Abstract
In most sexual, diploid eukaryotes, at least one crossover occurs between each pair of homologous chromosomes during meiosis, presumably in order to ensure proper segregation. Well-known exceptions to this rule are species in which one sex does not recombine and specific chromosomes lacking crossover. We review other possible exceptions, including species with chromosome maps of less than 50 cM in one or both sexes. We discuss the idea that low recombination rates may favour sex–asex transitions, or, alternatively may be a consequence of it. We then show that a yet undescribed species of brine shrimp Artemia from Kazakhstan (A. sp. Kazakhstan), the closest known relative of the asexual Artemia parthenogenetica, has one of the shortest genetic linkage maps known. Based on a family of 42 individuals and 589 RAD markers, we find that many linkage groups are considerably shorter than 50 cM, suggesting either no obligate crossover or crossovers concentrated at terminal positions with little effect on recombination. We contrast these findings with the published map of the more distantly related sexual congener, A. franciscana, and conclude that the study of recombination in non-model systems is important to understand the evolutionary causes and consequences of recombination.
This article is part of the themed issue ‘Evolutionary causes and consequences of recombination rate variation in sexual organisms’.
Keywords: Artemia sp. (Kazakhstan), contagious asexuality, genetic map, obligate crossover, recombination rate meiosis
1. Introduction
Genetic linkage maps are invaluable tools for investigating genome structure and for quantitative trait loci (QTL) mapping. They are based on frequencies of recombinant maker genotypes, typically assessed in families or crosses. Genetic map distances directly reflect the number of crossovers per meiosis (with 50 cM corresponding to one crossover, ‘CO’, per meiosis), although this assumes that the survival of gametes and offspring does not differ between recombinant and non-recombinant genotypes. Next-generation sequencing techniques, such as RAD sequencing have rendered the construction of high-density maps relatively straightforward also in non-model species. As a consequence, many such maps have been published in recent years. In most diploid, sexual species, chromosomes have a minimum length of 50 cM, suggesting that at least one CO occurs per bivalent during each meiosis [1–4]. Indeed, such an ‘obligate’ CO may be required for proper segregation of homologues during the first meiotic division. COs establish physical connections between homologous chromosomes, which may provide the tension needed for the bipolar spindle to establish [5–7]. Evidence for the necessity of at least one CO per bivalent comes from studies indicating that bivalents without COs have an increased probability of non-disjunction, and often result in inviable or unfit aneuploid offspring [7,8]. Other indirect evidence comes from pseudo-autosomal regions of sex chromosomes. In many species, recombination between sex chromosomes is largely suppressed in the heterogametic sex (between X and Y or between W and Z). Yet in most cases, there is a pseudo-autosomal region, which can physically be very small, but still typically has one CO per meiosis [9,10].
Yet the obligate CO is not universal. In several species across a wide range of taxa, one sex does not undergo CO [11–13]. These species have alternative mechanisms to ensure proper disjunction of achiasmate bivalents [14,15]. Similar alternative mechanisms have also been identified in species that do usually have CO [16,17], where they may serve as back-up mechanisms. Furthermore, in some species, specific chromosomes do not undergo CO. Examples are the dot chromosome of Drosophila [18] and the absence of CO between X and Y in marsupials [19]. Again, alternative mechanisms ensure their proper segregation [14,17,20,21].
These examples indicate that it is not always obligatory to have at least one CO per bivalent and meiosis, which suggests that COs are maintained for reasons other than to ensure proper disjunction. Conversely, this also suggests that selection for low recombination is not necessarily constrained by a need of COs for proper segregation. Indeed, it has been suggested that the evolution of achiasmy may be the result of selection for reduced rates of recombination [11], which may occur for a wide variety of reasons (e.g. [22]).
Particularly strong selection for reduced recombination rates may occur during evolutionary transitions from sexual to asexual reproduction. Many asexual organisms produce diploid eggs that can develop without fertilization (‘parthenogenesis’ in the animal literature). They do so by either suppressing or by modifying meiosis. Under most forms of asexual reproduction, recombination leads to a loss of heterozygosity, with negative fitness consequences similar to inbreeding depression [2,23–30]. Recombination is thought to be rare under some forms of asexuality, especially under mitotic clonality (‘apomixis’ in the animal literature), where offspring are genetically identical copies of their mothers, barring mutation and rare mitotic recombination (e.g. [31]). Other forms of asexuality may more commonly involve recombination. An example is automixis, which involves the fusion of two haploid products of a single meiosis, such as fusion of two spores of a single tetrad (also called within-tetrad mating) or fusion between an egg cell and one of its polar bodies [32–34]. If this fusion occurs between products that have separated during the first meiotic division, one speaks of ‘central fusion’. This maintains maternal heterozygosity around the centromeres, but loss of heterozygosity may occur when there is recombination between a locus and the centromere.
Suppression of meiosis I, which occurs in some groups, has the same genetic effects as central fusion because homologues are simply never separated [26,35]. An example is abortive, non-reductive meiosis in Daphnia, where bivalents are started to be pulled apart in meiosis I, but the division never follows through. Rather, the half-bivalents move back to the equatorial plate, and sister chromatids are subsequently pulled apart as in normal meiosis II [36]. Ploidy is maintained during this process because one of the two meiotic division is aborted. We will group these cases under the name ‘central fusion’ for simplicity, even if, biologically, they do not include a fusion of cells.
Central fusion automixis is a particularly interesting form of asexual reproduction because it maintains maternal heterozygosity as long as there is no recombination. In fact, if recombination is fully suppressed, central fusion automixis is genetically indistinguishable from mitotic asexuality. Cytological studies have shown that this situation occurs in some species that have formerly been thought to reproduce by mitotic asexuality (e.g. [36]). Finally, some species that regularly reproduce by central fusion automixis do so with low to very low levels of recombination, thus maintaining maternal heterozygosity to a large degree [29,34,37–39], although strong selection against recombinants, such as found during automictic reproduction in the Cape honeybee [40–42], may also contribute to low observed recombination rates in some of these cases.
The fact that at least some asexuals have low recombination rates, much lower than typically seen in sexual species, has several puzzling implications. First, it is possible that recombination rate rapidly evolves to much reduced levels following the transition to asexuality. However, this poses a problem: Given that many asexual organisms, and automicts in particular, still segregate chromosomes during meiosis I, how could recombination be rapidly reduced if one obligate CO is required for proper disjunction? Even if many extant asexual organisms probably do not segregate homologues during meiosis I (e.g. suppressed or aborted meiosis I), which may reduce this constraint, normal segregation probably still occurred in many cases immediately following the transition [2,25,26]. Second, it has been suggested that very low rates of recombination in certain sexual species might function as a pre-adaptation to asexuality (i.e. make a successful transition to asexuality more likely [43]).
To understand these transitions to asexuality it seems essential to understand how universal and strong the constraint of the obligate CO really is. Many high-density linkage maps for a large variety of organisms have been published in recent years. Indeed, large numbers of genetic markers and cost-effective genotyping can be obtained with next-generation sequencing (NGS) even without the need of a reference genome. Here we review these studies, looking for evidence for exceptions from the obligate CO constraint. We restrict this analysis to genetic maps based on moderate to high numbers of markers (at least a few hundreds of markers per map) and to maps showing evidence of linkage groups that span less than 50 cM in chromosome-wide genetic length. We note, however, that a genetic map length of less than 50 cM does not necessarily imply the absence of an obligate CO, as terminal COs may be missed; evidence for this will also be discussed. Second, we present new data on a linkage map of a yet undescribed Artemia species from Kazakhstan (A. sp. Kazakhstan; [44]). This species is fully bisexual, and is the closest extant relative of the diploid Artemia parthenogenetica [45]. The latter represents a group of diploid and polyploid parthenogenetic Artemia. Polyploids reproduce clonally, but diploids are automicts: they combine central fusion and low levels of recombination [29]. We compare this new genetic map with the published map of a more distantly related, sexual species of the same genus [46], assessing whether reduced recombination probably has evolved before or after the separation of A. parthenogenetica. More generally, we assess the role of the obligate CO constraint for the evolution of low recombination in sexual species and during transitions to asexuality.
2. Review: low levels of recombination in sexual species
Above, we have briefly discussed two well-known exceptions from the requirement of an obligate CO: achiasmy in one of the two sexes and achiasmy of specific chromosomes. These have been discussed elsewhere in detail [11–13], and will not be reviewed further here. Besides this, some plants with holocentric chromosomes and inverted meiosis (separation of sister chromatids during meiosis I, followed by separation of homologues during meiosis II) appear to be achiasmate in both sexes [47]. Inverted meiosis is possibly an adaptation to holocentric chromosome structure [48,49], and alternative correct segregation may be achieved in different ways, although some species with inverted meiosis do have COs [47,50].
