Abstract
Meiotic recombination is necessary for successful gametogenesis in most sexually reproducing organisms and is a fundamental genomic parameter, influencing the efficacy of selection and the fate of new mutations. The molecular and evolutionary functions of recombination should impose strong selective constraints on the range of recombination rates. Yet, variation in recombination rate is observed on a variety of genomic and evolutionary scales. In the past decade, empirical studies have described variation in recombination rate within genomes, between individuals, between sexes, between populations and between species. At the same time, theoretical work has provided an increasingly detailed picture of the evolutionary advantages to recombination. Perhaps surprisingly, the causes of natural variation in recombination rate remain poorly understood. We argue that empirical and theoretical approaches to understand the evolution of recombination have proceeded largely independently of each other. Most models that address the evolution of recombination rate were created to explain the evolutionary advantage of recombination rather than quantitative differences in rate among individuals. Conversely, most empirical studies aim to describe variation in recombination rate, rather than to test evolutionary hypotheses. In this Perspective, we argue that efforts to integrate the rich bodies of empirical and theoretical work on recombination rate are crucial to moving this field forward. We provide new directions for the development of theory and the production of data that will jointly close this gap.
This article is part of the themed issue ‘Evolutionary causes and consequences of recombination rate variation in sexual organisms’.
Keywords: recombination rate, theory, meiotic recombination, evolution, genetic variation
1. Introduction
During meiosis, germ cells generate DNA double-strand breaks. A minority of these breaks are repaired as crossovers between homologous chromosomes. This process of recombination diversifies offspring genomes, interacting with other evolutionary forces to shape major features of the genome landscape, including nucleotide diversity [1–4], codon bias [5], base composition [6] and repetitive element density [7,8]. The number and placement of crossovers along chromosomes are tightly controlled, with aberrations reducing fertility and offspring viability [9]. Owing to the significance of recombination for evolution and reproduction, the rate at which this process occurs has long been of interest to biologists.
The observation that the recombination rate varies among individuals, among populations and among species (reviewed by [10–12]) raises the questions of how and why this fundamental genomic characteristic evolves. In this Perspective, we suggest that despite decades of research relevant to these questions, evolutionary biologists remain surprisingly far away from answering them. We argue that an important barrier to progress has been a lack of coordination between theoretical and empirical studies. Successes at documenting variation in recombination rate often proceed without an underlying theoretical framework, challenging the ability to test hypotheses and convert observed patterns into inferences about evolutionary process. Theoretical advances that reveal the population genetic conditions under which recombination is predicted to evolve ignore key biological aspects of recombination rate variation, including genetic complexity and genomic scale.
Our goal is to catalyse alignment and integration of theoretical and empirical efforts to understand recombination rate evolution. We begin with a short overview of existing theory on the evolution of recombination rate, highlighting the key predictions and ingredients of existing models. Next, we describe how empirical work can better evaluate theoretical predictions and we motivate the data-based examination of the role of natural selection in recombination rate evolution. Finally, we suggest new directions for theory that capture observed patterns of recombination rate variation.
2. General features of theoretical models of recombination rate evolution
Like other phenotypes, recombination rate has the potential to affect individual fitness and experience direct selection, in this case by impacting gamete viability (direct selection—table 1). In contrast with most other traits, recombination rate itself shapes offspring genotype frequencies, raising the possibility of indirect selection. This type of indirect selection on recombination rate can be mediated by short-term or long-term advantages [13]. Short-term benefits occur when recombination breaks apart deleterious gene combinations and immediately increases the mean fitness in the next generation. Long-term benefits accrue when recombination increases the additive genetic variance in a population, enabling selection (on other traits) to act more efficiently [13]. Understanding how recombination rate evolves requires knowledge of the magnitude and direction of direct selection, short-term indirect selection and long-term indirect selection. The sum of these effects determines whether alleles that modify recombination rate spread through a population [13,14].
Table 1.
An overview of nine hypotheses proposed to explain the evolution of recombination rate (direct selection, plus eight models of indirect selection). Each theoretical model differs in the key requirements that must be met for indirect selection on recombination rate to occur. There are no formal theoretical models underlying the direct selection hypothesis. We highlight the role of epistasis, linkage disequilibrium (LD) and drift in each model. Among models of indirect selection on recombination rate, there is also variation on whether the genetic modifier of recombination rate must be linked to other loci under direct selection. We describe a set of simple, testable predictions that arise from each model and describe the expected direction of selection on recombination rate. LD, linkage disequilibrium.
| hypothesis | key requirements | epistasis | linkage disequilibrium | drift | linked modifier | predictions | direction of effect |
|---|---|---|---|---|---|---|---|
| direct selection | an optimal number of crossovers per chromosome is required for successful meiosis | none | none | no | no prediction | upper and lower bounds on recombination rate | stabilizing (increases or decreases) around an optimal recombination rate |
| reduction principle | large, equilibrium populations with no mutation, no migration, random mating and constant viability selection | not required | initial LD is required | no | no requirement | no recombination; genetic modifiers that decrease recombination rate spread | decrease |
| negative epistasis | pervasive, weak, negative epistasis and recurrent deleterious mutations or selective sweeps (directional selection) | weak, negative epistasis | negative, arises due to epistatic selection; disadvantageous | no | linked modifier relaxes the requirement for weak epistasis | increase in recombination rate following episodes of strong directional selection | increase if requirements met, otherwise decrease |
| temporal heterogeneity | rapid, temporal fluctuations in the environment favour different combinations of alleles | the sign of epistasis changes every two to five generations | the sign of LD tracks changes in the sign of epistasis | no | linked modifiers | increase in recombination rate in environments with rapid and consistent temporal variation | increase if requirements met, otherwise decrease |
| host–parasite interactions | rare combinations of alleles have an advantage via resistance to parasites | the sign of epistasis changes every two to five generations | the sign of LD tracks changes in the sign of epistasis | no | linked modifiers | increased recombination in populations exposed to virulent parasites | increase if requirements met, otherwise decrease |
| inter-locus sexual conflict | inter-locus sexual antagonism is common and rare combinations of alleles are advantageous | the sign fluctuates at a frequency dependent on the strength of sexual selection | the sign of LD tracks changes in the sign of epistasis | no | unlinked modifiers | positive correlation between recombination rate and strength of sexual conflict | increase if requirements met, otherwise decrease |
| spatial heterogeneity | environmental selection pressures vary between populations with frequent migration | variable: none, positive, or negative | variable: positive or negative; generated by epistasis and/or spatial heterogeneity | no | variable: linked, loosely linked, or unlinked | more variation in recombination rate among populations in highly spatially variable environments | increase if requirements met, otherwise decrease |
| Hill–Robertson effect | population is finite and subject to selection | not required | negative, generated by drift | yes | linked modifiers | higher recombination rate with lower effective population size | increase if requirements met, otherwise decrease |
| fitness-associated recombination (FAR) | recurrent deleterious mutations, modifier sensitive to haplotype fitness | not required | positive, generated between fitness-determining loci | no | linked modifiers | negative correlation between fitness and recombination rate | stabilizing (increases or decreases) around an optimal recombination rate |
Identifying the conditions under which indirect selection favours increases in recombination has been a particular emphasis of theory treating the evolution of recombination rate. A first class of models, called optimality [15] or intrinsic models [16], examines how recombination rate optimizes group-level traits, such as equilibrium mean fitness [17–25] or mutational load [23,26–30]. This set of models compares populations that vary in recombination rate [15], but does not explicitly consider genetic modifiers of the trait. The second class of models, termed modifier [15] or extrinsic models [16], examines how various forms of individual-level selection change recombination rate [31,32]. In this group of models, a genetic modifier of recombination rate is treated as a single, Mendelian locus at which different alleles confer different recombination rates to individuals. The frequency of crossing-over in a specific genomic interval between two additional loci is considered. By varying selection pressures and tracking the change in frequency of modifier alleles, these models explicitly analyse the expected change in recombination rate within a single population. Modifier models have been favoured over optimality arguments because they invoke individual-level, rather than group-level, selection and they tend to reveal complex, short-term dynamics [15].
