Abstract
Pollen tube growth and reorientation is a prerequisite for fertilization and seed formation. Here we report imaging of cAMP distribution in living pollen tubes microinjected with the protein kinase A-derived fluorosensor. Growing tubes revealed a uniform distribution of cAMP with a resting concentration of ≈100–150 nM. Modulators of adenylyl cyclase (AC), forskolin, and dideoxyadenosine could alter these values. Transient elevations in the apical region could be correlated with changes in the tube-growth axis, suggesting a role for cAMP in polarized growth. Changes in cAMP arise through the activity of a putative AC identified in pollen. This signaling protein shows homology to functional motifs in fungal AC. Expression of the cDNA in Escherichia coli resulted in cAMP increase and complemented a catabolic defect in the fermentation of carbohydrates caused by the absence of cAMP in a cyaA mutant. Antisense assays performed with oligodeoxynucleotide probes directed against conserved motifs perturbed tip growth, suggesting that modulation of cAMP concentration is vital for tip growth.
Pollen tubes are cells with polarized growth that must find their way to the ovule to achieve fertilization. These cells have been extensively used for the study of signal transduction in plants, namely on the role of cytosolic free calcium ([Ca2+]c), cytoskeletal proteins, and Rop GTPases (1–4). [Ca2+]c seems to play a key role in connecting different pathways but processes downstream of it remain largely unknown.
cAMP is the prototypical second messenger and adenylyl cyclase (AC) its sole source. In cyanobacteria, cAMP levels respond rapidly to environmental changes, and in Anabaena cylindrica it has been shown to act as a second messenger of light signal transduction (5). In algae, many AC genes have been cloned (6–9), and in fungi, cAMP signaling is known to regulate tip growth (10). In the rice blast fungus, cAMP is involved in the pathogenic growth of the fungus (10), and in the barley powdery mildew, cAMP is involved in the formation of germ tubes (11).
Many reports have claimed the identification of cAMP-dependent processes in plant cells mainly by biochemical methods (12, 13). Nevertheless, the cAMP signaling pathway remains largely unknown because of the lack of evidence for its biological role and identification of a plant AC. Molecular techniques led to the identification of members of cAMP-dependent proteins like protein kinase A-like kinases (14) and CREBs (cAMP response element-binding proteins; ref. 15) but the diversity of known ACs does not facilitate their identification by homology search. The algal and fungal enzymes are soluble, and recently a new soluble form of AC was identified in rat with a catalytic domain very similar to ACs from cyanobacteria and myxobacteria (16, 17).
In this work, we obtained results that account for the presence of a cAMP signaling pathway in pollen tube growth along with molecular evidence for a signaling protein from pollen that is identified as an AC.
Materials and Methods
Plant Material.
Pollen of Agapanthus umbellatus (Liliaceae) was harvested and stored, and pollen tubes grown in vitro as described (18).
Effect of External Gradients.
External gradients were imposed by diffusion of solutions from a micropipette placed near the tip of the pollen tubes (4). We tested the effect of the following modulators of cAMP levels: dibutyryl cAMP, 20 μM (Sigma); forskolin, 20 μM (Sigma); Rp-Isomer-8-Br-cAMP monophosphorothioate, 40 μM (Calbiochem); 8-phenyltheophilline, 50 μM (Sigma); and 2′,5′-dideoxyadenosine, 1 mM (Calbiochem).
Loading and Photoactivation of Caged-cAMP.
Intracellular cAMP ([cAMP]i) was manipulated in discrete areas of the pollen tube as described (4) by photoactivation of the membrane-permeable caged cAMP [1-(2-nitrophenyl)ethyl adenosine 3′,5′-cyclicmonophosphate; Molecular Probes] (extracellular concentration: 0.25 mM).
Measuring [cAMP]i Levels.
Pollen tubes were microinjected (1) with ≈1 pl of a 20 μM solution of cAMP fluorosensor (Molecular Probes; ref. 19). Cells that recovered from microinjection (≈5%) were analyzed with an MRC-600 (Bio-Rad) confocal microscope. Images were collected with 3% laser intensity (excitation: 514 nm; barrier filter 1: 525–555 nm; barrier filter 2: >600 nm) and a ×3.0 zoom factor, using a Nikon ×60 plan apolipoprotein objective (numerical aperture = 0.95). The time interval between image acquisition was 20 sec because shorter intervals caused significant photobleaching. Ratios of 540/600 nm were obtained with Bio-Rad software (tcsm). The fluorosensor response was determined after addition of dibutyryl cAMP, forskolin, or dideoxyadenosine (20) at the concentrations indicated. Calibration of [cAMP]i was achieved with an in vitro method (5 nM-1 μM cAMP; ref. 19).
RNA and DNA Techniques.
