Abstract
Despite the progress in development of better AR-targeted therapies for prostate cancer (PCa), there is no curative therapy for castration-resistant prostate cancer (CRPC). Therapeutic resistance in PCa can be characterized in two broad categories of AR therapy resistance: the first and most prevalent one involves restoration of AR activity despite AR targeted therapy, and the second one involves tumor progression despite blockade of AR activity. As such AR remains the most attractive drug target for CRPC. Despite its oncogenic role, AR signaling also contributes to the maturation and differentiation of prostate luminal cells during development. Recent evidence suggests that AR cistrome is altered in advanced PCa. Alteration in AR may result from AR amplification, alternative splicing, mutations, post-translational modification of AR, and altered expression of AR co-factors. We reasoned that such alterations would result in the transcription of disparate AR target genes and as such may contribute to the emergence of castration-resistance. In the present study, we evaluated the expression of genes associated with canonical or non-canonical AR cistrome in relationship with PCa progression and prostate development by analyzing publicly available datasets. We discovered a transcription switch from canonical AR cistrome target genes to the non-canonical AR cistrome target genes during PCa progression. Using Gene Set Enrichment Analysis (GSEA), we discovered that canonical AR cistrome target genes are enriched in indolent PCa patients and the loss of canonical AR cistrome is associated with tumor metastasis and poor clinical outcome. Analysis of the datasets involving prostate development, revealed that canonical AR cistrome target genes are significantly enriched in prostate luminal cells and can distinguish luminal cells from basal cells, suggesting a pivotal role for canonical AR cistrome driven genes in prostate development. These data suggest that the expression of canonical AR cistrome related genes play an important role in maintaining the prostate luminal cell identity and might restrict the lineage plasticity observed in lethal PCa. Understanding the molecular mechanisms that dictate AR cistrome may lead to development of new therapeutic strategies aimed at restoring canonical AR cistrome, rewiring the oncogenic AR signaling and overcome resistance to AR targeted therapies.
Keywords: AR cistrome, prostate cancer, prostate development
Introduction
Prostate cancer (PCa) is the most commonly diagnosed cancer in men and is the direct cause of over 26,000 death in the USA last year [1]. Alterations in androgen receptor (AR) signaling is one of the master drivers in prostate cancer and tumor progression. Targeting AR signaling by current androgen deprivation therapies yields a high 5-year survival rate, however, most patients relapse with metastatic castration-resistant prostate cancer (mCRPC) for whom no effective therapy is available [2,3].
AR signaling plays an essential role not only in prostate development but also in prostate cancer progression, and AR signaling remains the most attractive target for intervention in PCa. In prostate tissue, androgens are involved in differentiation, development and normal functioning [4]. AR, a member of nuclear steroid family of receptors, is a ligand-dependent transcription factor and is essential for mediating actions of androgens [5]. AR is a 110 kDa protein with 4 functional domains: the amino-terminal transactivation domain, DNA-binding domain, the hinge region and the ligand-binding domain. As a classic steroid hormone receptor, it forms a complex with chaperones such as heat shock protein 90 when androgen is absent. Upon androgen binding to AR, it dimerizes and enters into the nucleus. Once in the nucleus, AR specifically binds to the androgen response elements (AREs) through its DNA binding domain, recruiting transcription co-factors initiating the transcription of AR target genes [6]. AR signaling is required for the development of normal prostate as well as PCa [7]. PCa rarely occurs in eunuchs or men with a deficiency in 5α-reductase, highlighting the importance of androgens and AR signaling in PCa. Normal prostate epithelial cells, as well as early-stage PCa cells, depend on androgens and other stromal-derived (paracrine) factors for growth and survival (reviewed by C. Abate-Shen in [8]); for example, epidermal growth factor (EGF) [9]. Advanced genomic studies of CRPC patients revealed that AR is the most common mutated gene and most CRPC patients harbor AR gene amplification and/or AR overexpression [10]. Despite its pivotal role in prostate cancer, the activity of AR is also crucial for the maturation and maintenance of prostate luminal cells [11].