The opportunity for indirect selection on recombination rate depends on the degree and form of non-random associations between alleles at different loci, or linkage disequilibrium (LD). LD between two loci harbouring alleles A, a and B, b (respectively) can be measured as the deviation (DAB) of the haplotype's frequency (pAB) from its expected frequency, given individual allele frequencies (pA and pB) and free recombination:
| 2.1 |
When the population is at linkage equilibrium (DAB = 0), recombination does not affect offspring genotype frequencies because the association between alleles cannot be further randomized and indirect selection on recombination rate cannot be generated. As a result, most population genetic theory on the evolution of recombination focuses on understanding how evolutionary processes generate and/or maintain LD.
Two potentially important determinants of LD that have received considerable attention from theoreticians are epistasis and genetic drift [14]. In this context, epistasis is usually defined as non-additive allelic effects across loci, such that the mean phenotype for a given multi-locus genotype does not match its expected value, given the mean phenotypes of the individual alleles [33–35]. Epistasis for fitness results in selection for beneficial combinations of alleles, increasing their frequency within a population and generating LD [36]. Epistatic scenarios range from pairwise interactions between alleles to nonlinear cumulative effects of new mutations; both are featured in the theory of recombination rate evolution [37,38]. In finite populations, genetic drift leads to non-random associations between beneficial and deleterious alleles, thereby delaying the response to selection [39].
Even when epistasis and/or genetic drift generate LD, it remains unclear whether more recombination should be generally beneficial. Reducing LD can increase ‘recombination load’ by breaking apart beneficial combinations of alleles that have accumulated due to selection [13,40,41]. Thus, recombination may impede adaptation and eventually be eliminated, as recombination-reducing modifiers become associated with beneficial allelic combinations and spread through the population [13,40]. This concept is formalized in the ‘Reduction Principle’, which states that only modifiers that reduce recombination rate can invade a population under equilibrium conditions (reduction principle—table 1) [15,41–43]. The widespread persistence of crossing-over despite these theoretical constraints is referred to as the paradox of recombination [41]. However, equilibrium conditions require idealized large populations with no mutation, no migration, random mating and constant viability selection [15]. Such populations are likely rare in nature. By identifying conditions under which sets of these assumptions are routinely violated, theoreticians have generated a rich body of work that describes sources of indirect selection on recombination rate. Eight of these theoretical models are outlined in table 1. Here, we briefly describe three main categories of hypotheses that predict indirect selection for increased recombination rate.
(a). Negative epistasis
Negative epistasis describes genetic interactions in which two beneficial alleles, when present in the same individual, increase fitness less than expected based upon their separate effects or conversely, when two deleterious alleles decrease fitness more than expected based upon their separate effects. As a result, negative epistasis maintains LD, characterized by a paucity of genotypes with the most extreme fitness values and an excess of individuals near the mean fitness value [44]. By reducing LD, recombination generates more extreme phenotypes, increasing the genetic variance in the population. Greater genetic variance allows populations to purge recurrent deleterious mutations more efficiently and respond to directional selection more rapidly (negative epistasis—table 1) [40,44,45]. As a result of these long-term benefits, genetic modifiers that increase the recombination rate may become associated with beneficial allele combinations and spread through the population [13,29,42,44,46].
(b). Heterogeneity in selection
Heterogeneity in selection pressure over time, across space, or between the sexes can generate LD (table 1). Fluctuating environments can favour increased recombination rate when allelic combinations that are advantageous at one time point become disadvantageous at another time point, resulting in an overabundance of deleterious allelic combinations when environmental conditions change (temporal heterogeneity—table 1) [13,37,40,47–49]. In this scenario, recombination produces short-term benefits by breaking up maladaptive allelic combinations and immediately increasing mean fitness among offspring [13]. However, the degree to which recombination is favoured is highly sensitive to the frequency of environmental fluctuation. To account for the high levels of recombination that are observed, fluctuations must occur every 2–5 generations and cycle with a period of 4–10 generations [13,47].
Whereas abiotic factors are unlikely to produce such rapid, consistent changes in the sign of epistasis, biotic factors offer clear potential [13,50]. In particular, coevolution between hosts and parasites can generate epistatic fluctuations of the form needed to favour recombination (host–parasite interactions—table 1) [48,51]. This occurs when under-represented combinations of alleles offer increased resistance to sufficiently virulent parasites, which adapt to the most abundant genotypes within a population [37,48]. In this scenario, recombination produces rare combinations of alleles that are less susceptible to parasites [37,48]. Inter-locus sexual conflict can also generate disadvantageous allelic combinations due to antagonistic coevolution between the sexes (inter-locus sexual conflict—table 1) [52]. In addition to varying over time, selection can also vary over space. Spatial variation in the strength or direction of selection among populations can generate differences in local allele frequencies. With migration, these differences produce LD among offspring (spatial heterogeneity—table 1) [14]. The direction and magnitude of the indirect selection on recombination rate produced by migration are determined by the similarity in selection pressures between the two populations [14,53,54]. For example, recombination modifiers that reduce the recombination rate are expected to frequently, but not always, spread in the face of maladaptive gene flow, potentially playing an important role in the process of speciation [53,54].
(c). Genetic drift
Within finite populations, the interaction between genetic drift and selection results in the build-up of LD [39,55,56]. Genetic drift generates positive LD, when a beneficial mutation arises on the background of another beneficial allele, and negative LD, when a beneficial mutation arises on the background of a deleterious allele. Haplotypes with double beneficial alleles can quickly fix, thereby eliminating positive LD. In contrast, the association between beneficial and deleterious alleles may persist and slow the response to selection [39,57]. The net effect is an accumulation of negative LD, referred to as the Hill–Robertson effect [55,58]. By breaking down LD, recombination modifiers can free beneficial mutations from their backgrounds and increase in frequency within populations due to their association with advantageous haplotypes (Hill–Robertson effect—table 1) [39,55–57].
3. Generating data that address existing theory
Theoretical work has provided an increasingly detailed picture of the evolutionary advantages to recombination, but the causes of natural variation in recombination rate remain poorly understood. Here, we highlight some empirical findings on the evolution of recombination rate, describe their current disconnect with existing theory and recommend empirical approaches to bridge this divide.