A cDNA library was constructed by using poly(A)+ RNA from maize pollen (inbred line A188). cDNA probes were prepared to pollen and shoot poly(A)+ RNA. All clones that showed hybridization to the radiolabeled pollen cDNA but not to the radiolabeled shoot cDNA were picked and sequenced (21). For Northern blot analysis, total RNA was extracted from germinated pollen by a phenol/SDS method, blotted onto nylon-positive membranes, and probed with digoxigenin (DIG)-labeled cDNA according to manufacturer's (Roche Molecular Biochemicals) instructions. Hybridization was performed at 65°C, and filters were washed three times for 30 min each at 65°C in 2 × SSC (1 × SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 0.1% SDS; 0.5 × SSC, 0.1% SDS; and 0.1 × SSC, 0.1% SDS. Filters were detected with disodium 3-(4-methoxyspiro/1,2-dioxetane-3,2′-(5′-chloro)-tricyclo[3.3.1.13,7] decan/-4-yl)phenylphosphate substrate, and chemiluminescence signals were recorded on Kodak BioMax film.
Activity Assay of Pollen-Signaling Protein (PSiP).
The PSiP coding region (nucleotides 1–2,691) was amplified by PCR and subcloned between the XhoI (5′ end) and NcoI (3′ end) sites of pRSET B (Invitrogen). This construct was transformed into Escherichia coli BL21(DE3) (Stratagene). Expression of the protein was induced by the addition of 0.5 mM isopropyl β-d-thiogalactoside (IPTG), and samples were harvested at regular time intervals. Total cAMP levels were measured in 100 μl of cellular extracts by using a BIOTRAK EIA kit (Amersham Pharmacia). In parallel, the bacterial extracts were fractionated by SDS/PAGE, and the expressed fusion protein was visualized from the gel stained with Coomassie blue.
Complementation of a cya Mutation in E. coli by PSiP.
E. coli SP850 [lam-, el4-, relA1, spoT1, cyaA1400(∷kan), thi-1] was obtained from the E. coli Genetic Stock Center (Yale University, New Haven, CT) where it is registered under the accession number 7200. Screening for the ability to ferment sugars was performed on MacConkey agar plates (Sigma) containing 1% lactose and 0.1 mM IPTG, grown at 30°C.
Antisense Assays.
Oligodeoxynucleotides (Genosys, The Woodlands, TX) having phosphorothioate modifications in the three bases adjacent to each terminus were resuspended in sterile water, incubated for 20 min with the appropriate volume of a stock solution of GS3815 cytofectin 0.2 mg⋅ml−1 (ref. 22; Glen Research, Sterling, VA), and diluted with growth medium to the final concentration of 30 μM for cationic delivery (23). Antisense sequences derived by back-translation of amino acid motifs were as follows: LRVLDLS (5′-GCTGAGGTCGAGCACCCTGAG-3′); DIPSSIG (5′-GCCGATGCTGCTTGGGATGTC-3′); LDDLKAL (5′-GAGGGCGTCCACGTCGCCGAT-3′); KPFLED (5′-GTCGATGAGGAAGGCCTT-3′); DETEKEE (5′-CTCCTCCTTCTCGGTCTCGTC-3′); KLEVLKL (5′-CTCGAACTCGTGGAGCTCGAA-3′); LSQLQSL (5′-CTCCGAGACCTCGACCGACTC-3′).
Results
Diffusion of cAMP Modulators Modifies Pollen Tube Growth Direction.
Pollen tubes frequently change their direction of growth, and we developed an assay to identify putative molecules important for the tip-directed growth. This procedure consists of placing micropipettes loaded with the molecule of study near the tips of growing tubes (4), creating a diffusion gradient across the tip where growth is located. When the membrane-permeable dibutyryl cAMP was used in this test, a strong attraction was observed (Fig. 1 and Table 1). A similar effect (but of less magnitude) was observed with the AC activator, forskolin, whereas antagonists of cAMP accumulation had the opposite effect (Table 1). The cell response to these chemicals thus suggests the involvement of a cAMP pathway in pollen tube growth.
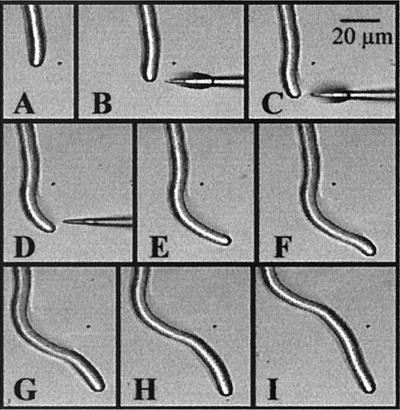
Figure 1.
Time course series of images taken when a micropipette containing dibutyryl cAMP (B–D) is placed near to the tip of a pollen tube. Of 19 tubes, 14 showed an equivalent response and 5 showed no effect.
Table 1.
