Abstract
In species with genetic sex-determination, the chromosomes carrying the sex-determining genes have often evolved non-recombining regions and subsequently evolved the full set of characteristics denoted by the term ‘sex chromosomes’. These include size differences, creating chromosomal heteromorphism, and loss of gene functions from one member of the chromosome pair. Such characteristics and changes have been widely reviewed, and underlie molecular genetic approaches that can detect sex chromosome regions. This review deals mainly with the evolution of new non-recombining regions, focusing on how certain evolutionary situations select for suppressed recombination (rather than the proximate mechanisms causing suppressed recombination between sex chromosomes). Particularly important is the likely involvement of sexually antagonistic polymorphisms in genome regions closely linked to sex-determining loci. These may be responsible for the evolutionary strata of sex chromosomes that have repeatedly formed by recombination suppression evolving across large genome regions. More studies of recently evolved non-recombining sex-determining regions should help to test this hypothesis empirically, and may provide evidence about whether other situations can sometimes lead to sex-linked regions evolving. Similarities with other non-recombining genome regions are discussed briefly, to illustrate common features of the different cases, though no general properties apply to all of them.
This article is part of the themed issue ‘Evolutionary causes and consequences of recombination rate variation in sexual organisms’.
Keywords: pseudo-autosomal region, sexual antagonism, inversion, translocation, Poecilia reticulata, stickleback
1. Introduction: non-recombining regions and sex chromosomes
Non-recombining regions of chromosomes have evolved many times, and theoretical models have shown that natural selection often favours low recombination rates, especially when gene interactions favour specific allele combinations of alleles maintained in populations. Lower survival or fertility of recombinants then favours closer linkage between the genes. Given heritable recombination rate differences, a response to selection may occur. Great efforts have therefore been devoted to understanding why recombination rates differ between different genome regions. Studying situations that are repeated in different organisms should be helpful. For example, the low-recombination centromeric regions of many organisms [1,2] may reflect selection to avoid recombination changing the numbers of functionally important repeats, or non-selected processes may have permitted chromatin states to evolve that resulted in low recombination rates [3]. Gene interactions of the kind just mentioned are probably not involved.
Sex chromosomes’ non-recombining regions are, however, of special interest because gene interactions of the kind predicted to generate selection for suppressed recombination are likely to be involved in the evolution of separate sexes, and can potentially be studied empirically to test the assumptions of the models involved. Recombination has clearly repeatedly become suppressed between sex chromosomes, as it is firmly established that, in many cases, including the very well-studied genus Drosophila, that they evolved from formerly recombining chromosomes (e.g. [4,5]). In male mammals, the XY pair do not cross over, except in a physically small parts at one or both ends of the pair, called the ‘pseudo-autosomal regions’, or PARs; in females, crossovers occur across the whole physical X pair. Restriction of crossing over to the physically small PAR causes exceptionally high recombination rates per megabase of DNA in male meiosis. In other genetically well-studied species with male heterogamety, part of the XY pair is also non-recombining. This includes several plants that have separate sexes [6]. Some species, including Diptera and some voles, have achiasmate male meiosis with no chromosome pairing. Their XY pairs are therefore wholly non-recombining, with no PAR [7]. In birds and Lepidoptera, females are the heterogametic sex (denoted by ZW, while males are ZZ), and the W chromosomes of many species are again largely or wholly non-recombining in females. In Lepidoptera, female meiosis is achiasmate, while in birds the recombining regions range from physically large PARs, of up to 2/3 of the total ZW pair in early-branching lineages (Paleognathous birds), to much reduced PARs in late-branching lineages (reviewed in [8]).
2. Do chromosomes carrying sex-determining genes evolve suppressed recombination more frequently than other chromosomes?
Sex chromosomes in the organisms just mentioned are very familiar, and biologists often take it for granted that suppressed recombination between chromosomes carrying sex-determining loci must inevitably evolve. However, it is not certain that chromosomes that carry sex-determining genes evolve suppressed recombination regions more frequently than other chromosomes. Systems with a single sex-determining gene are known (see §4 below), and some systems have not evolved extensive sex-linked regions [9]. Such sex-determining regions should be distinguished from one another, and from non-recombining, sex-chromosome-like regions that include multiple genes.
To infer the evolution of fully sex-linked regions, it is necessary to make comparisons with species that have retained the ancestral state. Dioecious species of flowering plants whose individuals are genetically male or female are excellent for such comparisons, because dioecy is often a recently derived state. Plants with separate sexes have evolved many times from ancestors that either had hermaphroditic flowers, or were monoecious (with male and female flowers produced by all individuals, and no genetic control of individuals' sex phenotypes). One can therefore potentially compare recombination rates for homologous chromosomes [10]. Other taxa where relevant comparisons may be possible are found among fish and lizards, where the chromosome involved in sex determination has changed over evolutionary time, and recombination can be studied for currently sex-determining chromosomes [11,12].