Activation of Androgen receptor signaling independent of androgens confers castration resistance in prostate cancer and is a hallmark of the majority of the CRPC patients. A high proportion of mutations are observed in the ligand-binding domain of AR in hormone-refractory and metastatic PCa, and mutant AR could be activated by other steroid hormones [7]. It is thought that androgen withdrawal therapy results in massive apoptosis of androgen-dependent malignant cells, thus selecting from a reservoir of cells which either are more dependent on polypeptide growth factors or cytokines and less dependent upon androgens for growth [12], or whose AR signaling pathway has experienced gain of function through heightened androgen sensitivity, alterations in co-factor expression, or other mechanisms [13]. In contrast to commonly used AR-independent cell lines, most prostatic cancers from patients failing hormone therapy over-express AR and many possess AR mutations [14,15]. Some evidence suggests that the androgen receptor can be transactivated independently of androgens by other growth factor and cytokine receptor pathways, notably EGF and interleukin-6 (IL-6) [16-18]. Mechanisms of AR activation independent of androgens include mutations to the ligand binding domain of the AR; AR activation by of phosphorylation, sumoylation and or acetylation; overexpression of AR; expression of the splice variants of AR that lack ligand binding domain, and/or; altered expression of AR co-factors. However, the impact of these AR-independent AR activation mechanisms on transcriptional targets of AR remains to be fully appreciated.
The activity of sequence-specific transcription factors requires not only the presence of specific DNA elements but also the accessibility of those DNA elements which are often modified by transcription co-factors. As in the case of the pioneer transcription factor FOXA1 and AR, various levels of FOXA1 and AR have been suggested to alter AR genomic binding sites (AR cistrome) which promotes the progression of prostate cancer [19]. Recently Pomerantz et al. discovered that AR cistrome is extensively altered in CRPC patients with the tumor-specific enrichment of HoxB13 and FoxA1 motifs near AR binding sites compared with normal prostate tissue [20]. Based on the classic genetic central dogma, disparate AR cistrome would lead to the transcription of divergent AR target genes which can be used as the markers for canonical or non-canonical AR cistrome respectively.
The burst of sequencing technology has greatly enhanced our ability to develop broader understanding of prostate organogenesis and the development of prostate cancer. With the decreasing sequencing expenses and the data sharing policy, growing number of genomic data from various experimental setups have been made available for the scientific community [21]. In this study, we analyzed the expression of AR-target genes in publicly available databases inferring the disparate AR cistrome to better understand the role of canonical and non-canonical AR cistromes in prostate development, prostate cancer progression, metastasis and therapeutic resistance.
Methods
Microarray and RNA-Seq datasets were downloaded from the GEO archive [22]. GEO hosts other categories of high-throughput functional genomic data, including those that examine genome copy number variations, chromatin structure, methylation status and transcription factor binding. These data are generated by the research community using high-throughput technologies like microarrays and, more recently, next-generation sequencing. The database has a flexible infrastructure that can capture fully annotated raw and processed data, enabling compliance with major community-derived scientific reporting standards such as ‘Minimum Information About a Microarray Experiment’ (MIAME. TCGA prostate cancer datasets were obtained from cBioPortal (http://www.cbioportal.org/). Statistical analysis of differences in expression levels for transcripts between the Gleason score 6-7 vs. Gleason score 8-10 patients were performed using the non-parametric Mann Whitney test. Gene set enrichment analysis was performed using default setting [23].
Results and discussion
Loss of canonical AR and enrichment of non-canonical AR cistrome during prostate cancer progression
To elucidate the role of AR cistrome in prostate cancer, we probed the expression level of a panel of genes involved in canonical or non-canonical AR cistrome in TCGA database. We discovered that expression of non-canonical AR cistrome related genes positively correlated with increasing Gleason score while expression of the canonical AR cistrome related genes negatively correlated with prostate cancer progression (Figure 1A and 1B). These data suggest that the loss of expression of canonical AR cistrome related genes and gain of expression of non-canonical AR cistrome associated genes correlate with prostate cancer progression.
Figure 1.