Effect of external gradients on pollen tube reorientation
| Agent | Reorientation toward (+)/away (−) | Growth arrest | No effect |
|---|---|---|---|
| Dibutyryl cAMP, 20 μM (n = 19) | 14 (+) | 0 | 5 |
| Forskolin, 20 μM (n = 21) | 14 (+) 1 (−) | 0 | 6 |
| Rp-8-Br-cAMPS, 40 μM (n = 20) | 12 (−) | 4 | 4 |
| Dideoxyadenosine, 1 mM (n = 18) | 1 (+) 6 (−) | 0 | 11 |
| Theophylline, 50 μM (n = 19) | 8 (−) | 6 | 5 |
An effect was considered only if accompanied by a transient decrease in growth rates; total arrest, even if followed by recovery, was considered random curvature. The concentration of the chemical refers to the value inside the micropipette.
Measurements of [cAMP]i in Pollen Tubes.
To further establish the role of cAMP in tube growth, the [cAMP]i was measured in living cells by using the 170-kDa cAMP fluorosensor. Full saturation of this fluorosensor lead to a 31% increase at 530 nm and a 13% decrease at 580 nm (≈1.5-fold increase; ref. 19). An in vitro calibration performed at 525–555 nm and >600 nm resulted in similar values and was used for in vivo estimates of cAMP. Imaging of the fluorosensor in pollen tubes showed that [cAMP]i is approximately uniform throughout the cell with a resting level of ≈100–150 nM (Fig. 2). These values, although regarded as estimates, are similar to measurements performed in animal cells (12).
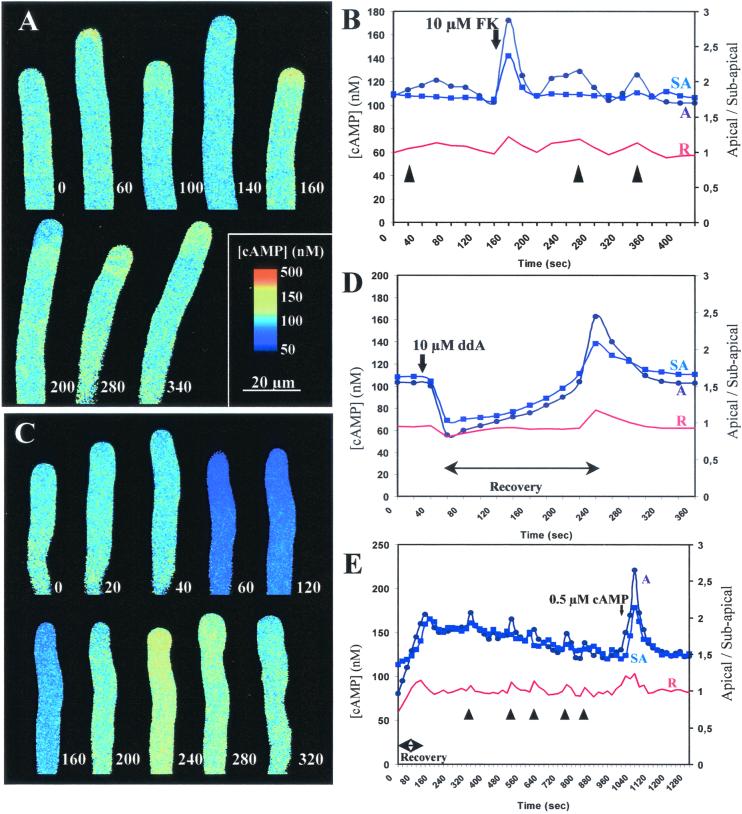
Figure 2.
Confocal time course ratio imaging of [cAMP]i. (A) Effect of 10 μM forskolin (FK) added at second 50. The times (seconds) at which images were taken are shown adjacent to the tip of the growing tube. The images are displayed with the color coding discussed next. (B) Plot of data in A. [cAMP]i in apical (A-●) and subapical (SA-■) regions. Equivalent data were obtained in four independent experiments. Red trace represents the ratio A/SA, and times at which pollen tubes changed growth direction are indicated by arrowheads. (C) Effect of 100 μM 2′,5′-dideoxyadenosine (ddA) added at second 30. (D) Plot of data in C. Equivalent data were obtained in four independent experiments. Symbols as in B. (E) Plot of data of a recovering pollen tube (0–120 sec) treated with 0.5 μM dibutyryl cAMP at sec 1,000. Equivalent data were obtained in six independent experiments. Symbols as in B.
Application of forskolin in a concentration as low as 10 μM caused a transient but significant increase in [cAMP]i that lasted for about 60 sec before returning to resting levels (Fig. 2 A and B). This response suggested the presence of cAMP-degrading phosphodiesterases. The increase in [cAMP]i was more pronounced in the apex, which might reflect AC distribution or the presence of a more permeable wall at the tip. Forskolin diffusing from a micropipette also acted as a chemo-attractant (Table 1).
