well as genetic, parameters influence the replication of autonomous replicons. These parameters include nuclear localization, chromatin structure and modifications, topology, nucleosome positioning, and transcriptional activity of adjacent DNA (34). The elements that we have mapped to the core enhancer region of HPV18 could all influence replication through these processes.
FIG 8 .
Map of the HPV18 URR. The diagram shows the three main regions of the HPV18 URR. The viral E1 and E2 sites are indicated on top (green and purple boxes). Binding sites for cellular factors that have been mapped to URR are indicated below by colored boxes (41, 42). The position of the del1, -2, and -3 deletions and the 7752 and 7817 5′ endpoints are shown.
Our data show that optimal viral genome maintenance replication requires the HPV replication origin (including three E2BS) and a region that overlaps the viral enhancer. The observation that three E2BS are sufficient for short-term replication is in agreement with previous data showing that the three promoter-proximal E2BS represent the origin of replication (27, 28, 35). Studies of BPV1 replication led to the model that the E2 protein tethers the viral genome to host chromosomes through multiple E2BS (>6), somewhat analogous to what occurs in the gammaherpesviruses (11, 14, 15). However, the studies presented here suggest that E2 can promote replication and partitioning of HPV18 replicons containing just the three E2 binding sites in the replication origin along with cis elements in the URR. These cis elements likely promote and stabilize DNA synthesis and plasmid partitioning through the epigenetic mechanisms described above. Although we did not directly test the role of the E2 protein in our complementation assay, Ustav and colleagues showed that E2 can partition replicons containing only two optimally spaced E2BS (24). Two E2 protein dimers must bind cooperatively to E2BS1 and -2 to facilitate partitioning (24). Taken together, we propose that the region containing the three E2BS surrounding the replication origin constitutes the core replicator (initiation and partitioning element), but this replicator requires additional cis elements from the URR to promote and stabilize these functions. The origin-bound E2 protein likely cooperates with, and promotes the function of, factors associated with the upstream cis element by promoting optimal nucleosomal positioning and formation of active chromatin (36) and by localizing the viral replicon to active regions of the nucleus (37, 38).
MATERIALS AND METHODS
Plasmids.
HPV18-based subgenomic replicons were generated by deleting large fragments from the full-length genome, as illustrated in Fig. 1. Specifically, fragments were deleted from HPV18 (cloned into the EcoRI site of pBR322) between restriction sites (18ΔURR replicon, NcoI [nt 6233] and BlpI [nt 823]; 18ΔER replicon, Blp1 [nt 824] and AleI [nt 1945]; 18ΔLR replicon, KpnI to KpnI [nt 4795 to 6273]). Specific mutations in E2BS were introduced into the full-length HPV18 genome by using the QuikChange XL site-directed mutagenesis kit (Stratagene). All mutations were sequence verified. Before transfection, wild-type HPV18 genomes and derived replicons were cleaved from the pBR322 prokaryotic backbone with EcoRI and recircularized by overnight incubation with T4 DNA ligase with 5 µg/ml DNA (39). In Fig. 4, 5A and B, 6, and 7, an HPV18 minicircle genome recircularized in bacteria was used, as described elsewhere (31). This minicircle genome was a generous gift from Mart Ustav.
Fragments of the HPV18 URR were cloned into a minimal plasmid, pCGΔneo, using standard procedures. An expression cassette derived from this modified CpG-free plasmid (pCpG-Neo) was described recently (22). These R6Kγ origin of replication-containing plasmids were propagated in E. coli GT115 bacteria (InvivoGen). Because of the propensity of these plasmids to multimerize in bacteria (data not shown), a ColE1 CER element (30) was inserted, as shown in Fig. 3. This was only partially successful in preventing multimer formation, and so monomeric supercoiled forms of CpG-Neo-derived plasmids were purified from agarose gels prior to transfection.
Cell culture.
Primary human keratinocytes were isolated from neonatal foreskins, as described previously (40) and with NIH Institutional Review Board approval. Cells were expanded in Rheinwald-Green F medium (3:1 Ham’s F-12/high-glucose Dulbecco’s modified Eagle’s medium [DMEM], 5% fetal bovine serum [FBS], 0.4 µg/ml hydrocortisone, 8.4 ng/ml cholera toxin, 10 ng/ml epidermal growth factor, 24 µg/ml adenine, 6 µg/ml insulin) on a layer of lethally irradiated J2-3T3 murine fibroblasts. For experiments using G418 selection, HFKs were cultured on G418-resistant J2-3T3 murine fibroblasts.
Transfection.
Electroporation was performed using the Amaxa electroporation system (Lonza) according to the manufacturer’s instructions. Briefly, 1 × 106 cells were mixed with 2 µg DNA and electroporated using the T-007 program defined by the manufacturer as optimal for enhanced keratinocyte survival. A total of 1.4 × 106 electroporated cells were plated on irradiated feeders for short-term replication, 0.4 × 106 cells were plated for long-term replication, and 0.1 × 106 were plated for colony formation experiments. The latter group of cells was plated on irradiated G418-resistant J2-3T3 cells, selected in 400 µg/ml G418 for 2 days, and then in 200 µg/ml G418 until colonies were fixed and stained with methylene blue (8 to 9 days).