Estimating and comparing genetic maps between species is laborious. However, if a heteromorphic chromosome pair is observed in one sex, this chromosome is probably non-recombining in that sex. Heteromorphism of a formerly homomorphic chromosome pair (not heteromorphic in related species) may evolve after a suppressed recombination region evolves, including through chromosomal changes including deletions or pericentric inversions; translocations between sex chromosomes and autosomes (including fusions between acrocentric chromosomes) can also create heteromorphism, by expanding a Y or a W chromosome's physical size [13,14]. Heteromorphism also evolves when repetitive sequences such as transposable element insertions accumulate. This is expected in non-recombining sex chromosomes or sex-linked regions, whereas insertions are disfavoured in recombining genome regions because they cause chromosome rearrangements [15]. TE accumulation can potentially detectably increase sex chromosome sizes [16], and create heterochromatic regions.
Suppressed recombination also reduces the efficacy of purifying selection, permitting an increase in deleterious substitutions in genes on Y and W chromosomes [17–20], including loss of function mutations, or deletion of entire genes from Y and W chromosomes (reviewed in [21]). After a non-recombining region has lost many genes, it may tolerate deletions reducing its physical size. There may thus be no overall consistent long-term trend in size, and bird W chromosome size changes indeed show both increases and decreases [22].
However, homomorphic sex chromosome pairs may also include non-recombining regions, often detectable only by refined cytogenetics, including banding methods that may show heterochromatin accumulation on a chromosome present only in one sex. Today, cytogenetic approaches are being supplemented by genome sequencing (see table 1 below).
Table 1.
Stages in the evolution of sex-linked regions and sex chromosomes, with approaches that can be used to detect them. For each stage, the approaches listed for earlier stages can also be helpful and are therefore not repeated (for example, discovering sex-linked genes that are not the sex-determining genes, e.g. visible colour markers, tells one that a sex-linked region exists). The table uses the case of male heterogamety, but the same basic stages can also be found in ZW systems.
| age of sex-determining system | type of sex chromosomes | non-recombining region | approaches for detection | references to examples of use of the approaches listed |
|---|---|---|---|---|
| very young | homomorphic | none (linked male- and female-determining loci, or single sex-determining locus) | genetic mapping in families to locate the sex-determining locus | [23] |
| young | usually homomorphic or micro-heteromorphic | offten small most genes still present on both homologues, only slightly diverged in sequence |
searches for male-specific variants at sites in genes other than the sex-determining genes but that show LD with the SEX locus, using males and females: (a) RAD-Seq of samples from populations or families (b) analysis of heterozygote frequencies to detect sequences with variants heterozygous in all males, but not females (Fis analysis) (c) genetic studies of RNA-Seq variants in families to detect sex-linked inheritance patterns |
(a) [11] (b) [24] (c) [25] |
| old | heteromorphic XY or ZW pair may be cytologically detectable in males | region includes multiple genes many genes present only in XX females due to loss from the Y chromosome Y-linked sequences diverged at many sites, sometimes making them ‘invisible’ (null alleles) |
analysis of coverage in whole genome sequences, to detect hemizygous regions in one sex, indicating degenerated Y-linked regions in X/Y males, or highly diverged Y-linked sequences that are not assembled with the X-linked gene (or similar differences in ZW systems) | [26] |
3. Old and young sex-linked systems in different organisms, evolutionary strata
High-throughput sequencing can now potentially be used to detect sex-linked regions (e.g. [27]), and this promises to uncover many more cases where non-recombining regions exist, including in species where no heteromorphism is cytologically detectable. Such data should refine our understanding of the evolutionary changes that have led to suppressed recombination. It is therefore helpful to distinguish between young sex chromosome systems and those with the well-developed properties familiar from mammals, birds and Drosophila.
Table 1 categorizes systems according to the development of these properties, and shows how different properties can be used to detect sex-determining regions. Systems at different evolutionary stages are so different that use of the term ‘sex chromosomes’ for all of them can be confusing. For instance, analyses of whole genome sequences can potentially detect a region where coverage in males is half that in females, even if a Y-linked region is physically small and includes few genes. However, a coverage difference is expected only if recombination stopped long enough ago for Y- (or W-) linked genes to have become deleted or diverged in sequence at multiple sites, so that they are not recognized as alleles of X- (or Z-) linked genes when assembling the genome. Also regions with few genes lose genes more slowly than large regions. Coverage differences alone may therefore fail to identify fully sex-linked regions in species with recently evolved sex chromosomes whose Y (or W) still carries many genes, such as the plant Silene latifolia [28], though a possible signal is detectable in the part of the threespine stickleback Y that stopped recombining most recently [29].
Sequence data also provide estimates of the time of recombination suppression, when Y- (or W-) linked sequences start to substitute variants independently of their X- or Z-linked alleles. Sequence divergence results show that, in the mammalian X and Y pair, regions furthest from the X chromosome PAR stopped recombining first, and regions nearer the PAR much more recently [5]. Such ‘evolutionary strata’, indicating repeated recombination suppression events, are also seen in birds [30], snakes [31,32], the plants Silene latifolia [28] and Carica papaya [33], and threespine sticklebacks [34]. More such analyses will illuminate the relationship between properties such as genetic degeneration and the systems' ages.