(a). Low chromosome numbers
Although not directly related to the obligate CO issue, the number of chromosomes itself is a strong determinant of genome-wide recombination: owing to independent segregation of chromosomes during meiosis, genes on different chromosomes assort independently into gametes, which is equivalent to free recombination between physically linked genes that are separated by at least 50 cM [51–53]. There is at least one animal species with just a single pair of chromosomes, the ant Myrmecia pilosula [54]. The number of COs during female meiosis is unknown, but this is almost certainly one of the species with the lowest genome-wide recombination level, especially given haplodiploidy and hence absence of recombination in males. A few animal and plant species are known to have just two pairs of chromosomes [55–57]. Very variable chromosome size may have similar effects, if a large majority of genes is found on just a single or a few chromosomes.
(b). Exactly one crossover per bivalent
Several species appear to have exactly one CO per bivalent, that is, only chromosomes with a genetic length of 50 cM. The most well-known example is Caenorhabditis elegans [58], but other potential examples exist, for instance the midge Clunio marinus [59] and the fish Hypoplectrus nigricans [60]. The C. elegans example is particularly interesting as it shows low recombination rates in chromosomal centres, but these are not associated with the presence of centromeres. Furthermore, regions with high gene densities correspond to low-recombination areas, and the single CO mostly occurs at chromosome ends, where it has relatively little effect on the recombination among genes on the same chromosome [61,62]. It might be that there is a general tendency for species with short genetic maps to have a particular concentration of COs at the chromosome ends.
A further example comes from a species combining a single CO with achiasmy. The planarian Mesostoma ehrenbergii has achiasmate meiosis in females [63]. During male meiosis three bivalents are formed, which have exactly one CO near the chromosome ends. However, the remaining two chromosome pairs are achiasmate in both male and female meiosis and are separated as univalents, similar to the dot chromosome in Drosophila [63–65].
(c). Low crossover numbers in a few chromosomes
In the large majority of species, the number of COs per chromosome varies with the physical size of the chromosomes (more COs on longer chromosomes), but with at least one CO even on the smallest ones [3,4]. Some chromosomes have a genetic map length of slightly less than 50 cM, but in many cases, this is most probably explained by sampling error, insufficient marker coverage, incomplete assemblies (for maps, in which single-nucleotide polymorphism (SNP) calls rely on alignment to a reference genome) and other sources of errors. However, some high-density maps do show a few linkage groups that are substantially below 50 cM. Examples are some linkage groups in the dog, particularly in males, in which the shortest linkage group is 27 cM [66,67], and in the Zebra Finch, particularly linkage group 4, in both sexes of [68–70]. Interestingly, both are also examples of species with generally shorter linkage maps compared to related species [4,71,72], though both have most chromosomes greater than 50 cM. Furthermore, both species have COs strongly concentrated towards the ends of chromosomes, more so than related species with longer maps [68,73]. A similar example may be the bluefin tuna, although this was analysed with a moderate number of markers [74]. Also the brown algae Undaria pinnatifida and Saccharina japonica have two to three linkage groups (of a total of 30–31) that are considerably less than 50 cM long, with the remaining linkage groups being rather compact [75,76]. A slightly different example is the European tree frog, which has strongly reduced recombination in chromosome centres only in males, and one linkage group with considerably less than 50 cM in males [77,78]. Avian ‘micro-chromosomes’ may also have less than 50 cM, though in this case it is more likely that the current data are explained by insufficient numbers of markers: These chromosomes show much higher rates of recombination (in cM/Mb) between the few existing markers than other chromosomes, an observation that is usually interpreted as evidence for the need of an obligate CO [3,72].
(d). Low crossover number in most chromosomes
Finally, very few species were found to have genetic maps with several or even the majority of linkage groups being less than 50 cM in total length. In some cases this is explained by a combination of a relatively compact map in one sex (but still with at least one CO per chromosome) and achiasmy in the other sex, leading to a sex-averaged map with chromosomes of at least 25 cM [79]. In some other cases, however, achiasmy in one sex does not seem to be the explanation. Two species of killifish, have genetic maps with 23 and 24 chromosomes, respectively, each with a total length of less than 50 cM [80]. Although the longest linkage groups are just slightly less than 50 cM, the shortest ones are considerably less than 25 cM, even less than 10 cM in one species. The maps are based on a moderate number of markers, but despite this, it is clear that the genetic map length of these chromosomes is exceptionally short.
Another species with an exceptionally compact genetic map is the fungus Agaricus bisporus. Based on a moderate number of markers, its linkage groups range between 4 and 58 cM [81]. Yet, whole-genome resequencing revealed results entirely consistent with an obligate CO. The reason why these COs were not detected was that the large majority of them occurred in the terminal 100 kb of the chromosomes [81].
Several species of the plant genus Oenothera do not form bivalents but multivalents, with chromosomes attached to each other at the ends. Separation is such that recombination and segregation are suppressed, except at chromosome ends. Hence, except for the recombined chromosome ends, meiosis results in parental haplotypes [43,82,83]. Similar systems occur in a few other plant and animal species [84–90]. Besides the functionally clonal Oneothera species, the genus also has bivalent-forming species, which reproduce by normal meiosis. Interestingly, their linkage groups (assessed with a moderate number of markers) have been found to be very short, between 2 and 20 cM, with recombination almost entirely restricted to chromosome ends [43].
3. Genetic map of Artemia sp. Kazakhstan
(a). Material and methods
(i). The mapping population
The mapping population was produced using a pseudo-testcross design [91], which involves a cross between two heterozygous parents and their F1 offspring. From such a cross, the male recombination map can be established using markers that are heterozygous only in the father, the female map using markers that are heterozygous only in the female, and the two maps can be linked using markers that are heterozygous in both parents [92].
Following [93], cysts were hatched by incubation in saline water (salinity 5 g l−1), and once hatched, the larvae (nauplii) were progressively transferred to higher salinity (100 g l−1) and fed every two days with a 50 : 50 mixture of marine algae and yeast. Throughout, the animals were kept at 20°C ± 1°C with a 12 L : 12 D cycle. Larvae were first cultured together, but were separated individually into 50 ml beakers at the last larval stage, in order to assure that the animals chosen for the crosses were unmated. Once adult, their sex was visually determined (based on the conspicuous claspers of adult males) and 36 pairs were placed in 50 ml beakers (one pair each) in order to start families. Adults were sampled once a substantial number of live-born larvae had been produced, and the heterozygosity of all adults was determined using a panel of 12 microsatellite markers [94,95]. The larvae were raised to adulthood so that their sex could be visually determined (sexual dimorphism is only visible at the adult stage). One family with two highly heterozygous parents (6 and 4 loci for the female and male, respectively) and a sufficient number of offspring was selected for genetic mapping. The two parents of that family and 40 of their F1 offspring (20 males and 20 females) were analysed using RAD sequencing.
(ii). DNA extraction
To minimize contamination of samples with non-specific DNA, live individuals were washed for 20 min in a 10% sodium hypochlorite solution, followed by 10 min in sterile salt water prior to sampling. Furthermore, following sampling, the animals were dissected, and their digestive tracts were discarded. Genomic DNA was extracted from the remaining tissue using the Qiagen ‘DNEasy Blood & Tissue’ kit with some modifications: before extraction, each sample was added to a vial containing glass beads, 180 µl of ATL buffer, and 20 µl of Proteinase K. Subsequently, each vial was vortexed vigorously for 15 s and then incubated for 8 h at 56°C on a 700 rpm shaker. After proteinase K digestion, extraction proceeded using the instructions provided with the kit.
(iii). RAD sequencing
We used the RAD-sequencing protocol developed by [96] with a few modifications. Genomic DNA was digested with SbfI (New England Biolabs), barcoded with individual-specific P1 adapters, and pooled to create a single library containing 2100 ng DNA. Each parent was added as two independent samples (i.e. with two different P1 adaptors), and hence the library contained a total of 44 samples. After pooling, the library was randomly sheared on a Bioruptor using six cycles (30 s ON, 1 min OFF per cycle), and fragments between 200 and 500 bp were selected using agarose gel electrophoresis. The fragments were then blunted, and a P2 adapter was ligated. The library was amplified by polymerase chain reaction (PCR) (30 s at 98°C, followed by 18 cycles of 10 s at 98°C, 30 s at 65°C and 30 s at 72°C; a final elongation step was performed at 72°C for 5 min.). A final electrophoresis was performed to purify fragments, and the library was sequenced on a single lane of an Illumina HiSeq 2000, using single-end 100 cycle sequencing by a commercial company (GenoScreen, Lille, France).