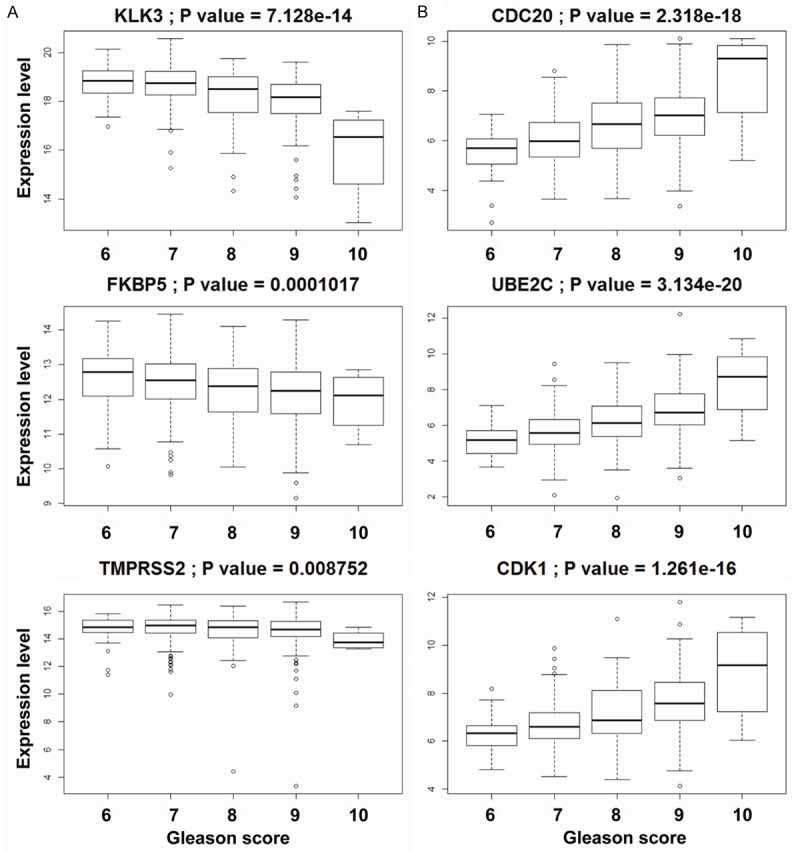
Loss of canonical AR cistrome and gain of non-canonical AR cistrome with increasing Gleason grade PCa: mRNA level of canonical (A) and non-canonical (B) AR cistrome target genes in TCGA. Patients were stratified according to their Gleason Score. P values are shown using Mann Whitney U test comparing patients with Gleason Score 6-7 VS. patients with Gleason Score 8-10.
Negative enrichment of canonical AR target genes during prostate cancer progression and metastasis
Next we performed Gene Set Enrichment Analysis (GSEA) in the Swedish-watchful waiting cohort (GSE16560). These results reveal that canonical AR target genes are enriched in indolent prostate cancer patients (Figure 2A). These data suggest that expression of canonical AR cistrome associated genes could be a predictor of better clinical outcome.
Figure 2.
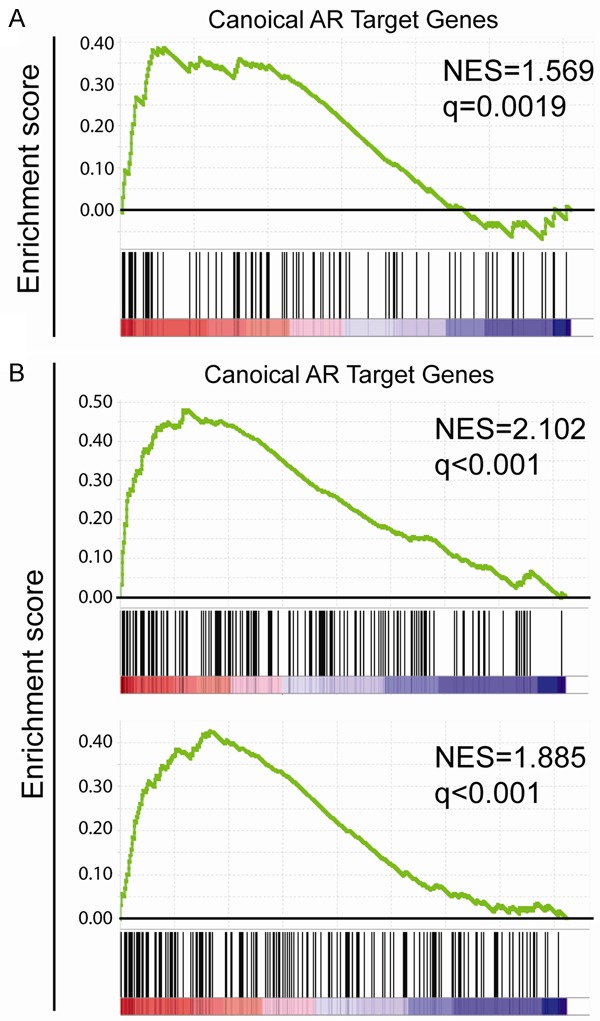
Enrichment of canonical AR cistrome related genes in indolent PCa, benign prostate and clinically localized PCa tissues: GSEA was performed with default settings. Canonical AR target genes were obtained from literature [29]. The normalized enrichment score (NES) and false discovery rate (q) were shown in the corresponding figure. Canonical AR target genes were enriched in indolent PCa patients (A) when compared with lethal PCa patients; in benign prostate tissue samples (B upper panel) and in clinically localized PCa tissue (B lower panel) when compared with metastatic tissue.