4. The ultimate cause of suppressed recombination
Two situations involving sexually antagonistic gene interactions that could select for suppressed recombination are likely to arise during the evolution of sex-linked regions. For simplicity, I outline the case of male heterogamety, where the ‘SEX’ locus is male-specific due to Y-linkage, and is often termed the MSY region. When a new sex-determining ‘SEX’ locus evolves, closely linked sexually antagonistic (SA) mutations that are advantageous in males, but disadvantageous in females, may establish polymorphisms that generate selection for closer linkage to the male-determining locus (figure 1). Unless sex-specific expression of the SA mutation evolves, enlarged SEX regions may thus evolve if sufficient time elapses without the SEX locus being replaced by a new one [9]. Some old-established systems have indeed retained large PARs. It has been suggested that Paleognathous birds' [35] failure to evolve new evolutionary strata might be explained by sex-specific expression [26]. However, it is unclear why sex-specific expression evolves so fast in these species that recombination modifiers do not also respond to the selective situation, while other species have evolved suppressed recombination.
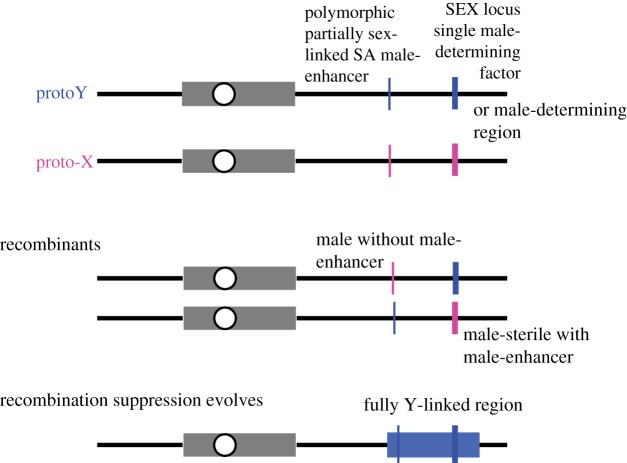
Figure 1.
The sexually antagonistic polymorphism model for the evolution of fully sex-linked regions. The top part of the figure shows an early stage in which a sex-determining region (or SEX locus, indicated by thick vertical lines) has evolved on a chromosome pair, either (i) as a closely linked region with genes promoting male functions on a proto-Y chromosome, and genes with male-sterility alleles on the proto-X, or (ii) as a single male-determining factor that has evolved by a turnover event that led to a new gene taking control of sex determination, or by movement of a sex-determining gene to this genome region. Most or all of the rest of the chromosome pair recombines (except in non-recombining regions near the centromeres, indicated by grey boxes). After a male-promoting allele has established a polymorphism on the same chromosome (thin vertical lines), recombination produces maladaptive combinations of alleles at the two loci (middle part of the figure). This creates selection for closer linkage between the loci, leading potentially to evolution of a genome region of suppressed recombination between the X and Y chromosomes (black box in the bottom diagram, blue in the online version in colour). This sex-linked region could contain many genes in addition to those affecting sex functions, but, for simplicity, the diagram does not indicate these. (Online version in colour.)
(a). De novo evolution of separate sexes from a functionally hermaphroditic ancestor
When a non-dioecious species evolves separate sexes from hermaphroditism (or from environmental sex-determination), at least two mutations are required to generate genetic males and females [36]. Under biologically plausible conditions, male-sterility (female-producing) mutations can invade populations of hermaphrodites, creating populations polymorphic for females alongside bisexual individuals (called gynodioecy). Although cytoplasmic male-sterility is often involved in plants (reviewed in [37]), many nuclear genes can generate male-sterility mutations [38,39], and nuclear femaleness polymorphisms are known in some plant populations [37,40,41]. In such gynodioecious populations, female-sterility mutations (producing males) may be favoured if they increase male function compared with the ancestral hermaphrodites [36]. However, expression of both male- and female-sterility mutations by the same individual is disadvantageous, so maleness mutations are sexually antagonistic unless expression is male-specific, and the same applies to mutations that increase maleness, if there are trade-offs exist between male and female functions. Moreover, new sex-specific mutations appear to be rare, even in organisms with long established separate sexes, such as Drosophila [42]. Male-producing and maleness-enhancing mutations are therefore most likely to invade gynodioecious populations if they are closely linked to the male-sterility gene; consequently, polymorphic male- and female-determining genes should cluster within a single chromosome region, and selection will favour closer linkage between them [43], potentially creating a fully sex-linked region genetically equivalent to a single-gene sex determination system.
However, evolution of suppressed recombination is not inevitable. If sex-limited maleness mutations do occur, they can create true single-gene sex-determination, as they can spread throughout populations, regardless of their genomic location, and should not establish polymorphisms linked to the SEX locus, or generate selection for closer linkage with it [36]. Alternatively, a completely dominant maleness mutation instantly creates a male-limited SEX locus; such a mutation may be involved in the persimmon Diospyros lotus, in the plant family Ebenaceae [44].
Finally, two mutations in the same gene could potentially produce males and females, generating a single-gene sex-determining locus and close linkage without selection for reduced recombination, though no such gene is currently known. Distinguishing between single-gene sex-determination, versus closely linked male- and female-determining genes, is difficult. However, genetic evidence in several plants with physically small sex-determining regions [45,46], suggests that two genes must be involved, because three gender phenotypes (males, females and hermaphrodites) are controlled by a single genetic locus [6].