(iv). Filtering, single-nucleotide polymorphism calling and genotype calling
The quality of the raw sequencing reads (library-wide and per-base) was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads with ambiguous barcodes and/or absent SbfI cut sites were removed, as were low-quality reads (average read quality < 10, assessed using a 15 bp sliding window). The retained reads (96% of total) were sorted according to the barcode and the barcode sequences, as well as the restriction site, were trimmed (leaving 89 bp of experimental sequence per read). Two progeny were removed because of considerably lower sequence depth. These procedures resulted in 65.5 million reads (mean: 1.6 million reads, standard deviation: 0.6 million reads per sample).
For SNP calling, we used the de novo pipeline of the program ‘Stacks’ (v. 1.20; [97]). For each individual, the ustacks procedure was used to identify identical reads and to group them together under a single ‘RAD locus’ if their alignment included two or fewer differences (Stacks options M = 2, m = 3, N = 2, n = 2). A catalogue of RAD loci and alleles was generated using data of the two parents, and progeny data were compared with this catalogue, determining which progeny inherited which parental alleles. These procedures resulted in a total of 13′809 RAD loci, sequenced at an average depth of 111× per locus and individual (without counting individuals with missing genotypes). However, a large number of these loci were either monomorphic or the two parents were homozygous for different alleles and all offspring heterozygous. The script ‘genotypes’ within stacks was used to remove these non-informative markers and to organize loci into a set of mappable markers. During further filtering, we discarded individual genotypes based on fewer than 20 reads and then removed markers with missing genotype information for more than seven offspring (out of 38). Furthermore, loci for which segregation patterns among offspring did not match the parental genotypes were removed (typically loci that were monomorphic among offspring despite being called heterozygous in one of the parents; note that alleles not present in the parents were not retained during the above genotype-calling procedure). There was a 7% genotype-calling mismatch between the two replicates of the father. These loci were only retained for further analysis when the correct genotype of the father could unambiguously be identified, based on the genotypes of the offspring. One of the replicates of the mothers was sequenced at much lower depth than the other samples (14% genotype-calling mismatch, mostly due to false homozygote calls in the replicate sequenced at lower depth). Hence the replicate sequenced at higher depth was used for all analyses. Markers were also tested for distorted segregation ratios (using χ2 tests). Segregation distortion was observed in 10.4% of male-informative and in 8.8% of female-informative markers (individual p-values < 0.05, testing for deviations from Mendelian 1 : 1 segregation ratios). However, in no case was distortion very strong (using sequential Bonferroni correction for multiple testing, not a single marker remained significantly distorted), and, according to current practice, distorted markers were kept in the maps. Altogether, these procedures resulted in a total of 733 informative markers, of which 333 female-informative markers were used to construct the female linkage map (genotype ab in the mother and aa or bb in the father). The male map was based on 315 male-informative markers, and the 84 biparental markers (genotype ab in both parents) were used to integrate the male and female maps.
(v). Linkage analysis and map construction
The linkage analysis and map construction was carried out using the R/qtl package [98]. First, to assign markers to linkage groups (done separately for male and female maps), pairwise recombination frequencies between all markers and the corresponding LOD scores were estimated using the function est.rf(). LOD scores correspond to the logarithm of the likelihood that the two markers are linked with a recombination fraction of <50% divided by the likelihood that they are unlinked. The maximum pairwise recombination frequency allowed between adjacent markers was 0.4 and the minimum LOD score was 4.0. Note that the relatively high maximum recombination frequency was chosen in order for linkage to be determined by LOD scores. The actually achieved maximum recombination frequency between adjacent markers with minimum LOD = 4 was 0.2. The female-specific and male-specific markers (type ab × aa) were analysed with the back-cross procedure, which infers phase automatically. The phase of the bi-parental markers was inferred from a correlation analysis of each of the bi-parental markers against the whole set of sex-specific markers.
We then used the order.markers() function to determine the most likely order of the markers along each linkage group. To improve the order of the markers, the ripple() function was used with a window size of 7 markers. Genetic distances were estimated using the Kosambi mapping function. For the final maps, we manually removed individual genotype calls suggesting a double recombination event (one immediately before and one immediately after a given marker). This occurred only in 1% of all cases and is probably explained by errors in genotype calling. Hence, these genotype calls were replaced by missing values. Finally, we estimated the length of the unmapped chromosome ends, using the mean value of the two methods described in [99]. These correction methods make many simplifying assumptions and should thus be interpreted with care.
(b). Results
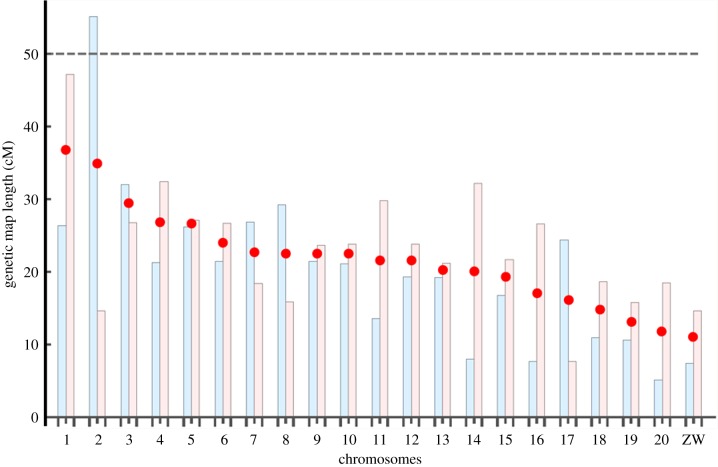
The female linkage map (electronic supplementary material, figure S1 and table S2) is based on 333 female-informative markers of which 273 grouped together in 21 linkage groups, which fits the karyotype of Artemia [100]. The remaining markers could not be assigned to any linkage group nor grouped together (based on a minimal LOD score of 4), and were, therefore, left unassigned. The number of markers per linkage group ranged from 4 to 22 (average: 12.8) with the size of linkage groups ranging from 4.9 cM to 47 cM (average: 23.12, figure 1). The total length of the 21 linkage groups is 478.6 cM with a median inter-marker distance of 2.7 cM (electronic supplementary material, table S2).
Figure 1.
Genetic map length of linkage groups in A. sp. Kazakhstan (in centimorgan, cM) as estimated from male (blue) and female (pink) meiosis. The red dot indicates the sex-average length. The dashed line corresponds to 50 cM (the ‘obligate CO’ baseline expectation). Linkage groups are ordered and numbered by their average map length. The shortest 21st linkage group corresponds to the ZW pair. Note that linkage group 2 of females is homologous either to linkage group 2 of males or to linkage group 18 (and vice versa).
The male linkage map (electronic supplementary material, figure S3 and table S2) is based on 315 male-informative markers, of which 268 grouped together in 21 linkage groups. The number of markers per linkage group varied from 5 to 23 (average: 12.8) with the length of linkage groups ranging from 5.1 to 55 cM (average: 20.1). The total size of the 21 linkage groups was 423.9 cM and the median distance between markers was 2.8 cM.
Of the 84 biparental markers, 44 could be associated to one of the linkage groups and allowed us to link male and female maps (electronic supplementary material, figure S4), except for linkage groups 2 and 18 in both sexes, for which the pairwise homology thus remains unknown (LG2 may be homologous to LG2 or LG 18 of the other sex). Integrating these biparental markers into the sex-specific linkage groups does, in some cases, increase map lengths of the linkage groups (electronic supplementary material, figure S4). However, the overall picture remains unchanged, with average map lengths still being much below 50 cM in both sexes. In addition, as these markers are informative only in some offspring, (in heterozygotes it is unknown which allele came from the father and which one from the mother), we could not apply the same correction techniques as in the sex-specific markers. Hence map length may be increased by genotyping error, and, therefore, figure 1 as well as electronic supplementary material, figures S1, S3 and table S2 are based on the sex-specific maps without the biparental markers. No cross-linkage was found between linkage groups, except for weak ‘pseudo-linkage’ [101] between linkage group 3 and the sex chromosome in females (electronic supplementary material, figure S5), which remains unexplained.