(a). Measuring recombination rate
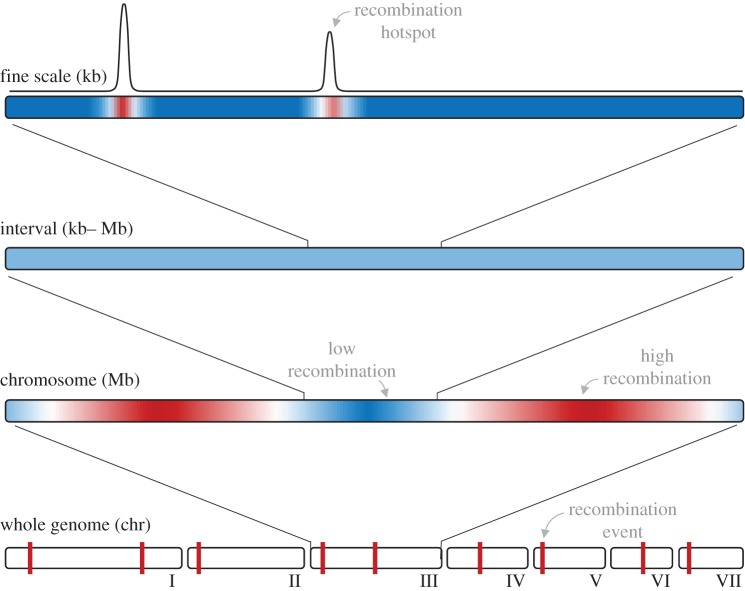
Multiple methods can be used to measure recombination rate, depending on the genomic scale of interest (figure 1). The total number of crossovers in a genome can be estimated by counting chiasmata (the physical bridges formed between homologous chromosomes) or in some species, by counting foci of the MLH1 mismatch repair protein (which localize to crossover sites and can be visualized with immunofluorescence) in meiotic cells [59]. These approaches characterize recombination in single cells; the recombination rate of an individual is estimated as the average number of crossovers among cells. A second method for estimating recombination rate is to examine the transmission of polymorphic DNA markers on the same chromosome in crosses or pedigrees. By comparing parent and offspring genotypes, the frequency of recombination between a pair of markers can be calculated and converted to a genetic distance. One centiMorgan (cM) is defined as the expected number of crossovers between markers in 100 meioses [60]. This linkage mapping approach enables the profiling of recombination rate variation along chromosomes. The genomic level of resolution depends on the number of meioses surveyed and is typically on the scale of megabases (Mb). Two methods enable estimation of recombination rate on finer (kilobase; kb) scales. The frequencies of marker haplotypes in very large numbers of sperm reveal recombination rates for individual males in targeted genomic regions [61]. Alternatively, patterns of LD among single nucleotide polymorphisms (SNPs) in samples of unrelated individuals can be converted to recombination rates using population genetic models [62]. This approach, which yields time- and sex-averaged recombination rate estimates for populations, can be applied to the entire genome. In all methods, the recombination rate between markers is typically standardized across interval sizes by dividing by the number of base pairs separating markers.
Figure 1.
Genomic scales on which recombination rate can be measured. (Online version in colour.)
(b). Towards hypothesis-based empirical studies
Empirical studies using methods described above have begun to describe how recombination rate varies along multiple evolutionary timescales. Individuals from the same species differ in the number and/or placement of crossovers [11,63–78]. Species pairs display divergent recombination rates in specific genomic intervals or across the genome [79–85]. Across the phylogeny of eutherian mammals, more closely related species tend to show more similar genome-wide recombination rates [10,86–88]. Nevertheless, these patterns remain disconnected from the theoretical hypotheses reviewed above, and few inferences about the underlying evolutionary processes have been reported.
Among existing theoretical hypotheses for the evolution of recombination rate, long-term indirect selection has received the most empirical attention (negative epistasis—table 1). The results of these studies have been largely inconclusive. Recombination rate increases in response to artificial selection targeting unrelated phenotypes in some experiments (reviewed by [41,57]). Although similar patterns might be expected to be associated with domestication [89], domesticated plants and animals show little to no evidence for expanded genetic maps compared to their wild relatives [90–92]. Another empirical approach that has been employed is to determine whether the conditions required for different theoretical hypotheses about the evolution of recombination rate exist. For example, theoretical models have clearly demonstrated that if indirect selection drives recombination rate evolution, the magnitude and sign of LD and epistasis must play key roles [14]. Although some empirical studies have attempted to quantify these variables (at least in experimental populations; reviewed in [93]), a connection to variation in recombination rate itself is still missing. We encourage empirical work that goes beyond evaluating the assumptions of existing models to directly testing their predictions.
From a theoretical perspective, the best models are those that make realistic assumptions and generate recombination rate evolution across the broadest parameter space. Based on these criteria, models that consider the combined action of genetic drift and selection suggest that Hill–Robertson effects may constitute a pervasive form of indirect selection on recombination rate. The major assumptions of these models—that populations are finite, are subject to recurrent mutation, and experience pervasive selection—likely apply to most natural populations [39]. But direct tests of predictions from this model are needed. Comparing recombination rates in conspecific pairs of populations with different effective population sizes (Hill–Robertson effect—table 1) and little to no evidence for gene flow would be one way to evaluate the importance of Hill–Robertson effects for recombination rate evolution [39].
(c). The role of selection
Although theoretical work is strongly biased towards selective explanations, there is limited empirical evidence for a role of selection in the evolution of recombination rate [12]. Three components must be present for the process of natural selection to occur: variation, inheritance and fitness differences. As mentioned above, empirical studies have documented extensive variation in recombination rate between individuals. Ample evidence indicates that phenotypic variation in recombination rate has a heritable component. Recombination rate shows resemblance among relatives in human pedigrees [69,94,95], differs among lines raised in a common environment [66,74,76,87,96,97] and responds to artificial selection in Drosophila melanogaster and other insects [63,65,98–113]. Broad-sense or narrow-sense heritability estimates from humans, mice, insects and maize range from 0.08 to 0.69 [94,95,99,100,102,112,114–116]. A few observations raise the prospect that recombination rate could affect fitness. Fecundity and recombination rate may be positively correlated in human mothers [69,117,118]. Phylogenetic comparative methods suggest that the genome-wide recombination rate has increased during mammalian evolution [88]. PRDM9, a protein that helps determine the position of crossovers in mice and humans, possesses one of the most rapidly evolving (zinc-finger) domains in mammals [119,120]. Finally, cellular needs to avoid non-disjunction (by generating at least one crossover per chromosome or chromosome arm) [117,121–124] and to minimize costs of double-strand break repair should impose selective bounds on the genome-wide recombination rate in nature [9,10,121,122].
A host of experiments using insects attempted to increase and/or decrease the recombination rate by direct artificial selection [63,65,98–113]. All but one study focused on crossover rates in individual genomic intervals monitored by visible markers. Ten of 15 studies that tried to increase the recombination rate were successful in at least one line; six of 15 studies were able to decrease crossing-over. Among the subset of experiments that applied both selection for higher and lower recombination rate, there was no obvious asymmetry in results. Although these reports are restricted to a few species of insects (mostly D. melanogaster) and results were highly variable both between and within experiments, this series at least demonstrates the potential for recombination rate to respond to directional selection in nature.
This summary underscores a few notable barriers to understanding the role of selection in recombination rate evolution. Whereas most models focus on indirect selection, patterns of variation in recombination rate and the functional role of recombination in meiosis suggest that direct selection may contribute to the evolution of this trait. In addition, empirical evidence for a relationship between recombination rate and fitness in natural populations is lacking [12]. Measuring both direct and indirect selection pressures on recombination rate in nature is a necessary next step to connecting data with existing theoretical predictions.
We offer several suggestions to elucidate the contributions of selection to recombination rate evolution. First, we encourage empirical work that better defines the lower and (especially) the upper bounds on recombination rate that reflect meiotic constraints. Widely cited lower bounds—one crossover per chromosome or one crossover per chromosome arm—are still based on data from a small number of species, and it is possible that this limit itself evolves [122]. Potential ceilings on the recombination rate remain poorly defined. Better characterization of the bounds on recombination rate and their meiotic causes would identify potential sources of purifying and stabilizing selection. Second, researchers should strive to connect variation in recombination rate with the natural environment. The question of whether recombination rate shows clines across gradients of geography and other environmental variables remains to be addressed. Third, quantitative genetic methods can be used to determine whether other patterns of recombination rate variation are consistent with neutral expectations. Genetic variance in recombination rate can be partitioned between and within populations using laboratory crosses. If recombination rate evolves neutrally, this partitioning should match levels of population structure at neutral molecular markers [125,126]; departures from this prediction could indicate directional or stabilizing selection on recombination rate. Finally, quantifying the distribution of mutational effects on recombination rate (using mutation-accumulation experiments and other approaches) ultimately will be necessary in order to draw firm conclusions about the role of selection in recombination rate evolution.