(b). Turnover events
New SEX genes can also arise by ‘turnover events’ that replace a sex-determining locus by a different one (reviewed in [9]). Turnovers produce homozygotes for the ancestral population's SEX locus, and are unlikely to occur in species with large non-recombining regions carrying many genes whose Y (or W) linked genes are deleted or have lost functions [43, p. 75], causing lethal or highly detrimental effects when homozygous. Turnovers are indeed rare in lizard taxa with heteromorphic sex chromosomes [11]. Turnovers are also less likely if the region includes sexually antagonistic genes essential for the heterogametic sex, although replacement of sex-determining genes by new ones can be promoted by genetic degeneration, under certain conditions of weak, but not very weak, selection [47]. The mutation in persimmon (mentioned above) could reflect a turnover event, rather than de novo evolution of maleness.
(c). Evidence for sexually antagonistic polymorphism
The sexually antagonistic polymorphism hypothesis for the evolution of sex-linked regions is difficult to test. SA variation is plausible and is supported by quantitative genetic data (reviewed in [48]). However, much SA genetic variation may be irrelevant for selection affecting recombination rates, because SA polymorphisms creating selection for linkage to sex-determining loci generally require strong selective differences between alleles, while (other things being equal) weakly selected SA mutations are more readily fixed, unless they are extremely closely linked to the SEX locus [49,50]. The existence of loci under strong SA selection is suggested by the recent discovery of a relationship between sex differences in expression and in allele frequencies at human autosomal genes [51].
Observing enrichment of SA polymorphisms in genome regions close to SEX loci would lend support to the SA polymorphism hypothesis for recombination suppression. This could not be examined in the human study just outlined, because processes other than SA can cause sex differences in allele frequencies at sex linked loci, restricting the analysis to autosomes. Such a situation has, however, long been known in the guppy, Poecilia reticulata, a fish whose conspicuous male coloration is sexually antagonistic, conferring an advantage to males during mating, despite being disadvantageous though increased predation. Genetic factors controlling male coloration in natural populations show a remarkable distribution in the guppy genome (reviewed in [52]). The partially sex-linked region(s) of guppy chromosome 12, on which the guppy SEX gene is located [53], are strikingly enriched for these factors, carrying twice as many such genes as the autosomes (reviewed in [52,54]), although LG12 represents only at most 6% of the genome [55]. As explained above, sex-specific mutations can become fixed regardless of their genomic location, and should therefore be distributed across many different genome regions. The over-representation of these factors in the guppy PAR(s), and the probable rarity of sex-specific mutations (see above, though more studies are needed), argue that they probably did not arise as sex-specific mutations, but that they initially had deleterious effects in females. If so, then, before male-limited expression evolved, resolving the conflict, reduced recombination between these loci and the guppy MSY would also have been favoured. Male guppies from natural high-predation populations in the Aripo river showed strict Y linkage and paternal inheritance of one coloration factor (Sb) more often than males from low-predation regions of the same river [56], suggesting that recombination rates have indeed changed.
Although the guppy X and Y chromosomes are small [57,58], about half the 47 coloration traits studied show father-to-son transmission, implying complete Y-linkage. These factors could represent mutations in genes that were already fully Y-linked when they arose (allowing SA mutations to increase in frequency in populations without counter-selection in females). The number of genes in the guppy MSY region within which multiple coloration mutations might have arisen is not yet known, though a heterochromatic region is cytologically detectable [57]. The chromosome carries more than 900 protein-coding genes [55], and an MSY region of 3 Mb, has been inferred [59], perhaps representing approximately 100 genes. Alternatively, Y-linkage could be an evolved situation, and these factors may formerly have been partially sex linked SA polymorphisms. If different coloration factors evolved in different low-predation populations, followed by evolution of close linkage, MSY regions might differ between populations, as has indeed been inferred [59].
Hybrids between populations with different male traits can help to test whether male traits were formerly expressed in both sexes [60]. If hybrid females express male traits, the populations must have fixed different modifiers suppressing expression in females. However, absence of this clear signal is not decisive, because male-specificity may have evolved within different populations through dominant cis-regulatory changes; foreign population coloration alleles inherited from a male parent might then remain unexpressed in hybrid females.
5. Proximate mechanisms of recombination suppression between sex chromosomes
So far, I have concentrated on the selective forces that may be responsible for suppressed recombination, and on suppression specifically between the sex chromosome pair. The SA selection described above can promote any mechanism that reduces recombination.
One mechanism for recombination suppression after separate sexes have evolved is sex-specific achiasmy, suppressing recombination genome-wide in one sex; for example, in Drosophila, chromosomes do not recombine in males, and in Lepidoptera females do not undergo crossing over (reviewed in [43]). However, this may not have evolved through selection for linkage to a SEX locus.
Suppression specific to the SEX region could involve either systems that control the locations of crossover events, or chromosome rearrangements, such as inversions (e.g. [33,61]). Inversions such as those in the human Y chromosome may, however, be consequences, not causes, of recombination suppression (reviewed in [62]), because recombination suppression allows rearrangements to occur more freely than formerly, since it prevents the generation of deleterious duplications and deletions that are caused when crossover events occur (reviewed in [15]). Moreover, rearrangements suppressing recombination often impair fertility of female heterozygotes [63], requiring a large advantage of restricting recombination.