Eighteen of the female-informative markers but none of the male-informative markers were strongly linked with phenotypic sex, confirming that females are the heterogametic sex [46]. One bi-parental marker mapped to the same linkage group and thus allowed identification of the Z-linkage group in the male map. Both male and female maps of the sex chromosomes are short, but non-zero (figure 1). Non-zero recombination between W and Z was also found in A. franciscana [46].
When corrections for chromosome ends without marker coverage were employed (see Material and Methods), the total corrected female map length was estimated at 564 cM and the male map length at 478 cM. Considering that the estimated haploid genome size of A. sp. Kazakhstan is 2.4 Gbp [102], the recombination rate corresponds to 0.23 cM/Mbp in females and to 0.20 cM/Mbp in males, on average.
4. Discussion
Despite the moderate number of markers employed, the map of A. sp. Kazakhstan is clearly an exceptionally short map for a fully sexual species. With an average genetic map length of just over 20 cM per linkage group in both sexes (and just one linkage group in each sex being greater than 40 cM, figure 1), A. sp. Kazakhstan is similar to the few known species with exceptionally short linkage maps discussed under point 2.4 above. By contrast, the map of the congener, A. franciscana, is considerably longer [46]. Even though it is based on a smaller number of markers, the linkage map of A. franciscana has average map lengths per linkage group of 52 cM in males and 60 cM in females [46] and is thus more than twice as long as the map of A. sp Kazakhstan. This map length might be inflated because the methodology used is typically associated with high genotyping errors. However, this difference in methodology is unlikely to explain the large difference between the two maps. First, the A. franciscana map length is typical of many sexual species. Second, many genotyping errors would be required to explain a two-fold difference. This is contradicted by the observation that the male and female maps in A. franciscana show full marker collinearity for bi-parental markers and good correlation of homologous inter-marker distances [46]. Note that A. franciscana also has some linkage groups that are considerably shorter than 50 cM, but this may well be explained by insufficient numbers of markers on these linkage groups [46].
It is possible that in our A. sp. Kazakhstan map, some of the short linkage groups are explained by insufficient numbers of markers. In addition, a substantial number of markers did not show significant linkage to any of the 21 linkage groups nor among them. Some of these markers may represent missing chromosome ends. However, this group may also contain markers that are not truly segregating, for instance paralogs that were lumped together during the stacks analysis (they may appear to be segregating for instance if lumping only occurred in a fraction of individuals). Also genotype-calling errors may have contributed both to the inability to map these markers, and also to some degree of uncertainty about map length. Erroneously called genotypes often resemble recombinants and have the tendency to upwardly bias map lengths. We removed genotypes that suggested a double recombination event (one immediately before and one immediately after the marker), but this procedure cannot be used for unmapped markers or for terminal markers. Removal of these genotypes may have led to the removal of some true double recombinants, especially if there was some degree of negative interference [103]. However, only 1% of genotypes were removed, and a possible inflation of map length due to remaining errors in genotype calling is conservative with respect to the main conclusion that the map is exceptionally short (i.e. it would be even shorter without those errors). Hence it seems unlikely that the very short map of A. sp. Kazakhstan can be explained by technical issues.
For similar technical issues, the ‘obligate CO’ rule is often difficult to evaluate with precision in most other available maps. There are many errors and filtering biases in NGS-based maps that can lead to upwards or downwards biases in map length [104,105], and in most cases it is impossible to judge this from the published data, especially as most of these studies were not conducted with the aim of precisely estimating map length. Apart from these technical issues, a biological difficulty is that in some species most COs occur at chromosome ends. With genetic mapping, these terminal COs can be easily missed. This is best exemplified by the Agaricus map [81] discussed above, which has chromosomes of similar map length as A. sp. Kazakhstan. However, whole-genome resequencing showed that an obligate CO occurs on all or almost all chromosomes, but is located in the large majority of cases in the terminal approximately 100 kb portion of chromosomes [81]. The occurrence of an obligate CO at such terminal positions cannot be safely excluded, neither in A. sp. Kazakhstan, in any of the other maps discussed above, nor in automictic species with central fusion (e.g. [40]). It can be excluded in some of the achiasmate examples, where the absence of CO was confirmed using cytological methods. Such terminally positioned COs also largely invalidate correction methods for unmapped chromosome ends, such as the two methods employed here (see above).
It is thus clear that the constraint of an obligate CO cannot be assessed with genetic maps alone and that cytological and other alternative methods are needed to reach a final answer. However, COs near the tips of the chromosomes have very little influence on the levels of recombination within the chromosomes and even on genome-wide recombination rate. Their only effect with respect to recombination is that chromosome tips recombine relatively freely with the rest of the genome. Hence, the recombination levels of a large majority of the genome, probably including also a large majority of genes, are still well characterized by maps based on a moderate-to-large number of markers. Conversely, this means that the total map length of a genome (even if 50 cM is added per chromosome pair to account for between-chromosome recombination) is not always a good measure of genome-wide recombination [106]. From an evolutionary point of view, a better measure of average genome-wide recombination would account for the average likelihood that a CO occurs between two randomly chosen genes. Such a measure could be derived from current maps from the distribution of recombination frequencies between pairs of markers (either making assumptions on uniform gene densities along chromosomes if this distribution is unknown, or restricting the analysis to markers within genes). If markers on different chromosomes as well as markers on the same chromosome were included, this would allow combining the effects of random assortment with those of recombination within chromosomes. It seems likely that such a measure could be relatively easily derived from the mean of all pairwise recombination frequencies, though its statistical properties would need to be studied in more detail.
Selection for reduced recombination may often result in a more terminal placement of COs rather than a reduction of chromosome map length below 50 cM. Consistent with this, more terminal placement of COs has been found in many of the examples of species with low recombination rates (either of single chromosomes or genome-wide) discussed above. In species without a reference genome, a terminal placement of COs is more difficult to assess. However, we do note that the A. sp. Kazakhstan map does contain on almost every chromosome one or a few groups of markers without recombination, flanked by a low number of markers with high genetic distances. This appears to be much less pronounced in the A. franciscana map [46]. A more terminal placement of COs might also play a role in the evolution of sex differences in recombination rates and recombination landscapes [107].
Another striking observation from the above literature review and from the two Artemia maps is that several of the species with particularly short maps are close sexual relatives of asexual species. Besides automictic parthenogenesis in A. parthenogenetica, asexual reproduction is known in killifish [119], and the unusual reproduction of Oenothera involving chromosome rings also leads to largely clonal genome transmission [43]. Even some Agaricus strains are apparently maintained by within-strain mating, which, in these fungi, is equivalent to central fusion automixis [81]. Whether or not low recombination in these cases and asexuality in closely related species are causally related is currently difficult to determine. These observations could be consistent with the hypothesis that a low recombination rate in sexual species may represent a pre-adaptation for a transition to asexuality [43]. In other words, transitions to asexuality might occur more easily in species with already-low recombination rates because, in these cases, the deleterious effects of recombination in asexuals (e.g. loss of heterozygosity) are expected to occur less often than in species with higher rates of recombination. Nonetheless, there are counter-examples: either of sexual species with low recombination rates but without close asexual relatives or of asexual species whose sexual relatives have high recombination rates. First, not all sexual species with low recombination rates have asexual relatives. For instance, no asexual species are known in the genus Tigriopus, and although a number of moths and at least one species of Drosophila do reproduce parthenogenetically, asexuals do not appear to be particularly common among relatives of achiasmate species (note though that substantial amounts of recombination usually occur in the other sex in these species). Second, not all close relatives of asexual species have short linkage maps. For instance, Daphnia species have substantial amounts of recombination during sexual reproduction [99,120], though obligate asexuals exist in the genus [121]. Finally, one of the most striking examples is the honeybee. Sexual honeybees, Apis mellifera, have one of highest numbers of COs per chromosomes known among all organisms [4,122,123]. Nonetheless, Cape honeybees, Apis mellifera capensis, which reproduce by central fusion automixis, have reduced levels of recombination. This is in part explained by strong selection against recombinants [40–42]. Hence, this example shows two important points. First, reduced levels of recombination are likely to have evolved during the transition to asexuality in this sub-species. Second, the strong selection against recombinants is an illustration of the strong costs that recombination may induce in asexuals due to loss of heterozygosity.