DNA extraction.
Whole-cell DNA extraction was carried out using the Qiagen blood and tissue kit according to the manufacturer’s instructions. Low-molecular-weight DNA was extracted using a modified Hirt extraction method (11).
Southern blotting.
Low-molecular-weight or total cellular DNA was digested with either a restriction enzyme to cleave the viral DNA and derived replicons (BstXI; 2 cuts) or a noncutter (HindIII or NdeI) to linearize cellular DNA. For transient DNA replication analysis, samples were also digested with DpnI to remove unreplicated viral DNA. After digestion, samples were separated on Tris-acetate-EDTA (TAE)–agarose gels. DNA was visualized with 10 µg/ml ethidium bromide and transferred to Nytran SPC membranes with a TurboBlotter downward transfer system (Whatman). Membranes were UV cross-linked, dried, incubated with prehybridization blocking buffer, and then incubated overnight with 25 ng [32P]dCTP-labeled HPV18 DNA or pCpG vector in hybridization buffer (3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2% SDS, 5× Denhardt’s solution, 0.2 mg/ml sonicated salmon sperm DNA). Radiolabeled probe was generated from twice-gel-purified linear HPV18 DNA or pCpG vector DNA by using a Random Prime DNA labeling kit (Roche). Hybridized DNA was visualized using a Typhoon scanner (GE Bioscience).
Quantitative PCR for copy number determinations.
Total cellular DNA was digested as described above. After digestion, 15 ng was analyzed by qPCR using 300 nM primers and SYBR green master mix (Roche). The reaction conditions consisted of a 15-min 95°C activation cycle, 40 cycles of 10 s at 95°C for denaturation and 30 s at 60°C for annealing, and elongation. Copy number was calculated by comparing data for the unknown samples to standard curves of linearized HPV18 or CpG vector DNA. β-Actin DNA copy number was used as a normalization control. HPV18 or CpG vector primers spanned DpnI sites. The primer sequences used were as follows: HPV18 sense (5′-CACAATACTATGGCGCGCTTT-3′) and antisense (5′-CCGTGCACAGATCAGGTAGCT-3′), CpG vector sense (5′-GGCAGTTTTTCGGGTGGTT-3′) and antisense (5′-CACCGCTAAAACGCGTTCA-3′), and β-actin sense (5′-TCGTCCACCGCAAATGC-3′) and antisense (5′-CGCAAGTTAGGTTTTGTCAAGAAA-3′).
ACKNOWLEDGMENTS
This work was funded by the Intramural Research Program of the NIAID, NIH (ZIA AI001073 to Alison McBride).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Van Doorslaer K, Chen D, Chapman S, Khan J, McBride AA. 2017. Persistence of an oncogenic papillomavirus genome requires cis elements from the viral transcriptional enhancer. mBio 8:e01758-17. https://doi.org/10.1128/mBio.01758-17.
REFERENCES
- 1.Parkin DM, Bray F. 2006. The burden of HPV-related cancers. Vaccine 24(Suppl 3):11–25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 2.Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, Mohamoud Y, Huyen Y, McBride AA. 2013. The papillomavirus episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res 41:D571–D578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, Liou D, Sun Q, Kaur R, Huyen Y, McBride AA. 2017. The papillomavirus episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res 45:D499–D506. doi: 10.1093/nar/gkw879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 5.Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride AA. 2017. Mechanisms and strategies of papillomavirus replication. Biol Chem 398:919–927. doi: 10.1515/hsz-2017-0113. [DOI] [PubMed] [Google Scholar]
- 7.Franco EL, Villa LL, Sobrinho JP, Prado JM, Rousseau MC, Désy M, Rohan TE. 1999. Epidemiology of acquisition and clearance of cervical human papillomavirus infection in women from a high-risk area for cervical cancer. J Infect Dis 180:1415–1423. doi: 10.1086/315086. [DOI] [PubMed] [Google Scholar]
- 8.McBride AA. 2008. Replication and partitioning of papillomavirus genomes. Adv Virus Res 72:155–205. doi: 10.1016/S0065-3527(08)00404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yates JL, Camiolo SM, Bashaw JM. 2000. The minimal replicator of Epstein-Barr virus oriP. J Virol 74:4512–4522. doi: 10.1128/JVI.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanda T, Otter M, Wahl GM. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol Cell Biol 21:3576–3588. doi: 10.1128/MCB.21.10.3576-3588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J 15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 12.Ustav M, Stenlund A. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J 10:449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ustav M, Ustav E, Szymanski P, Stenlund A. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J 10:4321–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skiadopoulos MH, McBride AA. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol 72:2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilves I, Kivi S, Ustav M. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J Virol 73:4404–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349–360. doi: 10.1016/S0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 17.Baxter MK, McPhillips MG, Ozato K, McBride AA. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J Virol 79:4806–4818. doi: 10.1128/JVI.79.8.4806-4818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPhillips MG, Ozato K, McBride AA. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J Virol 79:8920–8932. doi: 10.1128/JVI.79.14.8920-8932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira JG, Colf LA, McBride AA. 2006. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes. Proc Natl Acad Sci U S A 103:1047–1052. doi: 10.1073/pnas.0507624103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol 80:9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helfer CM, Wang R, You J. 2013. Analysis of the papillomavirus E2 and bromodomain protein Brd4 interaction using bimolecular fluorescence complementation. PLoS One 8:e77994. doi: 10.1371/journal.pone.0077994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride AA. 2013. The papillomavirus E2 proteins. Virology 445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stubenrauch F, Lim HB, Laimins LA. 1998. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J Virol 72:1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ustav M Jr, Castaneda FR, Reinson T, Männik A, Ustav M. 2015. Human papillomavirus type 18 cis-elements crucial for segregation and latency. PLoS One 10:e0135770. doi: 10.1371/journal.pone.0135770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubert WG, Kanaya T, Laimins LA. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J Virol 73:1835–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thierry F, Howley PM. 1991. Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol 3:90–100. [PubMed] [Google Scholar]
- 27.Lee D, Kim H, Lee Y, Choe J. 1997. Identification of sequence requirement for the origin of DNA replication in human papillomavirus type 18. Virus Res 52:97–108. doi: 10.1016/S0168-1702(97)00114-7. [DOI] [PubMed] [Google Scholar]
- 28.Remm M, Brain R, Jenkins JR. 1992. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res 20:6015–6021. doi: 10.1093/nar/20.22.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Doorslaer K, Porter S, McKinney C, Stepp WH, McBride AA. 2016. Novel recombinant papillomavirus genomes expressing selectable genes. Sci Rep 6:37782. doi: 10.1038/srep37782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams JA, Carnes AE, Hodgson CP. 2009. Plasmid DNA vaccine vector design: impact on efficacy, safety and upstream production. Biotechnol Adv 27:353–370. doi: 10.1016/j.biotechadv.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinson T, Toots M, Kadaja M, Pipitch R, Allik M, Ustav E, Ustav M. 2013. Engagement of the ATR-dependent DNA damage response at the human papillomavirus 18 replication centers during the initial amplification. J Virol 87:951–964. doi: 10.1128/JVI.01943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lace MJ, Anson JR, Turek LP, Haugen TH. 2008. Functional mapping of the human papillomavirus type 16 E1 cistron. J Virol 82:10724–10734. doi: 10.1128/JVI.00921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyde SC, Pringle IA, Abdullah S, Lawton AE, Davies LA, Varathalingam A, Nunez-Alonso G, Green AM, Bazzani RP, Sumner-Jones SG, Chan M, Li H, Yew NS, Cheng SH, Boyd AC, Davies JC, Griesenbach U, Porteous DJ, Sheppard DN, Munkonge FM, Alton EW, Gill DR. 2008. CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat Biotechnol 26:549–551. doi: 10.1038/nbt1399. [DOI] [PubMed] [Google Scholar]
- 34.Hagedorn C, Lipps HJ, Rupprecht S. 2010. The epigenetic regulation of autonomous replicons. Biomol Concepts 1:17–30. doi: 10.1515/bmc.2010.009. [DOI] [PubMed] [Google Scholar]
- 35.Del Vecchio AM, Romanczuk H, Howley PM, Baker CC. 1992. Transient replication of human papillomavirus DNAs. J Virol 66:5949–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefebvre O, Steger G, Yaniv M. 1997. Synergistic transcriptional-activation by the papillomavirus E2 protein occurs after DNA binding and correlates with a change in chromatin structure. J Mol Biol 266:465–478. doi: 10.1006/jmbi.1996.0807. [DOI] [PubMed] [Google Scholar]
- 37.Jang MK, Kwon D, McBride AA. 2009. Papillomavirus E2 proteins and the host BRD4 protein associate with transcriptionally active cellular chromatin. J Virol 83:2592–2600. doi: 10.1128/JVI.02275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helfer CM, Yan J, You J. 2014. The cellular bromodomain protein Brd4 has multiple functions in E2-mediated papillomavirus transcription activation. Viruses 6:3228–3249. doi: 10.3390/v6083228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson R, Laimins LA. 2005. Differentiation of HPV-containing cells using organotypic “raft” culture or methylcellulose. Methods Mol Med 119:157–169. doi: 10.1385/1-59259-982-6:157. [DOI] [PubMed] [Google Scholar]
- 40.Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. 2010. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J Clin Invest 120:2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernard HU. 2013. Regulatory elements in the viral genome. Virology 445:197–204. doi: 10.1016/j.virol.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 42.O’Connor M, Chan SY, Bernard HU. 1995. Transcription factor binding sites in the long control region of the genital HPVs, p III-21–III-40. In Myers G, Bernard H, Baker C, Halpern A, Delius H, Icenogel J (ed), Human papillomaviruses. Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]