The AC inhibitor 2′,5′-dideoxyadenosine transiently reduced [cAMP]i (Fig. 2 C and D). At 100 μM, this inhibitor caused temporary growth arrest and a reduction of [cAMP]i followed by a recovery period (≈200 sec), during which [cAMP]i returned to resting levels. Just before growth recovery, [cAMP]i in the apical region rose above the subapical. This transient apical elevation was also observed in cells recovering from the microinjection procedure (Fig. 2E), suggesting their importance in the establishment of polarity. Similar asymmetric elevations of [cAMP]i could be frequently correlated with changes in tube growth direction (Fig. 2 B, D, and E; arrowheads). From the 24 changes in growth direction observed in the cells under study, 17 (≈70%) registered an apical [cAMP]i transient. The magnitude of these transients was quite variable (up to 200 nM) but usually did not exceed 160 nM. The measurements performed did not allow us to precisely determine whether, within the apical dome, the [cAMP]i was higher in the side to which the cell bends. Although the diffusion experiments suggest this to be the case, it is likely that the fluorosensor was not sensitive enough to detect gradients across the short diameter of the tube.
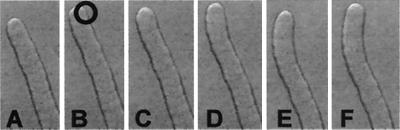
After challenge with dibutyryl cAMP at an external concentration of 0.5 μM, [cAMP]i was elevated to 220–300 nM but rapidly decreased to resting levels, indicating that [cAMP]i is homeostatically regulated (Fig. 2E). These [cAMP]i elevations were more pronounced in the apical region and led to reorientation of the growth axis. It was also found that if [cAMP]i was maintained between ≈70–350 nM, growth was not arrested. Outside those limits and depending on the intensity of the treatment, growth was permanently or transiently arrested. To help confirm a role for cAMP in reorientation, loaded caged cAMP was photolysed in one side of the apical dome. From the 20 cells where the experiment was performed, 14 showed curvature of the growth axis toward side of release within 5–15 sec (Fig. 3).
Figure 3.
Time course series of a pollen tube loaded with caged cAMP. A flash of UV light was given at image B in the circled area.
Identification of a Novel Signaling Protein from Pollen.
To further study the role of cAMP, we resorted to a maize cDNA library from which 300 classes of pollen-specific clones had been isolated and sequenced (21). Sequence analysis and database searching unveiled a signaling protein with homology to AC from fungi investigated further as a putative AC fragment (GenBank accession no. AJ307886). From genomic Southern analysis, the gene encoding this protein, which we named PSiP, seems to be single-copy (data not shown). This cDNA with 3,051 base pairs, although not full-length, contained a large ORF that encoded 897 amino acids (Fig. 4). This protein showed homology to fungal AC (e.g., Neurospora crassa), and highest similarity was found in a region characterized by periodic leucine-rich motifs. Similarly to the fungal enzymes, this sequence predicts a soluble protein. PSiP blast query in the Arabidopsis database prompted nucleotide binding site-leucine-rich repeat disease-resistance proteins as putative homologs (Supplemental Material, which is published on the PNAS web site, www.pnas.org).
Figure 4.
Predicted amino acid sequence of PSiP. A consensus ATP-binding site is indicated in bold. A leucine-zipper motif is identified in positions 317–338 (LRSLARALIADESILIYGQQKL). Underlined sequences were used to design ODN probes for antisense assays, by back-translation, using the Liliaceae codon usage.
PSiP Produces cAMP when Expressed in E. coli.
Because there are no other credible sequences identified in plants, only the clone-functional characterization could provide definite evidence for AC activity. The coding region of PSiP was cloned into the expression vector pRSET and used to transform E. coli BL21(DE3) cells. Expression was induced with IPTG, and total cAMP was measured over time in bacterial extracts (Fig. 5). When compared with the levels from noninduced cells (Fig. 5B), [cAMP] consistently increased with time after induction, which was already noticeable after 2 h. Addition of 100 μM forskolin to induced bacteria significantly increased the production of cAMP (Fig. 5B). This result suggests that the cDNA is competent in synthesizing cAMP although missing the 5′-end terminal region.
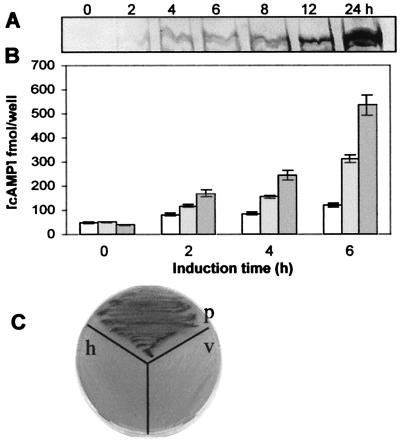
Figure 5.
Functional characterization of PSiP. (A) SDS protein gel stained with Coomassie blue showing the PSiP band in bacterial extracts collected at the indicated times after IPTG induction. (B) cAMP production in E. coli transformed with pRSET containing the PSiP coding region. The values refer to total [cAMP] in 100 μl of bacterial extracts (with corrections made for the different OD600 values) at the indicated time intervals: (□) noninduced cells, ( ) IPTG-induced cells, and (
) IPTG-induced cells, and ( ) IPTG-induced cells in the presence of 100 μM forskolin. Error bars indicate standard deviation from a minimum of three experiments. (C) Complementation of the SP850 E. coli cyaA mutation by PSiP. Nontransformed host cells (h) and bacteria transformed with the pRSET vector (v) remained colorless, whereas colonies with the vector containing the PSiP insert (p) showed a strong purple color (darker in image).