Since tumor metastasis is the leading cause of prostate cancer deaths, we asked the question whether canonical AR cistrome related genes were downregulated in metastasis. For these studies we interrogated the microarray data from GSE3325 which includes 4 benign prostate tissues samples, 5 clinically localized primary prostate cancer samples and 4 metastatic prostate cancer samples [24]. Results reveal that canonical AR target genes were significantly enriched in benign and clinically localized prostate cancer tissues when compared with metastatic tissues (Figure 2B). Moreover, comparison of the benign prostate tissue with clinically localized prostate cancer tissue did not yield significant enrichment of canonical AR target genes (data not shown). These collectivedata provide further evidence to support the notion that loss of canonical AR cistrome related genes is associated with prostate cancer metastasis.
Induction of canonical AR cistrome in prostate organogenesis
Due to the fact that canonical AR cistrome target gene set is produced in LNCaP, a prostate cancer cell line, we asked the question whether it is also upregulated during prostate organogenesis especially in the differentiation of prostate luminal cells. We used the dataset GSE81439 where RWPE1 cells were transfected with NKX3.1 to induce its luminal differentiation [25]. We observed the significant enrichment of canonical AR cistrome target genes in RWPE1 cells overexpressing NKX3.1 compared with vector control group (Figure 3A). Furthermore, the loss of function mutation in NKX3.1 disrupts the enrichment of canonical AR cistrome target (Figure 3A). Notably, we also observed the significant enrichment of canonical AR cistrome target genes in human prostate luminal cells when compared with prostate basal cells using the dataset GSE67070 (Figure 3B) [26]. These association studies suggest that canonical AR-cistrome plays a key role in luminal differentiation of prostate epithelium and as such may reflect luminal phenotype in prostate cancer.
Figure 3.
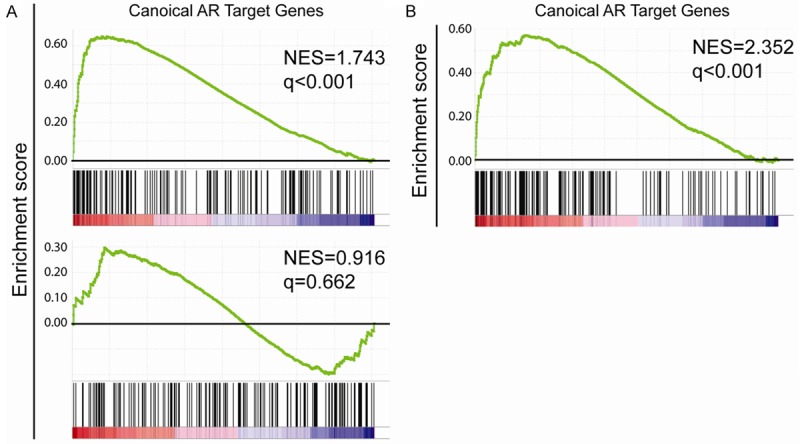
Role of AR cistrome in prostate organogenesis. A: Canonical AR target genes were enriched in NKX3.1 expressing RWPE1 cells when comparing with when vector control (upper panel) but not in RWPE1 cells expressing mutant form of NKX3.1 (lower panel). B: Canonical AR target genes were enriched in human prostate luminal cells when compared with prostate basal cells.