A possible alternative explanation is that short genetic maps in sexual relatives of asexual species may be a consequence of the transition to asexuality rather than a pre-adaption. Many asexual species retain some residual capacity of sexual reproduction, and this may even be more common during early stages of sex–asex transitions [2,26]. Residual sexual reproduction occurs in particular in species with so-called contagious asexuality, such as A. sp. Kazakhstan, where parthenogenetically reproducing females rarely produce sons, which are able to fertilize closely related sexual females [124]. In this way, asexuality-determining genes are transmitted to new genetic backgrounds, and new asexual lineages are formed [125–130]. Though details depend on the exact mode of inheritance of asexuality (especially the number of genes involved and the dominance of the asexuality-conferring alleles), a part of the offspring of such back-crosses are sexuals [127]. Hence, males produced by asexual females are also able to transfer parts of genomes with an asexual history back to a sexual background. If there is strong selection for reduced recombination in asexuals, this may lead to repeated introgression of low-recombination alleles into the sexual lineage. Selection against recombination is for instance expected to be strong in central-fusion automicts, as reduced recombination allows preserving heterozygosity and the masking of deleterious mutations [26,128,131]. This may provide an alternative scenario explaining, why sexual species that are closely related to asexuals have low recombination. This scenario is particularly intriguing as gene flow is usually thought to occur exclusively in the other direction, from sexuals to asexuals. Hence low recombination might facilitate transitions to asexuality, but selection for low recombination in asexuals and gene flow back to sexuals via rare sex seems to be a viable alternative hypothesis for the observations that many species with short genetic chromosome maps are closely related to asexual species.
5. Conclusion
The currently available data do not allow for conclusive tests, neither of the obligate CO constraint, nor of the correlation between low levels of recombination and the evolution of asexuality. Our review and the A. sp. Kazakhstan map show that recombination is sometimes very low in chiasmate sexual species; and lower than expected from the ‘obligate CO constraint’. In addition to the many achiasmate species, these cases tend to support the view that the obligate CO constraint is not universal, although the occurrence of distal COs appears to explain short chromosome maps in at least some cases. Another conclusion is that the placement of the CO seems to be less constrained: our review tentatively suggests that low intra-chromosome recombination evolves by moving COs towards the tips. Hence it appears that the one-CO constraint should better be assessed by alternative methods (e.g. cytological studies) that do not have a reduced likelihood of detection of terminal COs. A broad phylogenetic analysis of the occurrence of species with no ‘obligate CO’ may also help determining the degree to which the evolution of alternative mechanisms to ensure proper segregation is constrained. For the correlation between low levels of recombination and the evolution of sexuality, we see several ways forward: First, data on a larger number of species are needed for a meaningful comparative analysis. Ideally, these data should be based on more meaningful measures of genome-wide recombination than the genetic map length of chromosomes. Second, different modes of asexuality are often lumped, though the fitness effects of recombination may strongly differ among different kinds of asexuality. Moreover, the different forms may not be static, but evolve via evolutionary intermediates, and selection pressures during these intermediate stages are not well understood. Third, contagious asexuality may offer a way to study whether the decreased rates of recombination observed in many asexuals are governed by different genes than asexuality. This may also help to elucidate whether there has been secondary evolution of reduced recombination in asexuals following the transition to asexuality. Fourth, similar insights might be obtained from studying recombination within asexual species, but using lineages of different ages. Finally, more data similar to the studies in the Cape honeybee are needed to better distinguish between reduced numbers of COs and selection against recombinants.
Supplementary Material
Acknowledgements
We thank D. Berner, F. Brunet, D. Charlesworth, L.-M. Chevin, A. Cutter, B. De Massy, T. de Meeûs, J.-F. Flot, D. Jeffries, T. Laurentino, R. Mercier, T. Kaiser, A. Korol, S. Otto, C. Peichel, C. Reisser, R. Rose, L. Ross, J. Stapley, H. Teotonio, and three anonymous reviewers for insightful comments and hints on the existing literature. We thank G. Van Stappen and C. Mahieu from the Artemia Reference Center (Ghent), for providing A. sp. Kazakhstan cysts. We gratefully acknowledge support by M-P. Dubois, C. Reisser, the platform Service des Marqueurs Génétiques en Écologie at CEFE, and the genotyping and sequencing facilities of the Institut des Sciences de l'Evolution-Montpellier and the Labex Centre Méditerranéen Environnement Biodiversité (CeMEB). We thank the Montpellier Bioinformatic Biodiversity platform and the Labex CeMEB for access to high-performance computing clusters.
Data accessibility
Data available on Dryad, doi:10.5061/dryad.hn777.
Authors' contributions
C.R.H. and T.L. conceived and coordinated the review and the empirical study. R.Z. carried out the crosses and prepared the library. L.T. analysed the RAD-sequencing data. All authors contributed to and approved this manuscript.
Competing interests
We have no competing interests.
Funding
We acknowledge funding from the European Union (Marie Curie Career Integration Grant PCIG13-GA-2013-618961, DamaNMP), the Institut Écologie et Environnement of the CNRS (APEGE grant ARTASEX, and the Swiss National Science Foundation (grant no. 31003A_138203).
References
- 1.Dumont BL. 2017. Variation and evolution of the meiotic requirement for crossing over in mammals. Genetics 205, 155–168. ( 10.1534/genetics.116.192690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenormand T, Engelstädter J, Johnston SE, Wijnker E, Haag CR, Christoph R. 2016. Evolutionary mysteries in meiosis. Phil. Trans. R. Soc. B 371, 20160001 ( 10.1098/rstb.2016.0001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Freudenberg J. 2009. Two-parameter characterization of chromosome-scale recombination rate. Genome Res. 19, 2300–2307. ( 10.1101/gr.092676.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. 2015. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327. ( 10.1146/annurev-arplant-050213-035923) [DOI] [PubMed] [Google Scholar]
- 5.Petronczki M, Siomos MF, Nasmyth K, Correns C, Vries HD, Tscher E. 2003. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell 112, 423–440. ( 10.1016/S0092-8674(03)00083-7) [DOI] [PubMed] [Google Scholar]
- 6.Hirose Y, Suzuki R, Ohba T, Hinohara Y, Matsuhara H, Yoshida M, Itabashi Y, Murakami H, Yamamoto A. 2011. Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet. 7, e1001329 ( 10.1371/journal.pgen.1001329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter N, Flemming W, Boveri T. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 ( 10.1101/cshperspect.a016618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagaoka SI, Hassold TJ, Hunt PA. 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504. ( 10.1038/nrg3245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kauppi L, Jasin M, Keeney S. 2012. The tricky path to recombining X and Y chromosomes in meiosis. Ann. N.Y. Acad. Sci. 1267, 18–23. ( 10.1111/j.1749-6632.2012.06593.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto SP, et al. 2011. About PAR: The distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 27, 358–367. ( 10.1016/j.tig.2011.05.001) [DOI] [PubMed] [Google Scholar]
- 11.Lenormand T, Dutheil J. 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3, 396–403. ( 10.1371/journal.pbio.0030063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burt A, Bell G, Harvey PH. 1991. Sex differences in recombination. J. Evol. Biol. 4, 259–277. ( 10.1046/j.1420-9101.1991.4020259.x) [DOI] [Google Scholar]
- 13.Bell G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. Berkeley, CA: University of California Press. [Google Scholar]
- 14.Hawley S, Theurkauf W. 1993. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet. 9, 310–316. ( 10.1016/0168-9525(93)90249-H) [DOI] [PubMed] [Google Scholar]
- 15.Wolf KW. 1994. How meiotic cells deal with non-exchange chromosomes. Bioessays 16, 107–114. ( 10.1002/bies.950160207) [DOI] [PubMed] [Google Scholar]