Translocations between autosomes and sex chromosomes may also create linkage to sex-linked regions. At present, no evidence convincingly demonstrates that they have spread due to selection for such linkages [13,64], and other possibilities exist, including meiotic drive favouring either acro- or metacentric chromosomes in female meiosis [65–67], or advantages of altering genes' expression near breakpoints [13]. These alternatives cannot explain repeated suppression of recombination specifically between sex chromosome pairs. So far, however, only one study has included information on the numbers of events involving only autosomes, which could occur as often as fusions involving sex chromosomes [68]. Moreover, translocations will generally create new recombination-suppressed genome regions of sex chromosomes only near the breakpoints, and perhaps only when autosomal regions are added onto non-recombining regions. Genome sequencing and assembly will add more information about such rearrangements, and their effects, in the future. It will also be interesting to investigate whether non-breakpoint parts of formerly autosomal regions evolve lower recombination. It is already known that autosomal regions added onto PARs sometimes subsequently evolved suppressed recombination, in mammals [69] and the plant Silene latifolia [10]. Future, genome sequencing and assembly should provide more information about these situations.
6. Some further conclusions and topics for future research
Despite being plausible, the SA polymorphism hypothesis for recombination suppression is not yet well supported. One way to test it is to examine alternative possibilities, to ask if they can be excluded. For example, chromatin structure adjacent to an existing non-recombining region might change to a less recombining state, gradually expanding the region of full sex linkage [70]. Single gene sex-determination systems would not experience such effects, so taxa with frequent turnovers could help test whether recombination suppression subsequently evolves, which would support the SA hypothesis. Another alternative explanation suggested for recombination suppression in sex chromosomes and other genome regions is that it permanently maintains deleterious alleles as heterozygotes (reviewed in [71]). However, homozygotes are not prevented from forming until after closely linked polymorphisms for deleterious alleles have become common, which indeed requires recombination suppression, but does not explain why this evolved in the first place.
However it should not be assumed that interactions must necessarily be involved in selecting for recombination suppression, or that similarities to sex chromosomes [72] imply antagonistic polymorphisms. In the case of fungi with large non-recombining mating type regions, interactions may not be involved. Such species, including Microbotrium violaceum [73] and Neurospora tetrasperma [74] seem mostly to be highly inbreeding, and co-segregation of the mating type alleles with the centromere is crucial for inbreeding to occur, because the mating-type system remains functional [75]. The lack of recombination probably evolved to ensure this co-segregation.
Other loci are thought to have evolved suppressed recombination under selection created by interactions between genes that may, in general terms, resemble those involved in sex chromosome evolution. These include the putative ‘supergenes’ controlling Batesian mimicry and heterostyly in plants. Like sex chromosomes controlling male versus female traits, these genetic loci control multiple trait differences between morphs that coexist as polymorphisms within single populations—the mimetic colour morphs in the butterflies (reviewed in [76]), and the two different flower morphologies in distylous plants (reviewed in [77,78]). However, it is not yet certain that selection for suppressed recombination was involved in these systems (see the reviews just cited and [79]).
Genome sequencing can now reveal supergenes, even in non-model species, because one ‘morph’ in a population or species is more heterozygous in the genome region than in the rest of the genome, suggesting some form of balancing selection maintaining different haplotypes with diverged sequences. Sequencing has revealed non-recombining regions including large regions controlling colour morphs in two birds, the white-throated sparrow [80] and the ruff [81,82], and two haplotypes of the fire ant ‘social chromosome’ determine single- versus multiple-queen colony types [83]. However, neither the maintenance of these polymorphisms is yet fully understood for these, nor the form of interactions that may have selected for suppressed recombination, and these too are interesting questions for future research.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
I received no funding for this study.
References
- 1.Nambiar M, Smith G. 2016. Repression of harmful meiotic recombination in centromeric regions. Semin. Cell Dev. Biol. 54, 188–197. ( 10.1016/j.semsdb.2016.01.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charlesworth B, Stephan W, Langley CH. 1986. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112, 947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petes T. 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369. ( 10.1038/35072078) [DOI] [PubMed] [Google Scholar]
- 4.Muller HJ. 1918. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3, 422–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahn BT, Page DC. 1999. Four evolutionary strata on the human X chromosome. Science 286, 964–967. ( 10.1126/science.286.5441.964) [DOI] [PubMed] [Google Scholar]