) IPTG-induced cells in the presence of 100 μM forskolin. Error bars indicate standard deviation from a minimum of three experiments. (C) Complementation of the SP850 E. coli cyaA mutation by PSiP. Nontransformed host cells (h) and bacteria transformed with the pRSET vector (v) remained colorless, whereas colonies with the vector containing the PSiP insert (p) showed a strong purple color (darker in image).
To further confirm the ability of PSiP to produce cAMP, we resorted to a cyaA mutant host strain, where the presence of this signaling nucleotide gave rise to a selectable phenotype. E. coli SP850 is a cyaA deletion mutant that lacks AC. Because of the cyaA mutation, this strain cannot use lactose (Lac−) and produces colorless colonies on MacConkey agar. The PSiP coding region cloned in the pRSET vector was transformed into SP850 cells, and the recombinant colonies were screened on MacConkey agar containing 0.1 mM IPTG. When compared with the colorless colonies produced by bacteria transformed with the pRSET vector alone, colonies containing PSiP stained deep purple. To confirm the complementation, the positive colonies were restreaked on fresh plates (Fig. 5C), and the presence of insert was confirmed by PCR analysis and restriction digestion (data not shown).
Antisense Oligodeoxynucleotides (ODNs) from PSiP Perturb Pollen Tube Growth.
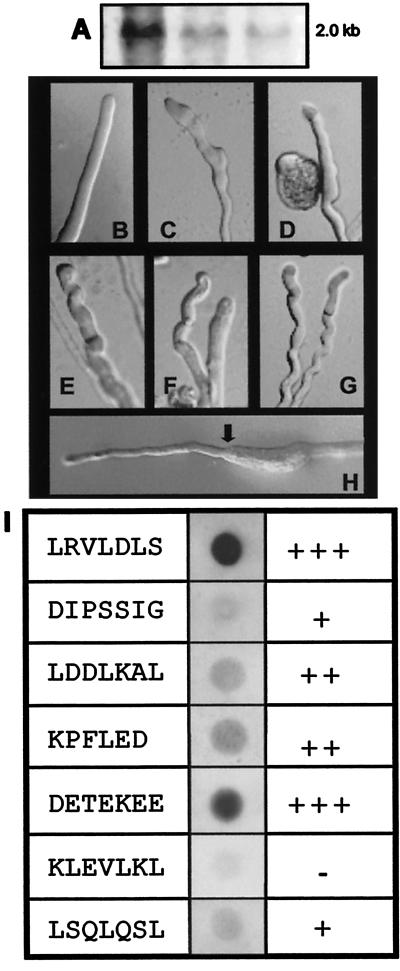
To characterize the putative role of PSiP in pollen, we resorted to pollen of Agapanthus umbellatus, in which intensive studies have been performed (1, 3, 4, 18, 23). The putative presence of a PSiP homolog was tested by Northern blot analysis in A. umbellatus pollen RNA. By using a DIG-labeled PSiP cDNA probe (coding region and 3′ untranslated region), a transcript of ≈2.0 kb was identified (Fig. 6A).
Figure 6.
Antisense perturbation of PSiP function. (A) Northern analysis showing positive hybridization of PSiP/DIG labeled with 25, 10, and 5 μg of total RNA from pollen of A. umbellatus. (B) Control cell, germinated in liquid medium supplemented with 15 μg⋅ml−1 of cytofectin in the absence of antisense probes. (C–G) Effect of ODNs after 3 h of germination. A clear perturbation of tip growth led to abnormal morphology. The examples were chosen from a gallery of images representative of altered behavior. C and D germinated in DETEKEE-antisense; E–G germinated in LRVLDLS-antisense. (H) Recovery of normal tip morphology in an ODN-treated cell induced by 20 μM dibutyryl cAMP; arrow indicates addition of cAMP (≈3 h and 30 min after germination). (I) Dot-blot hybridization of 5 μg of total RNA from pollen of A. umbellatus probed with different DIG-labeled ODNs. The hybridization signals correlated with the antisense effect observed in the germinating cells. Estimates from countings were +++, more than 70% of cells perturbed; ++, between 40 and 70%; +, less than 40%; and −, less than 10% (which can be considered normal for germination periods longer than 3 h).