- 16.Krishnaprasad GN, Anand MT, Lin G, Tekkedil MM, Steinmetz LM, Nishant KT. 2015. Variation in crossover frequencies perturb crossover assurance without affecting meiotic chromosome segregation in Saccharomyces cerevisiae. Genetics 199, 399–412. ( 10.1534/genetics.114.172320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fledel-Alon A, Wilson DJ, Broman K, Wen X, Ober C, Coop G, Przeworski M. 2009. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 5, 1–7. ( 10.1371/journal.pgen.1000658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddle NC, Elgin SC. R. 2006. The dot chromosome of Drosophila: insights into chromatin states and their change over evolutionary time. Chromosom. Res. 14, 405–416. ( 10.1007/s10577-006-1061-6) [DOI] [PubMed] [Google Scholar]
- 19.Graves JAM. 1996. Mammals that break the rule: genetics of marsupials and monotremes. Annu. Rev. Genet. 30, 233–260. ( 10.1146/annurev.genet.30.1.233) [DOI] [PubMed] [Google Scholar]
- 20.De La Fuente R, Parra MT, Viera A, Calvente A, Gómez R, Suja JÁ, Rufas JS, Page J. 2007. Meiotic pairing and segregation of achiasmate sex chromosomes in eutherian mammals: The role of SYCP3 protein. PLoS Genet. 3, 2122–2134. ( 10.1371/journal.pgen.0030198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grell RF. 1964. Distributive pairing: the size-dependent mechanism for regular segregation of the fourth chromosomes in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 52, 226–232. ( 10.1073/pnas.52.2.226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenormand T. 2003. The evolution of sex dimorphism in recombination. Genetics 163, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omilian AR, Cristescu MEA, Dudycha JL, Lynch M. 2006. Ameiotic recombination in asexual lineages of Daphnia. Proc. Natl Acad. Sci. USA 103, 18 638–18 643. ( 10.1073/pnas.0606435103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svendsen N, et al. 2015. Uncovering cryptic asexuality in Daphnia magna by RAD-sequencing. Genetics 201, 1143–1155. ( 10.1534/genetics.115.179879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelstädter J. 2008. Constraints on the evolution of asexual reproduction. Bioessays 30, 1138–1150. ( 10.1002/bies.20833) [DOI] [PubMed] [Google Scholar]
- 26.Archetti M. 2010. Complementation, genetic conflict, and the evolution of sex and recombination. J. Hered. 101, 1–13. ( 10.1093/jhered/esq009) [DOI] [PubMed] [Google Scholar]
- 27.Stenberg P, Saura A. 2009. Cytology of asexual animals. In Lost sex. The evolutionary biology of parthenogenesis (eds Schön I, Martens K, van Dijk P), pp. 63–74. Dordrecht, The Netherlands: Springer Science. [Google Scholar]
- 28.Neiman M, Schwander T. 2011. Using parthenogenetic lineages to identify advantages of sex. Evol. Biol. 38, 115–123. ( 10.1007/s11692-011-9113-z) [DOI] [Google Scholar]
- 29.Nougué O, et al. 2015. Automixis in Artemia: Solving a century-old controversy. J. Evol. Biol. 28, 2337–2348. ( 10.1111/jeb.12757) [DOI] [PubMed] [Google Scholar]
- 30.Pearcy M, Hardy OJ, Aron S. 2006. Thelytokous parthenogenesis and its consequences on inbreeding in an ant. Heredity 96, 377–382. ( 10.1038/sj.hdy.6800813) [DOI] [PubMed] [Google Scholar]
- 31.Mandegar MA, Otto SP. 2007. Mitotic recombination counteracts the benefits of genetic segregation. Proc. R. Soc. B 274, 1301–1307. ( 10.1098/rspb.2007.0056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mogie M. 1986. Automixis: its distribution and status. Biol. J. Linn. Soc. 28, 321–329. ( 10.1111/j.1095-8312.1986.tb01761.x) [DOI] [Google Scholar]
- 33.Suomalainen E, Saura A, Lokki J. 1987. Cytology and evolution in parthenogenesis. Boca Raton, FL: CRC Press. [Google Scholar]
- 34.Zakharov IA. 2005. Intratetrad mating and its genetic and evolutionary consequences. Russ. J. Genet. 41, 402–411. ( 10.1007/s11177-005-0103-z) [DOI] [PubMed] [Google Scholar]
- 35.Asher JH. 1970. Parthenogenesis and genetic variability. II. One-locus models for various diploid populations. Genetics 66, 369–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiruta C, Nishida C, Tochinai S. 2010. Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex. Chromosom. Res. 18, 833–840. ( 10.1007/s10577-010-9159-2) [DOI] [PubMed] [Google Scholar]
- 37.Kellner K, Heinze J. 2011. Mechanism of facultative parthenogenesis in the ant Platythyrea punctata. Evol. Ecol. 25, 77–89. ( 10.1007/s10682-010-9382-5) [DOI] [Google Scholar]
- 38.Rey O, et al. 2011. Meiotic recombination dramatically decreased in thelytokous queens of the little fire ant and their sexually produced workers. Mol. Biol. Evol. 28, 2591–2601. ( 10.1093/molbev/msr082) [DOI] [PubMed] [Google Scholar]
- 39.Oxley PR, et al. 2014. The genome of the clonal raider ant Cerapachys biroi. Curr. Biol. 24, 451–458. ( 10.1016/j.cub.2014.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baudry E, Kryger P, Allsopp M, Koeniger N, Vautrin D, Mougel F, Cornuet J, Solignac M. 2004. Whole-genome scan in thelytokous-laying workers of the Cape honeybee (Apis mellifera capensis): central fusion, reduced recombination rates and centromere mapping using half-tetrad analysis. Genetics 167, 243–252. ( 10.1534/genetics.167.1.243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goudie F, Oldroyd BP. 2014. Thelytoky in the honey bee. Apidologie 45, 306–326. ( 10.1007/s13592-013-0261-2) [DOI] [Google Scholar]
- 42.Goudie F, Allsopp MH, Beekman M, Oxley PR, Lim J, Oldroyd BP. 2012. Maintenance and loss of heterozygosity in a thelytokous lineage of honey bees (Apis mellifera capensis). Evolution 66, 1897–1906. ( 10.1111/j.1558-5646.2011.01543.x) [DOI] [PubMed] [Google Scholar]
- 43.Rauwolf U, Greiner S, Mráček J, Rauwolf M, Golczyk H, Mohler V, Herrmann RG, Meurer J. 2011. Uncoupling of sexual reproduction from homologous recombination in homozygous Oenothera species. Heredity 107, 87–94. ( 10.1038/hdy.2010.171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilla EJS, Beardmore JA. 1994. Genetic and morphometric differentiation in old world bisexual species of Artemia (the brine shrimp). Heredity 73, 47–56. ( 10.1038/hdy.1994.97) [DOI] [Google Scholar]
- 45.Muñoz J, Gómez A, Green AJ, Figuerola J, Amat F, Rico C. 2010. Evolutionary origin and phylogeography of the diploid obligate parthenogen Artemia parthenogenetica (Branchiopoda: Anostraca). PLoS ONE 5, e11932 ( 10.1371/journal.pone.0011932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Vos S, Bossier P, van Stappen G, Vercauteren I, Sorgeloos P, Vuylsteke M. 2013. A first AFLP-based genetic linkage map for brine shrimp Artemia franciscana and its application in mapping the sex locus. PLoS ONE 8, 1–10. ( 10.1371/journal.pone.0057585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cabral G, Marques A, Schubert V, Pedrosa-Harand A, Schlögelhofer P. 2014. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Commun. 5, 5070 ( 10.1038/ncomms6070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heckmann S, Jankowska M, Schubert V, Kumke K, Ma W, Houben A. 2014. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat. Commun. 5, 4979 ( 10.1038/ncomms5979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogdanov YF. 2016. Inverted meiosis and its place in the evolution of sexual reproduction pathways. Russ. J. Genet. 52, 473–490. ( 10.1134/S1022795416050033) [DOI] [PubMed] [Google Scholar]
- 50.Melters DP, Paliulis LV, Korf IF, Chan SWL. 2012. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosom. Res. 20, 579–593. ( 10.1007/s10577-012-9292-1) [DOI] [PubMed] [Google Scholar]
- 51.Ross L, Blackmon H, Lorite P, Gokhman VE, Hardy NB. 2015. Recombination, chromosome number and eusociality in the Hymenoptera. J. Evol. Biol. 28, 105–116. ( 10.1111/jeb.12543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lynch M. 2007. The origins of genome architecture. Sunderland, MA: Sinauer Assocs., Inc. [Google Scholar]
- 53.White MJD. 1973. Animal cytology and evolution. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 54.Crosland MWJ, Crozier RH. 1986. Myrmecia pilosula, an ant with only one pair of chromosomes. Science 231, 1278 ( 10.1126/science.231.4743.1278) [DOI] [PubMed] [Google Scholar]
- 55.Blackmon H, Hardy NB, Ross L. 2015. The evolutionary dynamics of haplodiploidy: genome architecture and haploid viability. Evolution 69, 2971–2978. ( 10.1111/evo.12792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cremonini R. 2005. Low chromosome number angiosperms. Caryologia 58, 403–409. ( 10.1080/00087114.2005.10589480) [DOI] [Google Scholar]