4. Developing models that address empirical patterns
Just as empirical approaches have largely failed to address theory in the context of recombination rate evolution, existing models have struggled to produce empirically testable hypotheses. Here, we describe how to advance theoretical studies of recombination rate evolution by grounding them with established empirical knowledge.
(a). Recombination rate as a quantitative trait
Empirical evidence demonstrates that recombination rate is a quantitative trait [12,113], with variance among individuals reflecting the cumulative effects of many underlying mutations and environmental influences. Sequence variants in or near 21 known genes are associated with recombination rate variation within populations of humans [127–129], domesticated cattle [130,131], or wild Soay sheep [116] . Variants in five of these genes correlate with recombination rate in multiple species: Rnf212 [116,127,128,130], Cplx1 [116,128,131], Rec8 [116,130,131], Msh4 [128,131] and Prdm9 [128,131]. Repeated association with a common set of genes raises the prospect that recombination rate variation harbours moderate genetic complexity [116]: a handful of variants have large enough effects to be detected on top of a polygenic background. Furthermore, suites of quantitative trait loci (QTL) confer differences in the genome-wide recombination rate between inbred strains of house mice belonging to different subspecies [132–134]. Although some alleles have appreciable phenotypic effects [128,132], most variance in recombination rate remains unexplained in the examined populations and crosses, suggesting an important role for environmental factors [116]. Taken together, this empirical evidence strongly supports the conclusion that recombination rate is a complex trait.
By contrast, existing theory focuses on population genetic models, which either ignore the genetic architecture of recombination rate (optimality arguments) or assume that recombination rate is a simple Mendelian trait (modifier models) [15,16]. While quantitative genetic theory has been applied to understand how indirect selection on a recombination modifier can be generated by direct selection on a complex trait [46], models that assume recombination rate itself is a complex trait are missing. The disconnect between the complex genetic underpinnings of recombination rate and the simplicity of the genetic architecture assumed in theoretical models is striking. Modelling recombination rate as a quantitative trait is likely to uncover novel evolutionary dynamics and to produce predictions that are straightforward to test.
(b). Genomic scale of recombination rate evolution
Another empirical observation is that distinct patterns of variation in recombination rate are sometimes observed on different genomic scales. Thus, the tempo and mode of recombination rate evolution may depend on the genomic scale on which it is measured [11,135,136] (figure 1). Patterns of LD indicate that both humans [80,82,137] and bonobos [83] share few recombination hotspots with chimpanzees, implying rapid evolution of recombination rate on the fine (kb) scale. By contrast, recombination rates calculated across larger chromosomal regions (from linkage maps or patterns of LD) usually show higher correlations among closely related species or conspecific populations [11,81,83,135,136,138–140], with statistically divergent intervals comprising a minority of the genome [85]. Intriguingly, recombination hotspots and double-strand break hotspots appear to be evolutionarily stable in finches [141] and budding yeast (respectively) [142], suggesting that the rapid divergence of recombination rate on the fine scale observed in mammals could be the exception [143]. Importantly, it is unclear how the evolution of recombination rate—including the form and intensity of selection—translates across genomic scales.
Models that address the evolution of recombination rate were generated to explain the evolutionary advantage of recombination, rather than quantitative differences in rate among individuals. One consequence of this motivation is that existing theory does not directly address genomic scale, focusing instead on single intervals of undefined physical size. It is difficult to use theoretical results for selection on a single interval to understand genome-wide patterns of variation in recombination rate, which can be measured empirically. We encourage the development of theory that parses and synthesizes the evolution of recombination rate across genomic scales [144].
(c). Plasticity and recombination rate evolution
Temperature, nutrient availability, pheromones and other environmental variables are known to modulate crossover number [145–152]. Furthermore, D. melanogaster females produce a higher proportion of recombinant offspring after they are infected by parasites [153]. Although the evolution of recombination rate has not been directly tied to these factors, natural variation in this phenotype is likely to be shaped by the external environment.
Theoretical models have explored the evolutionary dynamics of plastic genetic modifiers that only increase the rate of recombination in low-fitness individuals (fitness-associated recombination (FAR)—table 1) [154,155]. Among haploid species, FAR generates selection for increased recombination over an expansive parameter space and does not require epistasis, initial LD, or finite population size, suggesting that it may be a powerful mechanism for the evolution of recombination rate [154]. By contrast, the evolution of FAR appears to be extremely restricted in diploid organisms because it requires that the modifier assess haplotype, rather than organismal, fitness [155]. Thus, these models are unable to provide a general adaptive explanation for the empirical observations described above.
(d). Sex differences in recombination rate
Recombination rates can also experience discordant evolutionary trajectories in the two sexes. In some species, only one sex recombines; the other (achiasmate) sex is the heterogametic one [156–159]. In most species that recombine, crossing-over happens in both sexes, but the degree of sexual dimorphism in the total number of crossovers (heterochiasmy) varies substantially among species [160–162]. There is limited evidence of higher genetic variance for recombination rate among females than among males within species [116,128]. Both genetic variants with sex-specific effects and variants with sex-shared effects on recombination rate have been identified [116,128,131]. Sexually dimorphic genomic patterns in crossover positioning—including higher recombination near centromeres in female vertebrates [67,72,116,131,161,163–167]—raise the prospect that the decoupling of recombination rate evolution in the two sexes could extend to finer genomic scales.
Theoretical models have also investigated the evolution of sex differences in recombination rate [168,169]. While selection for the suppression of recombination on sex chromosomes can be generated rapidly, the evolution of sex differences in recombination rate on autosomes has been more difficult to explain [32]. Similar to FAR, it is unlikely that sex differences in the strength of selection in diploid adults can generate selection for sex differences in recombination rate [32,168]. Selection for heterochiasmy is more likely to be caused by sex differences in the strength of haploid-level selection or in the strength of selection on imprinted genes, which are effectively haploid [160,168]. In such cases, it is expected that the sex experiencing strong haploid selection will exhibit less recombination [168]. Nevertheless, theory that explains the major empirical characteristics of heterochiasmy evolution is not yet available.
5. Conclusion
Forging stronger connections between theory and data in recombination rate evolution holds considerable potential to advance the field. The rich body of theory on the evolution of recombination rate provides a strong framework for generating hypothesis-driven empirical studies. We encourage biologists to collect data that can be used to directly test these hypotheses. Furthermore, established approaches from evolutionary biology should be applied to answer the basic question of whether recombination rate experiences selection in natural populations. Likewise, empirical investigations uncovering patterns of variation in recombination rate, as well as its genetic underpinnings, are generating exciting new avenues for theoretical development. Models that treat recombination rate as a quantitative trait and explicitly incorporate genomic scale are likely to provide novel and increasingly realistic predictions that will lend themselves to empirical examination. A meaningful integration between theoretical and empirical studies should be the next step towards understanding how recombination rate evolves.
Acknowledgements
We thank April Peterson, Michael Wade, Jessica Stapley, Daniel Ortiz-Barrientos and multiple anonymous reviewers for useful comments on drafts of this article. We thank the editors of this special issue for inviting us to contribute.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
A.L.D. was supported by NHGRI Training Grant to the Genomic Sciences Training Program 5T32HG002760. B.A.P. was supported by NIH grants R01GM120051 and R01GM100426, and NSF grant DEB 1353737.