Conclusion/Discussion
Although canonical AR cistrome target gene set was generated in prostate cancer cell lines, we discovered that canonical AR cistrome target genes correlate with luminal differentiation in the prostate, and canonical AR cistrome target genes are able to distinguish prostate luminal cells vs. basal cells. These considerations suggest a central role for activation of AR in prostate differentiation and prostate carcinogenesis. More importantly, our findings that canonical AR cistrome target genes were enriched in prostate cancer tissues from indolent prostate cancer patients while non-canonical AR target genes are enriched during disease progression, suggest that prostate cancer progression involves de-differentiation of the luminal epithelial phenotype. Interestingly non-canonical AR cistrome targets are enriched in cell-cycle-related genes suggesting the involvement in promoting tumor growth. In addition, canonical AR cistrome might function as transcription repressor inhibiting the transcription of metastasis-related transcription factors such as Snai1 [27,28]. These findings also raise the possibility that canonical AR target genes might be involved in maintaining the differentiated status of prostate cells, thereby playing tumor suppressive role, and disruption of the canonical AR cistrome might contribute to prostate cancer progression (Figure 4).
Figure 4.
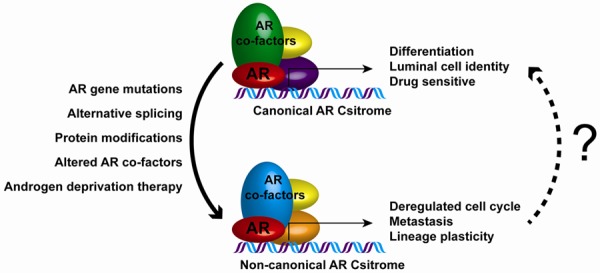
Hypothetical model of disparate actions of AR signaling: Canonical AR cistrome drives expression of genes associated with differentiation and luminal cell identity. This phenotype is highly sensitive to anti androgen therapies, as such PCa patients that display canonical AR signaling are highly responsive to AR targeted therapies and have prolonged survival. AR gene mutation, alternative splicing, protein modifications, altered level of AR co-factors and in response to androgen ablation therapies, non-canonical AR cistrome emerges which drives expression of genes associated with deregulated cell cycle, metastasis, lineage plasticity. Such changes result in androgen independent growth and proliferation in PCa and drive castration resistance. Research efforts aimed at developing therapeutic interventions that help restore canonical AR cistrome in CRPC are warranted.
As such, it is logical to predict that the restoration of canonical AR cistrome might convert lethal prostate cancer into indolent prostate cancer which is non-metastatic and sensitive to castration. Since AR co-factors play an important role in defining AR cistrome, more efforts are needed in studying which co-factors facilitate canonical AR cistrome and which co-factors facilitate non-canonical AR cistrome.Furthermore, such studies will offer a deeper understanding of how AR regulates different target genes and shed light on the development of possible therapeutic interventions targeting AR cistrome and for development of biomarkers for the early detection of lethal prostate cancer.