- 57.The Tree of Sex Consortium. 2014. Tree of sex: a database of sexual systems. Sci. Data 1, 1–8. ( 10.1007/s40745-014-0001-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barnes TM, Kohara Y, Coulson A, Hekimi S. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141, 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaiser TS, et al. 2016. The genomic basis of circadian and circalunar timing adaptations in a midge. Nature 540, 69–73. ( 10.1038/nature20151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theodosiou L, McMillan WO, Puebla O. 2016. Recombination in the eggs and sperm in a simultaneously hermaphroditic vertebrate. Proc. R. Soc. B 283, 20161821 ( 10.1098/rspb.2016.1821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cutter AD, Dey A, Murray RL. 2009. Evolution of the Caenorhabditis elegans genome. Mol. Biol. Evol. 26, 1199–1234. ( 10.1093/molbev/msp048) [DOI] [PubMed] [Google Scholar]
- 62.Rockman MV, Kruglyak L. 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5, e1000419 ( 10.1371/journal.pgen.1000419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oakley HA, Jones GH. 1982. Meiosis in Mesostoma ehrenbergii ehrenbergii (Turbellaria, Rhabdocoela) I. Chromosome pairing, synaptonemal complexes and chiasma localisation in spermatogenesis. Chromosoma 85, 311–322. ( 10.1007/BF00330355) [DOI] [Google Scholar]
- 64.Oakley HA. 1985. Meiosis in Mesostoma ehrenbergii ehrenbergii (Turbellaria, Rhabdocoela). III. Univalent chromosome segregation during the first meiotic division in spermatocytes. Chromosoma 91, 95–100. ( 10.1007/BF00294051) [DOI] [PubMed] [Google Scholar]
- 65.Oakley HA. 1982. Meiosis in Mesostoma ehrenbergii ehrenbergii (Turbellaria, Rhabdocoela) II. Synaptonemal complexes, chromosome pairing and disjunction in achiasmate oogenesis. Chromosoma 87, 133–147. ( 10.1007/BF00338485) [DOI] [Google Scholar]
- 66.Wong AK, et al. 2010. A comprehensive linkage map of the dog genome. Genetics 184, 595–605. ( 10.1534/genetics.109.106831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campbell CL, Bhérer C, Morrow BE, Boyko AR, Auton A. 2016. A pedigree-based map of recombination in the domestic dog genome. G3 6, 3517–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Backström N, et al. 2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 20, 485–495. ( 10.1101/gr.101410.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stapley J, Birkhead TR, Burke T, Slate J. 2010. Pronounced inter- and intrachromosomal variation in linkage disequilibrium across the zebra finch genome. Genome Res. 20, 496–502. ( 10.1101/gr.102095.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stapley J, Birkhead TR, Burke T, Slate J. 2008. A linkage map of the zebra finch Taeniopygia guttata provides new insights into avian genome evolution. Genetics 179, 651–667. ( 10.1534/genetics.107.086264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawakami T, Smeds L, Backström N, Husby A, Qvarnström A, Mugal CF, Olason P, Ellegren H. 2014. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol. Ecol. 23, 4035–4058. ( 10.1111/mec.12810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groenen MAM, et al. 2009. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 19, 510–519. ( 10.1101/gr.086538.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K. 2012. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 22, 51–63. ( 10.1101/gr.124123.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uchino T, et al. 2016. Constructing genetic linkage maps using the whole genome sequence of pacific bluefin tuna (Thunnus orientalis) and a comparison of chromosome structure among teleost species. Adv. Biosci. Biotechnol. 7, 85–122. ( 10.4236/abb.2016.72010) [DOI] [Google Scholar]
- 75.Shan T, Pang S, Li J, Li X, Su L. 2015. Construction of a high-density genetic map and mapping of a sex-linked locus for the brown alga Undaria pinnatifida (Phaeophyceae) based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Genomics 16, 902 ( 10.1186/s12864-015-2184-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang N, et al. 2015. Construction of a high density SNP linkage map of kelp (Saccharina japonica) by sequencing Taq I site associated DNA and mapping of a sex determining locus. BMC Genomics 16, 189 ( 10.1186/s12864-015-1371-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berset-Brändli L, Jaquiéry J, Broquet T, Ulrich Y, Perrin N. 2008. Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc. R. Soc. B 275, 1577–1585. ( 10.1098/rspb.2008.0298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brelsford A, Dufresnes C, Perrin N. 2015. High-density sex-speci fi c linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity 116, 177–181. ( 10.1038/hdy.2015.83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foley BR, Rose CG, Rundle DE, Leong W, Moy GW, Burton RS, Edmands S. 2011. A gene-based SNP resource and linkage map for the copepod Tigriopus californicus. BMC Genomics 12, 568 ( 10.1186/1471-2164-12-568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berdan EL, Kozak GM, Ming R, Rayburn AL, Kiehart R, Fuller RC. 2014. Insight into genomic changes accompanying divergence: Genetic linkage maps and synteny of Lucania goodei and L. parva reveal a Robertsonian fusion. G3 4, 1363–1372. ( 10.1534/g3.114.012096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sonnenberg AS. M., et al. 2016. A detailed analysis of the recombination landscape of the button mushroom Agaricus bisporus var. bisporus. Fungal Genet. Biol. 93, 35–45. ( 10.1016/j.fgb.2016.06.001) [DOI] [PubMed] [Google Scholar]
- 82.Johnson MTJ, Fitzjohn RG, Smith SD, Rausher MD, Otto SP. 2011. Loss of sexual recombination and segregation is associated with increased diversification in evening primroses. Evolution 65, 3230–3240. ( 10.1111/j.1558-5646.2011.01378.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ranganath RM. 2008. Meiotic chromosome pairing and recombination take refuge in the telomeres. Nat. Rev. Genet. 9, 318 ( 10.1038/nrg2224-c1) [DOI] [PubMed] [Google Scholar]
- 84.Holsinger KE, Ellstrand NC. 1984. The evolution and ecology of permanent translocation heterozygotes. Am. Nat. 124, 48–71. ( 10.1086/284251) [DOI] [Google Scholar]
- 85.Grützner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O'Brien PCM, Jones RC, Ferguson-Smith MA, Marshall GJA. 2004. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432, 913–917. ( 10.1038/nature03021) [DOI] [PubMed] [Google Scholar]
- 86.Syren RM, Luykx P. 1977. Permanent segmental interchange complex in the termite Incisitermes schwarzi. Nature 266, 167–168. ( 10.1038/266167a0) [DOI] [PubMed] [Google Scholar]
- 87.Wiens D, Barlow BA. 1975. Permanent translocation heterozygosity and sex determination in East African Mistletoes. Science 187, 1208–1209. ( 10.1126/science.187.4182.1208) [DOI] [PubMed] [Google Scholar]
- 88.Lin YJ. 1982. Chiasma failures and chromosome association in Rhoeo spathacea var. variegata. Cytobios 33, 7–14. [PubMed] [Google Scholar]
- 89.Gross MC, Feldberg E, Cella DM, Schneider MC, Schneider CH, Porto JIR, Martins C. 2009. Intriguing evidence of translocations in Discus fish (Symphysodon, Cichlidae) and a report of the largest meiotic chromosomal chain observed in vertebrates. Heredity 102, 435–441. ( 10.1038/hdy.2009.3) [DOI] [PubMed] [Google Scholar]
- 90.Kenton A, Davies A, Jones K. 1987. Identification of Renner complexes and duplications in permanent hybrids of Gibasis pulchella (Commelinaceae). Chromosoma 95, 424–434. ( 10.1007/BF00333994) [DOI] [Google Scholar]
- 91.Grattapaglia D, Sederoff R. 1994. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137, 1121–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu R, Ma C-X, Wu SS, Zeng Z-B. 2002. Linkage mapping of sex-specific differences. Genet. Res. 79, 85–96. ( 10.1017/S0016672301005389) [DOI] [PubMed] [Google Scholar]
- 93.Rode NO, Charmantier A, Lenormand T. 2011. Male-female coevolution in the wild: evidence from a time series in Artemia franciscana. Evolution 65, 2881–2892. ( 10.1111/j.1558-5646.2011.01384.x) [DOI] [PubMed] [Google Scholar]
- 94.Nougué O, Flaven E, Jabbour-Zahab R, Rode NO, Dubois M-P, Lenormand T. 2015. Characterization of nine new polymorphic microsatellite markers in Artemia parthenogenetica. Biochem. Syst. Ecol. 58, 59–63. ( 10.1016/j.bse.2014.10.014) [DOI] [Google Scholar]
- 95.Muñoz J, Green AJ, Figuerola J, Amat F, Rico C. 2008. Characterization of polymorphic microsatellite markers in the brine shrimp Artemia (Branchiopoda, Anostraca). Mol. Ecol. Resour. 9, 547–550. ( 10.1111/j.1755-0998.2008.02360.x) [DOI] [PubMed] [Google Scholar]
- 96.Etter PD, Preston JL, Bassham S, Cresko WA, Johnson EA. 2011. Local de novo assembly of RAD paired-end contigs using short sequencing reads. PLoS ONE 6, e18561 ( 10.1371/journal.pone.0018561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3 1, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. ( 10.1093/bioinformatics/btg112) [DOI] [PubMed] [Google Scholar]
- 99.Cristescu MEA, Colbourne JK, Radivojac J, Lynch M. 2006. A microsatellite-based genetic linkage map of the waterflea, Daphnia pulex: On the prospect of crustacean genomics. Genomics 88, 415–430. ( 10.1016/j.ygeno.2006.03.007) [DOI] [PubMed] [Google Scholar]
- 100.Abatzopoulos TJ, Beardmore JA, Clegg JS, Sorgeloos P. 2002. Artemia: basic and applied biology.