References
- 1.Maynard-Smith J, Haigh J. 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23, 23–35. ( 10.1017/S0016672300014634) [DOI] [PubMed] [Google Scholar]
- 2.Begun DJ, Aquadro CF. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356, 519–520. ( 10.1038/356519a0) [DOI] [PubMed] [Google Scholar]
- 3.Cutter AD, Payseur BA. 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14, 262–274. ( 10.1038/nrg3425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlesworth B, Morgan MT, Charlesworth D. 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134, 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comeron JM, Kreitman M, Aguadé M. 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duret L, Arndt PF. 2008. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4, e1000071 ( 10.1371/journal.pgen.1000071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlesworth B, Jarne P, Assimacopoulos S. 1994. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. III. Element abundances in heterochromatin . Genet. Res. 64, 183–197. ( 10.1017/S0016672300032845) [DOI] [PubMed] [Google Scholar]
- 8.Kent TV, Uzunović J, Wright SI. 2017. Coevolution between transposable elements and recombination. Phil. Trans. R. Soc. B 372, 20160458 ( 10.1098/rstb.2016.0458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassold T, Hunt P. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291. ( 10.1038/35066065) [DOI] [PubMed] [Google Scholar]
- 10.Coop G, Przeworski M. 2007. An evolutionary view of human recombination. Nat. Rev. Genet. 8, 23–34. ( 10.1038/nrg1947) [DOI] [PubMed] [Google Scholar]
- 11.Smukowski CS, Noor MAF. 2011. Recombination rate variation in closely related species. Heredity 107, 496–508. ( 10.1038/hdy.2011.44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritz KR, Noor MA, Singh ND. 2017. Variation in recombination rate: adaptive or not? Trends Genet. 33, 364–374. ( 10.1016/j.tig.2017.03.003) [DOI] [PubMed] [Google Scholar]
- 13.Barton NH. 1995. A general model for the evolution of recombination. Genet. Res. 65, 123–144. ( 10.1017/S0016672300033140) [DOI] [PubMed] [Google Scholar]
- 14.Lenormand T, Otto SP. 2000. The evolution of recombination in a heterogeneous environment. Genetics 156, 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman MW, Otto SP, Christiansen FB. 1996. Population genetic perspectives on the evolution of recombination. Annu. Rev. Genet. 30, 261–295. ( 10.1146/annurev.genet.30.1.261) [DOI] [PubMed] [Google Scholar]
- 16.Kondrashov AS. 1993. Classification of hypotheses on the advantage of amphimixis. J. Hered. 84, 372–387. ( 10.1093/oxfordjournals.jhered.a111358) [DOI] [PubMed] [Google Scholar]
- 17.Muller HJ. 1932. Some genetic aspects of sex. Am. Nat. 66, 118–138. ( 10.1086/280418) [DOI] [Google Scholar]
- 18.Kimura M. 1956. A model of a genetic system which leads to closer linkage by natural selection. Evolution 10, 278–287. ( 10.1111/j.1558-5646.1956.tb02852.x) [DOI] [Google Scholar]
- 19.Crow JF, Kimura M. 1965. Evolution in sexual and asexual populations. Am. Nat. 99, 439–450. ( 10.1086/282389) [DOI] [Google Scholar]
- 20.Bodmer WF. 1970. The evolutionary significance of recombination in prokaryotes. Symp. Soc. General Microbiol. 20, pp. 279–294. [Google Scholar]
- 21.Lewontin RC. 1971. The effect of genetic linkage on the mean fitness of a population. Proc. Natl Acad. Sci. USA 68, 984–986. ( 10.1073/pnas.68.5.984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlin S. 1973. Sex and infinity: a mathematical analysis of the advantages and disadvantages of genetic recombination. In The mathematical theory of the dynamics of biological populations (eds Bartlett MS, Hiorns RW), pp. 155–194. New York, NY: Academic Press. [Google Scholar]
- 23.Felsenstein J. 1974. The evolutionary advantage of recombination. Genetics 78, 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otto SP, Feldman MW, Christiansen FB. 1994. Some advantages and disadvantages of recombination. In Frontiers in mathematical biology (ed. SA Levin), pp. 198–211. Berlin, Germany: Springer. [Google Scholar]
- 25.Christiansen FB, Otto SP, Bergman A, Feldman MW. 1998. Waiting with and without recombination: the time to production of a double mutant. Theor. Popul. Biol. 53, 199–215. ( 10.1006/tpbi.1997.1358) [DOI] [PubMed] [Google Scholar]
- 26.Muller HJ. 1964. The relation of recombination to mutational advance. Mutat. Res. Mol. Mech. Mutagen. 1, 2–9. ( 10.1016/0027-5107(64)90047-8) [DOI] [PubMed] [Google Scholar]
- 27.Kimura M, Maruyama T. 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54, 1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondrashov AS. 1982. Selection against harmful mutations in large sexual and asexual populations. Genet. Res. 40, 325–332. ( 10.1017/S0016672300019194) [DOI] [PubMed] [Google Scholar]
- 29.Kondrashov AS. 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336, 435–440. ( 10.1038/336435a0) [DOI] [PubMed] [Google Scholar]
- 30.Lynch M, Gabriel W. 1990. Mutation load and the survival of small populations. Evolution 44, 1725–1737. ( 10.1111/j.1558-5646.1990.tb05244.x) [DOI] [PubMed] [Google Scholar]
- 31.Nei M. 1967. Modification of linkage intensity by natural selection. Genetics 57, 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nei M. 1969. Linkage modification and sex difference in recombination. Genetics 63, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher RA. 1919. XV.—The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52, 399–433. ( 10.1017/S0080456800012163) [DOI] [Google Scholar]
- 34.Wade MJ. 1992. Epistasis. In Keywords in evolutionary biology (eds EF Keller, EA Lloyd), pp. 87–91. Cambridge, MA: Harvard University Press. [Google Scholar]
- 35.Phillips PC. 1998. The language of gene interaction. Genetics 149, 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. 1965. The effect of linkage on directional selection. Genetics 52, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto SP, Michalakis Y. 1998. The evolution of recombination in changing environments. Trends Ecol. Evol. 13, 145–151. ( 10.1016/S0169-5347(97)01260-3) [DOI] [PubMed] [Google Scholar]
- 38.Peters AD, Lively CM. 2000. Epistasis and the maintenance of sex. In Epistasis and the evolutionary process (eds Wolf JB, Brodie ED III, Wade MJ.), pp. 99–112. Oxford, UK: Oxford University Press. [Google Scholar]
- 39.Barton NH, Otto SP. 2005. Evolution of recombination due to random drift. Genetics 169, 2353–2370. ( 10.1534/genetics.104.032821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barton NH, Charlesworth B. 1998. Why sex and recombination? Science 281, 1986–1990. ( 10.1126/science.281.5385.1986) [DOI] [PubMed] [Google Scholar]
- 41.Otto SP, Lenormand T. 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3, 252–261. ( 10.1038/nrg761) [DOI] [PubMed] [Google Scholar]
- 42.Feldman MW, Christiansen FB, Brooks LD. 1980. Evolution of recombination in a constant environment. Proc. Natl Acad. Sci. USA 77, 4838–4841. ( 10.1073/pnas.77.8.4838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldman MW, Liberman U. 1986. An evolutionary reduction principle for genetic modifiers. Proc. Natl Acad Sci. USA 83, 4824–4827. ( 10.1073/pnas.83.13.4824) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto SP, Feldman MW. 1997. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol. 51, 134–147. ( 10.1006/tpbi.1997.1301) [DOI] [PubMed] [Google Scholar]
- 45.Kouyos RD, Silander OK, Bonhoeffer S. 2007. Epistasis between deleterious mutations and the evolution of recombination. Trends Ecol. Evol. 22, 308–315. ( 10.1016/j.tree.2007.02.014) [DOI] [PubMed] [Google Scholar]
- 46.Charlesworth B. 1990. Mutation–selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55, 199–221. ( 10.1017/S0016672300025532) [DOI] [PubMed] [Google Scholar]