Acknowledgements
Financial support from Carrol W. Fiest Endowed Chair (H. Koul). VA Merit Award-01BX001258 (H. Koul), and NIH/NCI R01CA161880 (H. Koul).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: emerging therapeutic strategies. Nat Rev Clin Oncol. 2011;8:357–68. doi: 10.1038/nrclinonc.2011.67. [DOI] [PubMed] [Google Scholar]
- 3.Loblaw DA, Virgo KS, Nam R, Somerfield MR, Ben-Josef E, Mendelson DS, Middleton R, Sharp SA, Smith TJ, Talcott J, Taplin M, Vogelzang NJ, Wade JL, Bennett CL, Scher HI. Initial hormonal management of androgen-sensitive metastatic, recurrent, or progressive prostate cancer: 2007 update of an american society of clinical oncology practice guideline. J. Clin. Oncol. 2007;25:1596–605. doi: 10.1200/JCO.2006.10.1949. [DOI] [PubMed] [Google Scholar]
- 4.Culig Z, Bartsch G. Androgen axis in prostate cancer. J Cell Biochem. 2006;99:373–81. doi: 10.1002/jcb.20898. [DOI] [PubMed] [Google Scholar]
- 5.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–95. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–44. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 7.Culig Z, Hobisch A, Hittmair A, Peterziel H, Cato AC, Bartsch G, Klocker H. Expression, structure, and function of androgen receptor in advanced prostatic carcinoma. Prostate. 1998;35:63–70. doi: 10.1002/(sici)1097-0045(19980401)35:1<63::aid-pros9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 8.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–34. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 9.Peehl DM, Wong ST, Bazinet M, Stamey TA. In vitro studies of human prostatic epithelial cells: attempts to identify distinguishing features of malignant cells. Growth Factors. 1989;1:237–50. doi: 10.3109/08977198908998000. [DOI] [PubMed] [Google Scholar]
- 10.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toivanen R, Shen MM. Prostate organogenesis: tissue induction, hormonal regulation and cell type specification. Development. 2017;144:1382–98. doi: 10.1242/dev.148270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Scholes J, Hsieh JT. Signal transduction targets in androgen-independent prostate cancer. Cancer Metastasis Rev. 2001;20:351–62. doi: 10.1023/a:1015504015302. [DOI] [PubMed] [Google Scholar]
- 13.Litvinov IV, De Marzo AM, Isaacs JT. Is the Achilles’ heel for prostate cancer therapy a gain of function in androgen receptor signaling? J Clin Endocrinol Metab. 2003;88:2972–82. doi: 10.1210/jc.2002-022038. [DOI] [PubMed] [Google Scholar]
- 14.Hyytinen ER, Haapala K, Thompson J, Lappalainen I, Roiha M, Rantala I, Helin HJ, Jänne OA, Vihinen M, Palvimo JJ, Koivisto PA. Pattern of somatic androgen receptor gene mutations in patients with hormone-refractory prostate cancer. Lab Invest. 2002;82:1591–8. doi: 10.1097/01.lab.0000038924.67707.75. [DOI] [PubMed] [Google Scholar]
- 15.Linja MJ, Savinainen KJ, Saramäki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–5. [PubMed] [Google Scholar]
- 16.Culig Z, Hobisch A, Cronauer M V, Radmayr C, Trapman J, Hittmair A, Bartsch G, Klocker H. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res. 1994;54:5474–8. [PubMed] [Google Scholar]
- 17.Bogdanos J, Karamanolakis D, Tenta R, Tsintavis A, Milathianakis C, Mitsiades C, Koutsilieris M. Endocrine/paracrine/autocrine survival factor activity of bone microenvironment participates in the development of androgen ablation and chemotherapy refractoriness of prostate cancer metastasis in skeleton. Endocr Relat Cancer. 2003;10:279–89. doi: 10.1677/erc.0.0100279. [DOI] [PubMed] [Google Scholar]
- 18.LeRoith D, Roberts CT. The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–37. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 19.Yang YA, Yu J. Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis. 2015;2:144–51. doi: 10.1016/j.gendis.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, Cejas P, Vazquez F, Cook J, Shivdasani RA, Bowden M, Lis R, Hahn WC, Kantoff PW, Brown M, Loda M, Long HW, Freedman ML. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47:1346–51. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. Big data: astronomical or genomical? PLoS Biol. 2015;13:e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genomewide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Dutta A, Le Magnen C, Mitrofanova A, Ouyang X, Califano A, Abate-Shen C. Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science. 2016;352:1576–80. doi: 10.1126/science.aad9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Park D, Zhong Y, Lu Y, Rycaj K, Gong S, Chen X, Liu X, Chao HP, Whitney P, Calhoun-Davis T, Takata Y, Shen J, Iyer VR, Tang DG. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:10798. doi: 10.1038/ncomms10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Jänne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao L, Yang L, Li R, Rodrigues DN, Crespo M, Hsieh JT, Tilley WD, de Bono J, Selth LA, Raj GV. Disrupting androgen receptor signaling induces snail-mediated epithelial-mesenchymal plasticity in prostate cancer. Cancer Res. 2017;77:3101–12. doi: 10.1158/0008-5472.CAN-16-2169. [DOI] [PubMed] [Google Scholar]
- 29.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu YM, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]