To evaluate the function of PSiP in tube growth, transient antisense assays were performed by using ODN probes directed against AC-conserved motifs in the PSiP sequence (Fig. 4, underlined). The ODN probes were combined with a cationic liposome formulation (22, 23) and supplied to cells germinating in growth medium. ODNs encapsulated in the liposome successfully traversed the membrane to achieve inhibition of gene expression (23). In the control experiment, cells were treated with the correspondent sense ODNs, as well as antisense oligos targeted for plasmid sequences (M13 and universal reverse primers). In the presence of these ODNs, the pollen tubes showed no significant perturbation in growth or polarity. Antisense ODNs without cytofectin encapsulation also failed to evoke detectable responses. During the first 3 h of germination, antisense-treated cells behaved similarly to controls (Fig. 6B). After that period, signs of perturbation started to be observed (Fig. 6 C–G); tip morphology was altered, rendering growth slower and suggesting a fundamental role for PSiP in tip growth. This effect could be reverted by the external addition of cAMP (Fig. 6H) or mimicked by the addition of AC antagonists (e.g., dideoxyadenosine), confirming that the ODNs are interfering with AC activity. Not all of the antisense probes had the same rate of success in perturbing tip growth. Although the type of perturbation exerted by each ODN was similar, some sequences affected larger proportions of the cell population. These observations correlated to the hybridization results of each DIG-labeled ODN in pollen RNA dot blots (Fig. 6I); ODNs with stronger hybridization induced more growth perturbation.
Discussion
This study describes the imaging of intracellular cAMP and assigns a role for this molecule in pollen tube growth. By using caged probes and imposed external gradients, we show that manipulation of cAMP levels in the apical region modifies growth direction. A pollen-specific clone that codes for a soluble signaling protein (PSiP) with AC activity was identified, and inhibition of PSiP by antisense assays disturbs apical growth.
cAMP Is a Second Messenger in Plant Tip Growth.
cAMP signaling has been a controversial issue in plant cells despite occasional reports of gene homologs for elements of this pathway (24) and responses induced by modulators of AC activity (25, 26). Previous methodologies used to measure cAMP levels were often found to provide values close to or below the detection limit (12). These methods used mainly whole-tissue extracts that masked possible asymmetries in the production of cAMP by specific groups of cells (e.g., anthers). The development of a fluorescent probe (19) was thus essential for the single-cell approach undertaken in this work.
The finding that cAMP is important for plant tip growth is significant. cAMP was suggested to promote the growth of self-incompatible pollen tubes (27) and the importance of AC and cAMP in the regulation of tip growth is known in fungal hyphae (10). Our imaging data showed that [cAMP]i is approximately uniform throughout the cell with a resting level of ≈100–150 nM. These values are similar to those found in animal cells (12). Pollen [cAMP]i seems to be tightly regulated because values outside the 70–350 nM range were found to perturb or arrest growth; the addition of forskolin evoked a transient raise in cAMP and 2′,5′-dideoxyadenosine diminished this value. Moreover, growth recovery and reorientation of the growth axis, which are moments of intense signaling, are accompanied by a rise in cAMP. External addition of the membrane-permeable dibutyryl cAMP also evoked a rise in cAMP concentration but homeostasis was efficiently imposed, suggesting the presence of a catabolic pathway for cAMP signaling. We previously reported that modification of cAMP levels was accompanied by elevations in [Ca2+]c (1), suggesting that the two signaling pathways are interconnected. Most interestingly, cAMP has been shown to induce switching in turning direction of nerve growth cones (28) in a process that is intimately connected with Ca2+ signaling (29). The many similarities of neurons with pollen tubes (1) thus suggest strategies for future work.
Unlike [Ca2+]c, the distribution of [cAMP]i does not reveal a tip-focused gradient. PSiP is a soluble protein so its distribution is most likely even in the cytosol. Furthermore, cAMP is a readily diffusible molecule, and the formation of a tip-focused gradient is unlikely. It is possible that the apical environment results in a higher activity of PSiP but the same can apply to phosphodiesterase. Transient elevations in [cAMP]i can then be explained by different affinities of PSiP and phosphodiesterase to their regulatory elements. As in neurons, the action of cAMP in polarized growth resembles that of a switch that is activated by certain conditions.
Generally, the downstream target of cAMP is PKA, and several lines of evidence suggest its presence in plant cells (12, 14). CREBs (cAMP response element-binding factors), which have been found in plants (15), are also putative targets for cAMP signaling. In Arabidopsis, a plant homolog to the cyclic nucleotide-gated cation channels was cloned and its function characterized (30), whereas in tobacco, a plasma membrane calmodulin-binding channel protein with a putative cyclic nucleotide-binding domain was identified (31).
PSiP Is Involved in Apical Growth.
PSiP is a novel signaling protein specific to pollen tissues and is involved in the control of polarity. The perturbation of its function with ODNs suggests its fundamental role in the maintenance of transduction mechanisms operating in pollen tubes. The analysis of a PSiP amino acid sequence predicted a role in signaling pathways—the presence of a nucleotide binding-site motif A (also known as P loop) for binding ATP or GTP, and leucine-rich repeats.