- 6.Westergaard M. 1958. The mechanism of sex determination in dioecious plants. Adv. Genet. 9, 217–281. ( 10.1016/S0065-2660(08)60163-7) [DOI] [PubMed] [Google Scholar]
- 7.Borodin P, Basheva E, Torgasheva A, Dashkevich O, Golenishchev F, Kartavtseva I, Mekada K, Dumont B. 2012. Multiple independent evolutionary losses of XY pairing at meiosis in the grey voles. Chromosome Res. 20, 259–268. ( 10.1007/s10577-011-9261-0) [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MTP, Zhang G. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346, 1332 ( 10.1126/science.1246338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stöck M, et al. 2011. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 9, e1001062 ( 10.1371/journal.pbio.1001062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu S, Bergero R, Guirao-Rico S, Campos JL, Cezard T, Gharbi K, Charlesworth D. 2016. RAD-mapping reveals an evolving, polymorphic and fuzzy boundary of a plant pseudoautosomal region. Mol. Ecol. 25, 414–430. ( 10.1111/mec.13297) [DOI] [PubMed] [Google Scholar]
- 11.Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. 2015. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol Bio Evol. 32, 1296–1309. ( 10.1093/molbev/msv023) [DOI] [PubMed] [Google Scholar]
- 12.Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M. 2015. Turnover of sex chromosomes in celebensis group medaka fishes. G3 5, 2685–2691. ( 10.1534/g3.115.021543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennell M, Kirkpatrick M, Otto S, Vamosi J, Peichel C, Valenzuela N, Kitano J. 2015. Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 11, e1005237 ( 10.1371/journal.pgen.1005237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto T, Kitano J. 2016. The intricate relationship between sexually antagonistic selection and the evolution of sex chromosome fusions. J. Theoret. Biol. 404, 97–108. ( 10.1016/j.jtbi.2016.05.036) [DOI] [PubMed] [Google Scholar]
- 15.Charlesworth B, Sniegowski P, Stephan W. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220. [DOI] [PubMed] [Google Scholar]
- 16.Bachtrog D. 2003. Accumulation of Spock and Worf, two novel non-LTR retrotransposons, on the neo-Y chromosome of Drosophila miranda. Mol. Bio. Evol. 20, 173–181. ( 10.1093/molbev/msg035) [DOI] [PubMed] [Google Scholar]
- 17.Bartolomé C, Charlesworth B. 2006. Evolution of amino acid sequences and codon usage on the Drosophila miranda neo-sex chromosomes. Genetics 174, 2033–2044. ( 10.1534/genetics.106.064113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berlin S, Ellegren H. 2006. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J. Mol. Evol. 62, 66–72. ( 10.1007/s00239-005-0067-6) [DOI] [PubMed] [Google Scholar]
- 19.Chibalina M, Filatov D. 2011. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr. Biol. 21, 1475–1479. ( 10.1016/j.cub.2011.07.045) [DOI] [PubMed] [Google Scholar]
- 20.Bergero R, Charlesworth D. 2011. Preservation of the Y transcriptome in a 10MY old plant sex chromosome system. Curr. Biol. 21, 1470–1474. ( 10.1016/j.cub.2011.07.032) [DOI] [PubMed] [Google Scholar]
- 21.Bachtrog D. 2008. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 179, 1513–1525. ( 10.1534/genetics.107.084012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutkowska J, Lagisz M, Nakagawa S. 2012. The long and the short of W chromosomes: no evidence for avian W degeneration. Biol. Lett. 8, 636–638. ( 10.1098/rsbl.2012.0083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tennessen JA, Govindarajulu R, Liston A, Ashman T-L. 2016. Homomorphic ZW chromosomes in a wild strawberry show distinctive recombination heterogeneity but a small sex-determining region. New Phytol. 211, 1202–1208. ( 10.1111/nph.13983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherif E, Zehdi A, Castello K, Chabrillange N, Abdoulkader S, Santoni S, Sahli-Hannachi A, Glemin S, Aberlenc-Bertossi F. 2012. Male specific DNA markers provide genetic evidence of an XY chromosome system and recombination arrest and allow to trace paternal lineages in date palm. New Phytol. 197, 409–415. ( 10.1111/nph.12069) [DOI] [PubMed] [Google Scholar]
- 25.Hough J, Hollister JD, Wang W, Barrett SCH, Otto SP. 2014. Genetic degeneration of old and young Y chromosomes in the flowering plant Rumex hastatulus. Proc. Natl Acad. Sci. USA 111, 7713–7718. ( 10.1073/pnas.1319227111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vicoso B, Kaiser V, Bachtrog D. 2013. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl Acad. Sci. USA 110, 6453–6458. ( 10.1093/gbe/evr010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamble T, Geneva AJ, Glor RE, Zarkower D. 2014. Anolis sex chromosomes are derived from a single ancestral pair. Evolution 68, 1027–1041. ( 10.1111/evo.12328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papadopulos AST, Chester M, Ridout K, Filatov DA. 2015. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proc. Natl Acad. Sci. USA 112, 13 021–13 026. ( 10.1073/pnas.1508454112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultheiß R, Viitaniemi H, Leder E. 2015. Spatial dynamics of evolving dosage compensation in a young sex chromosome system. Genome Bio. Evol. 7, 581–590. ( 10.1093/gbe/evv013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright AE, Harrison PW, Montgomery SH, Pointer MA, Mank JE. 2014. Independent strata formation on the avian sex chromosomes reveals inter-chromosomal gene conversion and predominance of purifying selection on the W chromosome. Evolution 68, 3281–3295. ( 10.1111/evo.12493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl Acad. Sci. USA 103, 18 190–18 195. ( 10.1073/pnas.0605274103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin W, et al. 2016. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat. Commun. 7, 13107 ( 10.1038/ncomms13107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, et al. 2012. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc. Natl Acad. Sci. USA 109, 13 710–13 715. ( 10.1073/pnas.1207833109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White M, Kitano J, Peichel C. 2015. Purifying selection maintains dosage-sensitive genes during degeneration of the threespine stickleback Y chromosome. Mol. Bio. Evol. 32, 1981–1995. ( 10.1093/molbev/msv078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pigozzi M. 2011. Diverse stages of sex-chromosome differentiation in tinamid birds: evidence from crossover analysis in Eudromia elegans and Crypturellus tataupa. Genetica 139, 771–777. ( 10.1007/s10709-011-9581-1) [DOI] [PubMed] [Google Scholar]
- 36.Charlesworth B, Charlesworth D. 1978. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112, 975–997. ( 10.1086/283342) [DOI] [Google Scholar]
- 37.Ashman T-L, Tennessen JA, Dalton RM, Govindarajulu R, Koski MH, Liston A. 2015. Multilocus sex determination revealed in two populations of gynodioecious wild strawberry, Fragaria vesca subsp. bracteata. G3 5, 2759–2773. ( 10.1534/g3.115.023358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohnishi O. 1985. Population genetics of cultivated buckwheat, Fagopyrum esculentum Moench. III. Frequency of sterility mutants in Japanese populations. Jap. J. Genet. 60, 391–404. ( 10.1266/jjg.60.391) [DOI] [Google Scholar]
- 39.Timofejeva L, Skibbe D, Lee S, Golubovskaya I, Wang R, Harper L, Walbot V, Cande W. 2013. Cytological characterization and allelism testing of anther developmental mutants identified in a screen of maize male sterile lines. G3 3, 231–249. ( 10.1534/g3.112.004465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakai AK, Weller SG, Culley T, Campbell D, Dunbar-Wallis A, Andres A. 2008. Sexual dimorphism and the genetic potential for evolution of sex allocation in the gynodioecious plant, Schiedea salicaria. J. Evol. Biol. 21, 1–17. [DOI] [PubMed] [Google Scholar]
- 41.Kohn JR. 1989. Sex ratio, seed production, biomass allocation, and the cost of male function in Cucurbita foetidissima HBK (Cucurbitaceae). Evolution 43, 1424–1434. ( 10.1111/j.1558-5646.1989.tb02593.x) [DOI] [PubMed] [Google Scholar]
- 42.Connallon T, Clark AG. 2011. Association between sex-biased gene expression and mutations with sex-specific phenotypic consequences in Drosophila. Genome Bio. Evol. 3, 51–155. ( 10.1093/gbe/evr004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bull JJ. 1983. Evolution of Sex determining mechanisms. Menlo Park, CA: Benjamin/Cummings. [Google Scholar]
- 44.Akagi T, Henry IM, Tao R, Comai L. 2014. A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346, 646–650. ( 10.1126/science.1257225) [DOI] [PubMed] [Google Scholar]
- 45.Spigler RB, Lewers KS, Ashman TL. 2011. Genetic architecture of sexual dimorphism in a subdioecious plant with a proto-sex chromosome. Evolution 65, 1114–1126. ( 10.1111/j.1558-5646.2010.01189.x) [DOI] [PubMed] [Google Scholar]
- 46.Picq S, et al. 2014. A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol. 14, 229 ( 10.1186/s12870-014-0229-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blaser O, Grossen C, Neuenschwander S, Perrin N. 2013. Sex-chromosome turnovers induced by deleterious mutation load. Evolution 67, 635–645. ( 10.1111/j.1558-5646.2012.01810.x) [DOI] [PubMed] [Google Scholar]
- 48.Connallon T, Clark AG. 2014. Evolutionary inevitability of sexual antagonism. Proc. R. Soc. B 281, 20132123 ( 10.1098/rspb.2013.2123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordan C, Charlesworth D. 2012. The potential for sexually antagonistic polymorphism in different genome regions. Evolution 66, 505–516. ( 10.1111/j.1558-5646.2011.01448.x) [DOI] [PubMed] [Google Scholar]
- 50.Charlesworth B, Jordan CY, Charlesworth D. 2014. The evolutionary dynamics of sexually antagonistic mutations in pseudoautosomal regions of sex chromosomes. Evolution 68, 1339–1350. ( 10.1111/evo.12364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng C, Kirkpatrick M. 2016. Sex-specific selection and sex-biased gene expression in humans and flies. PLoS Genet. 12, e1006170 ( 10.1371/journal.pgen.1006170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindholm A, Breden F. 2002. Sex chromosomes and sexual selection in Poeciliid fishes. Am. Nat. 160, S214–S224. ( 10.1086/342898) [DOI] [PubMed] [Google Scholar]
- 53.Tripathi N, Hoffmann M, Willing E, Lanz C, Weigel D, Dreyer C. 2009. Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc. R. Soc. B 276, 2195–2208. ( 10.1098/rspb.2008.1930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon SP, López-Sepulcre A, Reznick DN. 2012. Predation-associated differences in sex-linkage of wild guppy coloration. Evolution 66, 912–918. ( 10.1111/j.1558-5646.2011.01495.x) [DOI] [PubMed] [Google Scholar]