- 47.Charlesworth B. 1976. Recombination modification in a fluctuating environment. Genetics 83, 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters AD, Lively CM. 1999. The red queen and fluctuating epistasis: a population genetic analysis of antagonistic coevolution. Am. Nat. 154, 393–405. ( 10.1086/303247) [DOI] [PubMed] [Google Scholar]
- 49.Gandon S, Otto SP. 2007. The evolution of sex and recombination in response to abiotic or coevolutionary fluctuations in epistasis. Genetics 175, 1835–1853. ( 10.1534/genetics.106.066399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondrashov AS, Yampolsky LY. 1996. High genetic variability under the balance between symmetric mutation and fluctuating stabilizing selection. Genet. Res. 68, 157–164. ( 10.1017/S0016672300034042) [DOI] [Google Scholar]
- 51.Nee S. 1989. Antagonistic co-evolution and the evolution of genotypic randomization. J. Theor. Biol. 140, 499–518. ( 10.1016/S0022-5193(89)80111-0) [DOI] [PubMed] [Google Scholar]
- 52.Dapper AL, Lively CM. 2014. Interlocus sexually antagonistic coevolution can create indirect selection for increased recombination. Evolution 68, 1216–1224. ( 10.1111/evo.12338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortiz-Barrientos D, Engelstädter J, Rieseberg LH. 2016. Recombination rate evolution and the origin of species. Trends Ecol. Evol. 31, 226–236. ( 10.1016/j.tree.2015.12.016) [DOI] [PubMed] [Google Scholar]
- 54.Reeve J, Ortiz-Barrientos D, Engelstädter J. 2016. The evolution of recombination rates in finite populations during ecological speciation. Proc. R. Soc. B 283, 20161243 ( 10.1098/rspb.2016.1243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill WG, Robertson A. 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, 226–231. ( 10.1007/BF01245622) [DOI] [PubMed] [Google Scholar]
- 56.Keightley PD, Otto SP. 2006. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 443, 89–92. ( 10.1038/nature05049) [DOI] [PubMed] [Google Scholar]
- 57.Otto SP, Barton NH. 2001. Selection for recombination in small populations. Evolution 55, 1921–1931. ( 10.1111/j.0014-3820.2001.tb01310.x) [DOI] [PubMed] [Google Scholar]
- 58.Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8, 269–294. ( 10.1017/S0016672300010156) [DOI] [PubMed] [Google Scholar]
- 59.Anderson LK, Reeves A, Webb LM, Ashley T. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151, 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haldane JBS. 1919. The combination of linkage values and the calculation of distances between the loci of linked factors. J. Genet. 8, 299–309. [Google Scholar]
- 61.Holloway K, Lawson VE, Jeffreys AJ. 2006. Allelic recombination and de novo deletions in sperm in the human β-globin gene region. Hum. Mol. Genet. 15, 1099–1111. ( 10.1093/hmg/ddl025) [DOI] [PubMed] [Google Scholar]
- 62.McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304, 581–584. ( 10.1126/science.1092500) [DOI] [PubMed] [Google Scholar]
- 63.Gowen JW. 1919. A biometrical study of crossing over. On the mechanism of crossing over in the third chromosome of Drosophila melanogaster. Genetics 4, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lawrence MJ. 1958. Genotypic control of crossing-over on the first chromosome of Drosophila melanogaster. Nature 182, 889–890. ( 10.1038/182889a0) [DOI] [PubMed] [Google Scholar]
- 65.Kidwell MG. 1972. Genetic change of recombination value in Drosophila melanogaster. II. Simulated natural selection. Genetics 70, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brooks LD, Marks RW. 1986. The organization of genetic variation for recombination in Drosophila melanogaster. Genetics 114, 525–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. 1998. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 63, 861–869. ( 10.1086/302011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ. 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296, 2222–2225. ( 10.1126/science.1071220) [DOI] [PubMed] [Google Scholar]
- 69.Kong A, et al. 2004. Recombination rate and reproductive success in humans. Nat. Genet. 36, 1203–1206. ( 10.1038/ng1445) [DOI] [PubMed] [Google Scholar]
- 70.Neumann R, Jeffreys AJ. 2006. Polymorphism in the activity of human crossover hotspots independent of local DNA sequence variation. Hum. Mol. Genet. 15, 1401–1411. ( 10.1093/hmg/ddl063) [DOI] [PubMed] [Google Scholar]
- 71.Bois PR. 2007. A highly polymorphic meiotic recombination mouse hot spot exhibits incomplete repair. Mol. Cell. Biol. 27, 7053–7062. ( 10.1128/MCB.00874-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SHS, Graber JH, Broman KW, Petkov PM.. 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet. 4, e1000119 ( 10.1371/journal.pgen.1000119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hassold T, Hansen T, Hunt P, VandeVoort C. 2009. Cytological studies of recombination in rhesus males. Cytogenet. Genome Res. 124, 132–138. ( 10.1159/000207519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Comeron JM, Ratnappan R, Bailin S. 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8, e1002905 ( 10.1371/journal.pgen.1002905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gruhn JR, Al-Asmar N, Fasnacht R, Maylor-Hagen H, Peinado V, Rubio C, Broman KW, Hunt PA, Hassold T. 2016. Correlations between synaptic initiation and meiotic recombination: a study of humans and mice. Am. J. Hum. Genet. 98, 102–115. ( 10.1016/j.ajhg.2015.11.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunter CM, Huang W, Mackay TF, Singh ND. 2016. The genetic architecture of natural variation in recombination rate in Drosophila melanogaster. PLoS Genet. 12, e1005951 ( 10.1371/journal.pgen.1005951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Levine RP, Levine EE. 1954. The genotypic control of crossing over in Drosophila pseudoobscura. Genetics 39, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang RJ, Gray MM, Parmenter MD, Broman KW, Payseur BA. 2017. Recombination rate variation in mice from an isolated island. Mol. Ecol. 26, 457–470. ( 10.1111/mec.13932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.True JR, Mercer JM, Laurie CC. 1996. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics 142, 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winckler W, et al. 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308, 107–111. ( 10.1126/science.1105322) [DOI] [PubMed] [Google Scholar]
- 81.Stevison LS, Noor MA. 2010. Genetic and evolutionary correlates of fine-scale recombination rate variation in Drosophila persimilis. J. Mol. Evol. 71, 332–345. ( 10.1007/s00239-010-9388-1) [DOI] [PubMed] [Google Scholar]
- 82.Auton A, et al. 2012. A fine-scale chimpanzee genetic map from population sequencing. Science 336, 193–198. ( 10.1126/science.1216872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stevison LS, Woerner AE, Kidd JM, Kelley JL, Veeramah KR, McManus KF, Bustamante CD, Hammer MF, Wall JD. 2016. The time scale of recombination rate evolution in great apes. Mol. Biol. Evol. 33, 928–945. ( 10.1093/molbev/msv331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ptak SE, Roeder AD, Stephens M, Gilad Y, Pääbo S, Przeworski M.. 2004. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2, e155 ( 10.1371/journal.pbio.0020155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dumont BL, White MA, Steffy B, Wiltshire T, Payseur BA. 2011. Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome Res. 21, 114–125. ( 10.1101/gr.111252.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dumont BL, Payseur BA. 2008. Evolution of the genomic rate of recombination in mammals. Evolution 62, 276–294. ( 10.1111/j.1558-5646.2007.00278.x) [DOI] [PubMed] [Google Scholar]