- 101.Sivagnanasundaram S, Broman KW, Liu M, Petronis A. 2004. Quasi-linkage: a confounding factor in linkage analysis of complex diseases? Hum. Genet. 114, 588–593. ( 10.1007/s00439-004-1109-7) [DOI] [PubMed] [Google Scholar]
- 102.Rode N. 2012. Microévolution en temps réel : étude quantitative dans les populations naturelles d’Artemia spp. PhD Thesis, Univ. Montpellier II. [Google Scholar]
- 103.Denell RE, Keppy DO. 1979. The nature of genetic recombination near the third chromosome centromere or Drosophila melanogaster. Genetics 93, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bowers JE, Pearl SA, Burke JM. 2016. Genetic mapping of millions of SNPs in safflower (Carthamus tinctorius L.) via whole-genome resequencing. G3 6, 2203–2211. ( 10.1534/g3.115.026690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ronin Y, Minkov D, Mester D, Akhunov E, Korol A. 2015. Building ultra-dense genetic maps in the presence of genotyping errors and missing data. In Advances in wheat genetics: from genome to field, pp. 127–133. Tokyo, Japan: Springer. [Google Scholar]
- 106.Colombo PC. 1992. A new index for estimating genetic recombination from chiasma distribution data. Heredity 69, 412–415. ( 10.1038/hdy.1992.144) [DOI] [Google Scholar]
- 107.Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM. 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Phil. Trans. R. Soc. B 372, 20160455 ( 10.1098/rstb.2016.0455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morgan CH, Zhang H, Bomblies K. 2017. Are the effects of elevated temperature on meiotic recombination and thermotolerance linked via the axis and synaptonemal complex? Phil. Trans. R. Soc. B 372, 20160470 ( 10.1098/rstb.2016.0470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stevison L, Sefick S, Rushton C, Graze R. 2017. Recombination rate plasticity: revealing mechanisms by design. Phil. Trans. R. Soc. B 372, 20160459 ( 10.1098/rstb.2016.0459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alves I, Houle AA, Hussin JG, Awadalla P. 2017. The impact of recombination on human mutation load and disease. Phil. Trans. R. Soc. B 372, 20160465 ( 10.1098/rstb.2016.0465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tiemann-Boege I, Schwarz T, Striedner Y, Heissl A. 2017. The consequences of sequence erosion in the evolution of recombination hotspots. Phil. Trans. R. Soc. B 372, 20160462 ( 10.1098/rstb.2016.0462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Comeron JM. 2017. Background selection as null hypothesis in population genomics: insights and challenges from Drosophila studies. Phil. Trans. R. Soc. B 372, 20160471 ( 10.1098/rstb.2016.0471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rybnikov SR, Frenkel ZM, Korol AB. 2017. What drives the evolution of condition-dependent recombination in diploids? Some insights from simulation modelling. Phil. Trans. R. Soc. B 372, 20160460 ( 10.1098/rstb.2016.0460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Latrille T, Duret L, Lartillot N. 2017. The Red-Queen model of recombination hot-spot evolution: a theoretical investigation. Phil. Trans. R. Soc. B 372, 20160463 ( 10.1098/rstb.2016.0463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Charlesworth D. 2017. Evolution of recombination rates between sex chromosomes. Phil. Trans. R. Soc. B 372, 20160456 ( 10.1098/rstb.2016.0456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dapper AL, Payseur BA. 2017. Connecting theory and data to understand recombination rate evolution. Phil. Trans. R. Soc. B 372, 20160469 ( 10.1098/rstb.2016.0469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haag CR, Theodosiou L, Zahab R, Lenormand T. 2017. Low recombination rates in sexual species and sex–asex transitions. Phil. Trans. R. Soc. B 372, 20160461 ( 10.1098/rstb.2016.0461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kent TV, Uzunović J, Wright SI. 2017. Coevolution between transposable elements and recombination. Phil. Trans. R. Soc. B 372, 20160458 ( 10.1098/rstb.2016.0458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dawley RM. 1992. Clonal hybrids of the common laboratory fish Fundulus heteroclitus. Proc. Natl Acad. Sci. USA 89, 2485–2488. ( 10.1073/pnas.89.6.2485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dukić M, et al. 2016. A high-density genetic map reveals variation in recombination rate across the genome of Daphnia magna. BMC Genet. 17, 137 ( 10.1186/s12863-016-0445-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hebert PDN, Crease T. 1980. Clonal coexistence in Daphnia pulex (Leydig): another planktonic paradox. Science 207, 1363–1365. ( 10.1126/science.207.4437.1363) [DOI] [Google Scholar]
- 122.Wallberg A, et al. 2015. Extreme recombination frequencies shape genome variation and evolution in the honeybee, Apis mellifera. PLoS Genet. 11, e1005189 ( 10.1371/journal.pgen.1005189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rueppell O, Kuster R, Miller K, Fouks B, Rubio Correa S, Collazo J, Phaincharoen M, Tingek S, Koeniger N. 2016. A new metazoan recombination rate record and consistently high recombination rates in the honey bee genus Apis accompanied by frequent inversions but not translocations. Genome Biol. Evol. 8, evw269 ( 10.1093/gbe/evw269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maccari M, Gómez A, Hontoria F, Amat F. 2013. Functional rare males in diploid parthenogenetic Artemia. J. Evol. Biol. 26, 1934–1948. ( 10.1111/jeb.12191) [DOI] [PubMed] [Google Scholar]
- 125.Simon J.-C., Delmotte F, Rispe C, Crease T. 2003. Phylogenetic relationships between parthenogens and their sexual relatives: the possible routes to parthenogenesis in animals. Biol. J. Linn. Soc. 79, 151–163. ( 10.1046/j.1095-8312.2003.00175.x) [DOI] [Google Scholar]
- 126.Paland S, Colbourne JK, Lynch M. 2005. Evolutionary history of contagious asexuality in Daphnia pulex. Evolution 59, 800–813. ( 10.1111/j.0014-3820.2005.tb01754.x) [DOI] [PubMed] [Google Scholar]
- 127.Innes DJ, Hebert PDN. 1988. The origin and genetic basis of obligate parthenogenesis in Daphnia pulex. Evolution 42, 1024–1035. ( 10.1111/j.1558-5646.1988.tb02521.x) [DOI] [PubMed] [Google Scholar]
- 128.Engelstädter J, Sandrock C, Vorburger C. 2011. Contagious parthenogenesis, automixis, and a sex determination meltdown. Evolution 65, 501–511. ( 10.1111/j.1558-5646.2010.01145.x) [DOI] [PubMed] [Google Scholar]
- 129.Xu S, Spitze K, Ackerman MS, Ye Z, Bright L, Keith N, Jackson CE, Shaw JR, Lynch M. 2015. Hybridization and the origin of contagious asexuality in Daphnia pulex. Mol. Biol. Evol. 32, 3215–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tucker AE, Ackerman MS, Eads BD, Xu S, Lynch M. 2013. Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc. Natl Acad. Sci. USA 110, 15 740–15 745. ( 10.1073/pnas.1313388110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Engelstädter J. 2017. Asexual but not clonal: evolutionary processes in automictic populations. Genetics 206, 993–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on Dryad, doi:10.5061/dryad.hn777.