- 55.Künstner A, Hoffmann M, Fraser BA, Kottler VA, Sharma E, Weigel D, Dreyer C. 2017. The genome of the Trinidadian guppy, Poecilia reticulata, and variation in the Guanapo population. PLoS ONE 11, e0169087 ( 10.1371/journal.pone.0169087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haskins C, Haskins EF, McLaughing J, Hewitt RE. 1961. Polymorphisms and population structure in Lebistes reticulatus, an ecological study. In Vertebrate speciation (ed. Blair WF.), pp. 320–395. Austin, TX: University of Texas Press. [Google Scholar]
- 57.Nanda I, Schories S, Tripathi N, Dreyer C, Haaf T, Schmid M, Schartl M. 2014. Sex chromosome polymorphism in guppies. Chromosoma 123, 373–383. ( 10.1007/s00412-014-0455-z) [DOI] [PubMed] [Google Scholar]
- 58.Lisachov A, Zadesenets K, Rubtsov N, Borodin P. 2015. Sex chromosome synapsis and recombination in male guppies. Zebrafish 12, 174–180. ( 10.1089/zeb.2014.1000) [DOI] [PubMed] [Google Scholar]
- 59.Wright A, et al. 2017. Convergent recombination suppression suggests a role of sexual conflict in guppy sex chromosome formation. Nat. Commun. 8, 14251 ( 10.1038/ncomms14251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coyne J, Kay E, Pruett-Jones S. 2008. The genetic basis of sexual dimorphism in birds. Evolution 62, 214–219. ( 10.1111/j.1558-5646.2007.00254.x) [DOI] [PubMed] [Google Scholar]
- 61.Roesti M, Moser D, Berner D. 2013. Recombination in the threespine stickleback genome-patterns and consequences. Mol. Ecol. 22, 3014–3027. ( 10.1111/mec.12322) [DOI] [PubMed] [Google Scholar]
- 62.Bergero R, Charlesworth D. 2009. The evolution of restricted recombination in sex chromosomes. Tr. Ecol. Evol. 24, 94–102. ( 10.1016/j.tree.2008.09.010) [DOI] [PubMed] [Google Scholar]
- 63.Swanson CP. 1957. Cytology and cytogenetics. New York, NY: Prentice Hall. [Google Scholar]
- 64.van Doorn G, Kirkpatrick M. 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449, 909–912. ( 10.1038/nature06178) [DOI] [PubMed] [Google Scholar]
- 65.Villena P-MFD, Sapienza C. 2001. Female meiosis drives karyotypic evolution in mammals. Genetics 159, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. 2014. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 24, 2295–2300. ( 10.1016/j.cub.2014.08.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshida K, Kitano J. 2012. The contribution of female meiotic drive to the evolution of neo-sex chromosomes. Evolution 66, 3198–3208. ( 10.1111/j.1558-5646.2012.01681.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maddison WP, Leduc-Robert G. 2013. Multiple origins of sex chromosome fusions correlated with chiasma localization in Habronattus jumping spiders (Araneae: Salticidae). Evolution 67, 2258–2272. ( 10.1111/evo.12109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waters PD, Duffy B, Frost CJ, Delbridge ML, Graves JAM. 2001. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet. Cell Genet. 92, 74–79. ( 10.1159/000056872) [DOI] [PubMed] [Google Scholar]
- 70.Reed K, Phillips R. 1997. Polymorphism of the nucleolus organizer region (NOR) on the putative sex chromosomes of Arctic char (Salvelinus alpinus) is not sex related. Chromosome Res. 5, 221–227. ( 10.1023/A:1018411417816) [DOI] [PubMed] [Google Scholar]
- 71.Hood ME, Antonovics J. 2000. Intra-tetrad mating, heterozygosity, and the maintenance of heterozygosity in Microbotryum violaceum (=Ustilago violacea). Heredity 85, 231–241. [DOI] [PubMed] [Google Scholar]
- 72.Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2, 2243–2255. ( 10.1371/journal.pbio.0020384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Badouin H, et al. 2015. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae. Genetics 200, 1275–1284. ( 10.1534/genetics.115.177709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellison C, et al. 2011. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics 189, 55–69. ( 10.1534/genetics.111.130690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacobson DJ. 2005. Blocked recombination along the mating-type chromosomes of Neurospora tetrasperma involves both structural heterozygosity and autosomal genes. Genetics 171, 839–843. ( 10.1534/genetics.105.044040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, Mullen SP, Kronforst MR. 2014. doublesex is a mimicry supergene. Nature 507, 229–232. ( 10.1038/nature13112) [DOI] [PubMed] [Google Scholar]
- 77.Huu C, et al. 2016. Presence versus absence of CYP734A50 underlies the style-length dimorphism in primroses. eLife 5, e17956 ( 10.7554/eLife.17956) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J, et al. 2016. Genetic architecture and evolution of the S locus supergene in Primula vulgaris. Nat. Plants 2, 16188 ( 10.1038/nplants.2016.188) [DOI] [PubMed] [Google Scholar]
- 79.Charlesworth D. 2016. The status of supergenes in the 21st century: recombination suppression in Batesian mimicry and sex chromosomes and other complex adaptations. J. Evol. Appl. 9, 74–90. ( 10.1111/eva.12291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tuttle EM, et al. 2016. Divergence and functional degradation of a sex chromosome-like supergene. Curr. Biol. 26, 344–350. ( 10.1016/j.cub.2015.11.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Küpper C, et al. 2015. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48, 79–83. ( 10.1038/ng.3443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamichhaney S, et al. 2015. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat. Genet. 48, 84–88. ( 10.1038/ng.3430) [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Wurm Y, Nipitwattanaphon M, Riba-Grognuz O, Huang Y, Shoemaker D, Keller L. 2013. A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493, 664–668. ( 10.1038/nature11832) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.