When the coding region of PSiP was subcloned into an expression vector in E. coli, induced cells show a robust increase in cAMP when compared with cells that were not induced but were carrying the competent cDNA. This result was confirmed by complementation of the SP850 cyaA mutant host strain, which lacks AC, with PSiP. In E. coli, cAMP bound to the transcriptional activator CAP (catabolite gene-activator protein) is a pleiotropic regulator of the expression of genes involved in the catabolism of carbohydrates such as lactose and maltose (32). Hence, E. coli strains lacking cAMP are unable to ferment these sugars, a condition that can be overcome by the expression of putative AC candidates (33). Therefore, PSiP has competence for producing cAMP and is considered to be a soluble AC. Recently, a distinct class of soluble ACs, most similar to the enzymes from cyanobacteria, was identified in mammals (16), and high levels of this AC were shown to be unique to male germ cells (34). The complete cDNA encoded a protein of 187 kDa, but the catalytically active purified form of the enzyme is only 48 kDa (16).
It is possible that the missing 5′ end in PSiP has a regulatory function. The original pollen transcript encoding PSiP in maize is ≈4.5 kb, but it is difficult to predict how many amino acids are missing from the 5′ end of PSiP. Nevertheless, addition of forskolin to PSiP-expressing bacteria had a positive effect in the production of cAMP, noticeable since the first hours of induction.
AC: Disguised by Diversity?
Polymorphism is high in the AC family and homologies between bacteria, fungi, and animal sequences are restrained to short motifs. The abundance of AC isoforms, from prokaryotes to eukaryotes, contrasts with the apparent poverty in plant cells. Nevertheless, indications are that signal transduction components have diverged considerably, and identification will not come readily from sequence comparison with animal genes alone (13).
The sequence resemblance of PSiP with disease-resistance proteins evokes the possibility that in plants AC enzymes might be disguised under the broad spectra of large gene families, like the R gene family, with recent estimates going from 200–300 in Arabidopsis to 1,500 in rice (35). R gene proteins directly activate multiple signaling pathways, and studies using various inhibitors suggest that Ca2+ channels, kinases, and phosphatases play important roles (36). Plant resistance responses to pathogenic fungi and bacteria are often highly localized, the responses occur within few minutes, and a rapid transient increase in cAMP has been reported (37). One of the first plant disease-resistance genes to be cloned, the RPS2 gene, contains leucine zipper, P loop domains, and leucine-rich repeats, an interaction domain similar to that of yeast AC (38). Leucine-rich repeats have been suggested to function as mediators of protein–protein interactions as a downstream step of signal transduction pathways (39). Indeed, a blast query with yeast AC in the Arabidopsis database reveals a great number of clones with sequence similarity to this enzyme, including many receptor protein kinases and disease-resistance proteins. This intimacy between AC and stress signaling in plants has been suggested (13), and the cellular responses driven by pathogens elicit a response similar to the one operating in pollen tubes: localized interaction with external cues at the very tip and cellular responses within minutes.
Supplementary Material
Acknowledgments
We thank G. Dönges for his help in the sequencing of PSiP. This work was supported by the Human Frontier Science Program and FCT Grant P/BIA/11016/98.
Abbreviations
- AC
adenylyl cyclase
- DIG
digoxigenin
- PSiP
pollen-signaling protein
- [cAMP]i
intracellular cAMP
- ODN
oligodeoxynucleotide
- IPTG
isopropyl β-d-thiogalactoside
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ307886).
References
- 1.Malhó R, Camacho L, Moutinho A. Ann Bot (London) 2000;85:59–68. [Google Scholar]
- 2.Zheng Z-L, Yang Z. Trends Plant Sci. 2000;5:298–303. doi: 10.1016/s1360-1385(00)01654-x. [DOI] [PubMed] [Google Scholar]
- 3.Malhó R, Read N D, Trewavas A J, Pais M S. Plant Cell. 1995;7:1173–1184. doi: 10.1105/tpc.7.8.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malhó R, Trewavas A J. Plant Cell. 1996;8:1935–1949. doi: 10.1105/tpc.8.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohmori M, Ohmori K, Hanasunuma K. Arch Microbiol. 1998;150:203–204. [Google Scholar]