- 87.Dumont BL, Payseur BA. 2011. Evolution of the genomic recombination rate in murid rodents. Genetics 187, 643–657. ( 10.1534/genetics.110.123851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Segura J. et al 2013. Evolution of recombination in eutherian mammals: insights into mechanisms that affect recombination rates and crossover interference. Proc. R. Soc. B 280, 20131945 ( 10.1098/rspb.2013.1945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burt A, Bell G. 1987. Mammalian chiasma frequencies as a test of two theories of recombination. Nature 326, 803–805. ( 10.1038/326803a0) [DOI] [PubMed] [Google Scholar]
- 90.Ross-Ibarra J. 2004. The evolution of recombination under domestication: a test of two hypotheses. Am. Nat. 163, 105–112. ( 10.1086/380606) [DOI] [PubMed] [Google Scholar]
- 91.Wilfert L, Gadau J, Schmid-Hempel P. 2007. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity 98, 189–197. ( 10.1038/sj.hdy.6800950) [DOI] [PubMed] [Google Scholar]
- 92.Muñoz-Fuentes V, et al. 2015. Strong artificial selection in domestic mammals did not result in an increased recombination rate. Mol. Biol. Evol. 32, 510–523. ( 10.1093/molbev/msu322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Visser JAG, Elena SF. 2007. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat. Rev. Genet. 8, 139–149. ( 10.1038/nrg1985) [DOI] [PubMed] [Google Scholar]
- 94.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319, 1395–1398. ( 10.1126/science.1151851) [DOI] [PubMed] [Google Scholar]
- 95.Fledel-Alon A, Leffler EM, Guan Y, Stephens M, Coop G, Przeworski M. 2011. Variation in human recombination rates and its genetic determinants. PLoS ONE 6, e20321 ( 10.1371/journal.pone.0020321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ. 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baier B, Hunt P, Broman KW, Hassold T. 2014. Variation in genome-wide levels of meiotic recombination is established at the onset of prophase in mammalian males. PLoS Genet. 10, e1004125 ( 10.1371/journal.pgen.1004125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Detlefsen JA, Roberts E. 1921. Studies on crossing over. I. The effect of selection on crossover values. J. Exp. Zool. Part Ecol. Genet. Physiol. 32, 333–354. ( 10.1002/jez.1400320206) [DOI] [Google Scholar]
- 99.Valentin J. 1973. Heritability of recombination frequency. Hereditas 75, 1–4. ( 10.1111/j.1601-5223.1973.tb01136.x) [DOI] [PubMed] [Google Scholar]
- 100.Dewees AA. 1975. Genetic modification of recombination rate in Tribolium castaneum. Genetics 81, 537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ebinuma H, Yoshitake N. 1981. The genetic system controlling recombination in the silkworm. Genetics 99, 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kidwell MG. 1972. Genetic change of recombination value in Drosophila melanogaster. I. Artificial selection for high and low recombination and some properties of recombination-modifying genes. Genetics 70, 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pritchard RH. 1955. The linear arrangement of a series of alleles of Aspergillus nidulans. Heredity 9, 343–371. ( 10.1038/hdy.1955.37) [DOI] [Google Scholar]
- 104.Calef E. 1957. Effect on linkage maps of selection of crossovers between closely linked markers. Heredity 11, 265–279. ( 10.1038/hdy.1957.23) [DOI] [Google Scholar]
- 105.Parsons PA. 1958. Selection for increased recombination in Drosophila melanogaster. Am. Nat. 92, 255–256. ( 10.1086/282033) [DOI] [Google Scholar]
- 106.Acton AB. 1961. An unsuccessful attempt to reduce recombination by selection. Am. Nat. 95, 119–120. ( 10.1086/282166) [DOI] [Google Scholar]
- 107.Mukherjee AS. 1961. Effect of selection on crossing over in the males of Drosophila ananassae. Am. Nat. 95, 57–59. ( 10.1086/282157) [DOI] [Google Scholar]
- 108.Allard RW. 1963. Evidence for genetic restriction of recombination in the lima bean. Genetics 48, 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kale PG. 1968. Spontaneous crossing over in the males of Drosophila ananassae: two-way selection for recombination values. Jpn. J. Genet. 43, 27–31. ( 10.1266/jjg.43.27) [DOI] [Google Scholar]
- 110.Abdullah NF, Charlesworth B. 1974. Selection for reduced crossing over in Drosophila melanogaster. Genetics 76, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Charlesworth B, Charlesworth D. 1985. Genetic variation in recombination in Drosophila. I. Responses to selection and preliminary genetic analysis. Heredity 54, 71–83. ( 10.1038/hdy.1985.10) [DOI] [Google Scholar]
- 112.Shaw DD. 1972. Genetic and environmental components of chiasma control. Chromosoma 37, 297–308. ( 10.1007/BF00319872) [DOI] [PubMed] [Google Scholar]
- 113.Chinnici JP. 1971. Modification of recombination frequency in Drosophila. I. Selection for increased and decreased crossing over. Genetics 69, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hadad RG, Pfeiffer TW, Poneleit CG. 1996. Repeatability and heritability of divergent recombination frequencies in the Iowa Stiff Stalk Synthetic (Zea mays L.). Theor. Appl. Genet. 93, 990–996. ( 10.1007/BF00224103) [DOI] [PubMed] [Google Scholar]
- 115.Dumont BL, Broman KW, Payseur BA. 2009. Variation in genomic recombination rates among heterogeneous stock mice. Genetics 182, 1345–1349. ( 10.1534/genetics.109.105114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johnston SE, Bérénos C, Slate J, Pemberton JM. 2016. Conserved genetic architecture underlying individual recombination rate variation in a wild population of Soay sheep (Ovis aries). Genetics 203, 583–598. ( 10.1534/genetics.115.185553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fledel-Alon A, Wilson DJ, Broman K, Wen X, Ober C, Coop G, Przeworski M.. 2009. Broad-scale recombination patterns underlying proper disjunction in humans. PLoS Genet. 5, e1000658 ( 10.1371/journal.pgen.1000658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stefansson H, et al. 2005. A common inversion under selection in Europeans. Nat. Genet. 37, 129–137. ( 10.1038/ng1508) [DOI] [PubMed] [Google Scholar]
- 119.Oliver PL, et al. 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5, e1000753 ( 10.1371/journal.pgen.1000753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ponting CP. 2011. What are the genomic drivers of the rapid evolution of PRDM9? Trends Genet. 27, 165–171. ( 10.1016/j.tig.2011.02.001) [DOI] [PubMed] [Google Scholar]
- 121.Mather K. 1938. Crossing-over. Biol. Rev. 13, 252–292. ( 10.1111/j.1469-185X.1938.tb00516.x) [DOI] [Google Scholar]
- 122.Dumont BL. 2017. Variation and evolution of the meiotic requirement for crossing over in mammals. Genetics 205, 155–168. ( 10.1534/genetics.116.192690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pardo-Manuel de Villena F, Sapienza C. 2001. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome 12, 318–322. ( 10.1007/s003350020005) [DOI] [PubMed] [Google Scholar]
- 124.Dutrillaux B. 1986. The evolutionary role of chromosomes—a new interpretation. Sem. Hop. 62, 3249–3255. [Google Scholar]
- 125.Spitze K. 1993. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135, 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prout T, Barker JS. 1993. F statistics in Drosophila buzzatii: selection, population size and inbreeding. Genetics 134, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kong A, et al. 2008. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319, 1398–1401. ( 10.1126/science.1152422) [DOI] [PubMed] [Google Scholar]
- 128.Kong A, et al. 2014. Common and low-frequency variants associated with genome-wide recombination rate. Nat. Genet. 46, 11–16. ( 10.1038/ng.2833) [DOI] [PubMed] [Google Scholar]