- 6.Katayama M, Wada Y, Ohmori M. J Bacteriol. 1995;177:3873–3878. doi: 10.1128/jb.177.13.3873-3878.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yashiro K, Sakamoto T, Ohmori M. Plant Mol Biol. 1996;31:175–181. doi: 10.1007/BF00020618. [DOI] [PubMed] [Google Scholar]
- 8.Kasahara M, Yashiro K, Sakamoto T, Ohmori M. Plant Cell Physiol. 1997;38:828–836. doi: 10.1093/oxfordjournals.pcp.a029241. [DOI] [PubMed] [Google Scholar]
- 9.Katayama M, Ohmori M. J Bacteriol. 1997;179:3588–3593. doi: 10.1128/jb.179.11.3588-3593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi W, Dean R A. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinane J, Dalvin S, Bindslev L, Hall A, Gurr S, Oliver R. Mol Plant–Microbe Interact. 2000;13:494–502. doi: 10.1094/MPMI.2000.13.5.494. [DOI] [PubMed] [Google Scholar]
- 12.Assman S M. Plant Physiol. 1995;108:885–889. doi: 10.1104/pp.108.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolwell G P. Trends Biochem Sci. 1995;20:492–495. doi: 10.1016/s0968-0004(00)89114-8. [DOI] [PubMed] [Google Scholar]
- 14.Biermann B, Johnson E M, Feldman L J. Plant Physiol. 1990;94:1609–1615. doi: 10.1104/pp.94.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katagini F, Lam E, Chua N-H. Nature (London) 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- 16.Buck J, Sinclair M L, Schapal L, Cann M J, Levin L R. Proc Natl Acad Sci USA. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Cahn M J, Litrin T N, Igurgenko V, Sinclair M L, Levin L R, Buch J. Science. 2000;289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- 18.Malhó R, Read N D, Pais M S, Trewavas A J. Plant J. 1994;5:331–341. [Google Scholar]
- 19.Adams S R, Harootunian A T, Buechler Y T, Taylor S S, Tsien R Y. Nature (London) 1991;349:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 20.Holgate S T, Lewis R A, Austen K F. Proc Natl Acad Sci USA. 1985;93:13389–13392. [Google Scholar]
- 21.Lopez I, Anthony R G, Maciver S K, Jiang C-J, Khan S, Weeds A G, Hussey P J. Proc Natl Acad Sci. 1996;93:7415–7420. doi: 10.1073/pnas.93.14.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis J G, Lin K Y, Kothavale A, Flanagan W M, Matteucci M D, DePrince R B, Mook R A, Hendren R W, Wagner R W. Proc Natl Acad Sci USA. 1996;93:3176–3181. doi: 10.1073/pnas.93.8.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moutinho, A., Camacho, L., Haley, A., Pais, M. S., Trewavas, A. J. & Malhó, R. (2001) Sex. Plant Reprod., in press.
- 24.Kawai M, Aotsuka S, Uchimiya H. Plant Cell Physiol. 1998;39:1380–1383. doi: 10.1093/oxfordjournals.pcp.a029346. [DOI] [PubMed] [Google Scholar]
- 25.Ehsan H, Reichheld J P, Roef L, Witters E, Lardon F, Van Bockstaele D, Van Montagu M, Inzé D, Van Onckelen H. FEBS Lett. 1998;422:165–169. doi: 10.1016/s0014-5793(97)01610-4. [DOI] [PubMed] [Google Scholar]
- 26.Volotovski I D, Sokolovsky S G, Molchan O V, Knight M R. Plant Physiol. 1998;117:1023–1030. doi: 10.1104/pp.117.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tezuka T, Hiratsuka S, Takahashi S Y. Plant Cell Physiol. 1993;34:955–958. [Google Scholar]
- 28.Song H-J, Ming G-L, Poo M-M. Nature (London) 1997;388:275–279. doi: 10.1038/40864. [DOI] [PubMed] [Google Scholar]
- 29.Petersen O H, Cancela J M. Curr Biol. 2000;10:311–318. doi: 10.1016/s0960-9822(00)00438-3. [DOI] [PubMed] [Google Scholar]
- 30.Leng Q, Mercier R W, Yao W, Berkowitz G A. Plant Physiol. 1999;121:753–761. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arazi T, Kaplan B, Fromm H. Plant Mol Biol. 2000;42:591–601. doi: 10.1023/a:1006345302589. [DOI] [PubMed] [Google Scholar]
- 32.Ullmann A, Danchin A. Advances in Cyclic Nucleotide Research. Vol. 15. New York: Raven; 1983. pp. 1–53. [Google Scholar]
- 33.Tang W-J, Gilman A G. Science. 1995;268:1769–1772. doi: 10.1126/science.7792604. [DOI] [PubMed] [Google Scholar]
- 34.Sinclair M L, Wang X Y, Mattia M, Conti M, Buck J, Wolgemuth D J, Levin L R. Mol Reprod Dev. 2000;56:6–11. doi: 10.1002/(SICI)1098-2795(200005)56:1<6::AID-MRD2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Young N D. Curr Opin Plant Biol. 2000;3:285–290. doi: 10.1016/s1369-5266(00)00081-9. [DOI] [PubMed] [Google Scholar]
- 36.Innes R W. Curr Opin Plant Biol. 1998;1:299–304. doi: 10.1016/1369-5266(88)80050-5. [DOI] [PubMed] [Google Scholar]
- 37.Bolwell G P. Phytochemistry. 1992;31:4081–4086. [Google Scholar]
- 38.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki N, Choe H R, Nishida Y, Yamawaki-Kataoka Y, Ohnishi S, Tamaoki T, Kataoka T. Proc Natl Acad Sci. 1990;87:8711–8715. doi: 10.1073/pnas.87.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.