- 129.Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. 2009. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 5, e1000648 ( 10.1371/journal.pgen.1000648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sandor C, Li W, Coppieters W, Druet T, Charlier C, Georges M. 2012. Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS Genet. 8, e1002854 ( 10.1371/journal.pgen.1002854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ma L, et al. 2015. Cattle sex-specific recombination and genetic control from a large pedigree analysis. PLoS Genet. 11, e1005387 ( 10.1371/journal.pgen.1005387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dumont BL, Payseur BA. 2011. Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet. 7, e1002116 ( 10.1371/journal.pgen.1002116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Murdoch B, Owen N, Shirley S, Crumb S, Broman KW, Hassold T. 2010. Multiple loci contribute to genome-wide recombination levels in male mice. Mamm. Genome 21, 550–555. ( 10.1007/s00335-010-9303-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Balcova M, et al. 2016. Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLoS Genet. 12, e1005906 ( 10.1371/journal.pgen.1005906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smukowski-Heil CS, Ellison C, Dubin M, Noor MAF. 2015. Recombining without hotspots: a comprehensive evolutionary portrait of recombination in two closely related species of Drosophila. Genome Biol. Evol. 7, 2829–2842. ( 10.1093/gbe/evv182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chan AH, Jenkins PA, Song YS. 2012. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 8, e1003090 ( 10.1371/journal.pgen.1003090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Pääbo S. 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37, 429–434. ( 10.1038/ng1529) [DOI] [PubMed] [Google Scholar]
- 138.Backström N, et al. 2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 20, 485–495. ( 10.1101/gr.101410.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meznar ER, Gadau J, Koeniger N, Rueppell O. 2010. Comparative linkage mapping suggests a high recombination rate in all honeybees. J. Hered. 101, S118–S126. ( 10.1093/jhered/esq002) [DOI] [PubMed] [Google Scholar]
- 140.McGaugh SE, Heil CSS, Manzano-Winkler B, Loewe L, Goldstein S, Himmel TL, Noor MAF. 2012. Recombination modulates how selection affects linked sites in Drosophila. PLoS Biol. 10, e1001422 ( 10.1371/journal.pbio.1001422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Singhal S, et al. 2015. Stable recombination hotspots in birds. Science 350, 928–932. ( 10.1126/science.aad0843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lam I, Keeney S. 2015. Nonparadoxical evolutionary stability of the recombination initiation landscape in yeast. Science 350, 932–937. ( 10.1126/science.aad0814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lichten M. 2015. Putting the breaks on meiosis. Science 350, 913 ( 10.1126/science.aad5404) [DOI] [PubMed] [Google Scholar]
- 144.Ubeda F, Wilkins JF. 2011. The Red Queen theory of recombination hotspots. J. Evol. Biol. 24, 541–553. ( 10.1111/j.1420-9101.2010.02187.x) [DOI] [PubMed] [Google Scholar]
- 145.Plough HH. 1917. The effect of temperature on crossingover in Drosophila. J. Exp. Zool. Part Ecol. Genet. Physiol. 24, 147–209. ( 10.1002/jez.1400240202) [DOI] [Google Scholar]
- 146.Law CN. 1963. An effect of potassium on chiasma frequency and recombination. Genetica 33, 313–329. ( 10.1007/BF01725768) [DOI] [Google Scholar]
- 147.Nolte DJ. 1968. The chiasma-inducing pheromone of locusts. Chromosoma 23, 346–358. ( 10.1007/BF02451006) [DOI] [PubMed] [Google Scholar]
- 148.Bennett MD, Rees H. 1970. Induced variation in chiasma frequency in rye in response to phosphate treatments. Genet. Res. 16, 325–331. ( 10.1017/S0016672300002585) [DOI] [Google Scholar]
- 149.Henderson SA. 1988. Four effects of elevated temperature on chiasma formation in the locust Schistocerca gregaria. Heredity 60, 387–401. ( 10.1038/hdy.1988.57) [DOI] [Google Scholar]
- 150.Bomblies K, Higgins JD, Yant L. 2015. Meiosis evolves: adaptation to external and internal environments. New Phytol. 208, 306–323. ( 10.1111/nph.13499) [DOI] [PubMed] [Google Scholar]
- 151.Grant V. 1953. Cytogenetics of the hybrid Gilia millefoliata×achilleaefolia. Chromosoma 5, 372–390. ( 10.1007/BF01271494) [DOI] [PubMed] [Google Scholar]
- 152.Stevison L, Sefick S, Rushton C, Graze R. 2017. Recombination rate plasticity: revealing mechanisms by design. Phil. Trans. R. Soc. B 372, 20160459 ( 10.1098/rstb.2016.0459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Singh ND, Criscoe DR, Skolfield S, Kohl KP, Keebaugh ES, Schlenke TA. 2015. Fruit flies diversify their offspring in response to parasite infection. Science 349, 747–750. ( 10.1126/science.aab1768) [DOI] [PubMed] [Google Scholar]
- 154.Hadany L, Beker T. 2003. On the evolutionary advantage of fitness-associated recombination. Genetics 165, 2167–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Agrawal AF, Hadany L, Otto SP. 2005. The evolution of plastic recombination. Genetics 171, 803–812. ( 10.1534/genetics.105.041301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Burt A, Bell G, Harvey PH. 1991. Sex differences in recombination. J. Evol. Biol. 4, 259–277. ( 10.1046/j.1420-9101.1991.4020259.x) [DOI] [Google Scholar]
- 157.Huxley JS. 1928. Sexual difference of linkage in Gammarus chevreuxi. J. Genet. 20, 145–156. ( 10.1007/BF02983136) [DOI] [Google Scholar]
- 158.Haldane JB. 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12, 101–109. ( 10.1007/BF02983075) [DOI] [Google Scholar]
- 159.Bell G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. Berkeley, CA: University of California Press, Google Scholar. [Google Scholar]
- 160.Lenormand T, Dutheil J.. 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3, e63 ( 10.1371/journal.pbio.0030063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brandvain Y, Coop G. 2012. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics 190, 709–723. ( 10.1534/genetics.111.136721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Charlesworth D. 2017. Evolution of recombination rates between sex chromosomes. Phil. Trans. R. Soc. B 372, 20160456 ( 10.1098/rstb.2016.0456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kong A, et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31, 241–247. ( 10.1038/ng917) [DOI] [PubMed] [Google Scholar]
- 164.Kochakpour N, Moens PB. 2008. Sex-specific crossover patterns in Zebrafish (Danio rerio). Heredity 100, 489–495. ( 10.1038/sj.hdy.6801091) [DOI] [PubMed] [Google Scholar]
- 165.Cox A, et al. 2009. A new standard genetic map for the laboratory mouse. Genetics 182, 1335–1344. ( 10.1534/genetics.109.105486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu EY, Morgan AP, Chesler EJ, Wang W, Churchill GA, de Villena FP-M. 2014. High-resolution sex-specific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics 197, 91–106. ( 10.1534/genetics.114.161653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Smeds L, Mugal CF, Qvarnström A, Ellegren H. 2016. High-resolution mapping of crossover and non-crossover recombination events by whole-genome re-sequencing of an avian pedigree. PLoS Genet. 12, e1006044 ( 10.1371/journal.pgen.1006044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lenormand T. 2003. The evolution of sex dimorphism in recombination. Genetics 163, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Trivers R. 1988. Sex differences in rates of recombination and sexual selection. In The evolution of sex: an examination of current ideas (eds Michod RE, Levin BR), pp. 270–286. Sunderland, MA: Sinauer Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.