Abstract
Identifying the genetic bases for colour patterns has provided important insights into the control and expression of pigmentation and how these characteristics influence fitness. However, much more is known about the genetic bases for traits based on melanin pigments than for traits based on another major class of pigments, carotenoids. Here, we use natural admixture in a hybrid zone between Audubon's and myrtle warblers (Setophaga coronata auduboni/S. c. coronata) to identify genomic regions associated with both types of pigmentation. Warblers are known for rapid speciation and dramatic differences in plumage. For each of five plumage coloration traits, we found highly significant associations with multiple single-nucleotide polymorphisms (SNPs) across the genome and these were clustered in discrete regions. Regions near significantly associated SNPs were enriched for genes associated with keratin filaments, fibrils that make up feathers. A carotenoid-based trait that differs between the taxa—throat colour—had more than a dozen genomic regions of association. One cluster of SNPs for this trait overlaps the Scavenger Receptor Class F Member 2 (SCARF2) gene. Other scavenger receptors are presumed to be expressed at target tissues and involved in the selective movement of carotenoids into the target cells, making SCARF2 a plausible new candidate for carotenoid processing. In addition, two melanin-based plumage traits—colours of the eye line and eye spot—show very strong associations with a single genomic region mapping to chromosome 20 in the zebra finch. These findings indicate that only a subset of the genomic regions differentiated between these two warblers are associated with the plumage differences between them and demonstrate the utility of reduced-representation genomic scans in hybrid zones.
Keywords: carotenoid, association mapping, scavenger receptor, pigmentation, hybrid zone, Setophaga coronata
1. Introduction
Coloration is among the most conspicuous phenotypes to differ among closely related species [1,2]. Using genetic crosses, association studies have identified a number of key genes involved in pigmentation pathways that give rise to such colour differences [2–5]. Identifying the genetic bases for pigmentation traits has provided important functional insights into the control and expression of animal pigmentation [6,7] and how these characteristics may subsequently influence fitness [8,9]. It has also facilitated key insights about the broader evolutionary patterns of pigmentation characters, such as how these traits are gained or lost [10,11], how often similar genes are used and how selection has shaped molecular variation at and around pigmentation genes [12].
In animals, this research has primarily focused on identifying the genes involved in the melanogenesis pathway, which produces melanin responsible for black or brown coloration [3,13,14]. However, in many species, particularly in birds, differences in red, orange and yellow coloration usually arise from variation in the distribution of carotenoid-derived pigments, the precursors of which cannot be synthesized endogenously and hence must be acquired from food [4]. We know much less about the genetic basis of carotenoid coloration, and only recently have candidate genes for carotenoid processing and transport been elucidated in animals [15–17]. These advances have largely been facilitated by anonymous genome scans, which avoid the need to focus on previously identified candidate genes (e.g. [15,18]).
Here, we use genomic data and admixture mapping to identify genomic regions (and possible candidate genes) involved in both melanin and carotenoid-based coloration traits, pigmentation molecules that are rarely studied in parallel in the same system. We focus on two New World warblers in North America that are part of a continental avian radiation that express a diversity of colours, presumably the result of strong, divergent sexual selection on different plumage patches in different species [19].
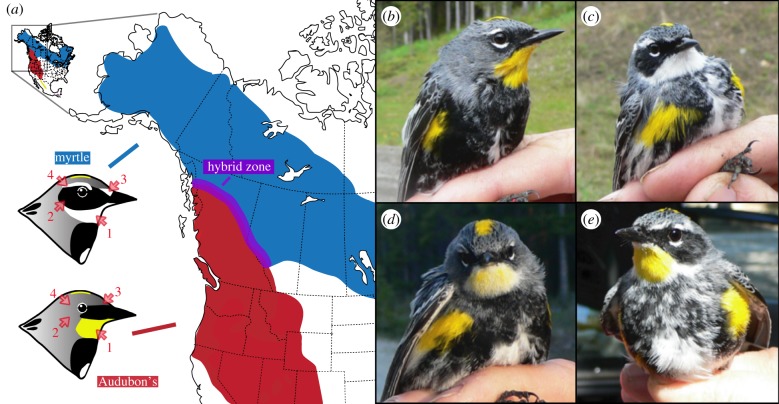
We take advantage of natural hybridization between two phenotypically distinct warblers—Audubon's (Setophaga coronata auduboni) and myrtle warblers (S. c. coronata)—to identify genotype–phenotype associations (we note the two groups are currently treated as a single species also called the ‘yellow-rumped warbler’ by the North American Classification Committee). Male breeding-season plumage is very different between the warblers, with melanistic differences in colour pattern in the face and wings (black or grey melanized feathers versus white non-melanized feathers) and carotenoid-based differences in their throats (yellow versus white feathers; figure 1; [20]).
Figure 1.
Breeding ranges and phenotypes. (a) Breeding range distributions for myrtle (blue) and Audubon's (red) warblers. The schematic also shows four of the five plumage characters quantified: (1) white versus yellow throat, (2) black versus grey auricular, (3) white versus grey eye spot, and (4) white versus grey eye line. The fifth plumage trait—the extent of white on the wing bars—is shown in figure 2. (b–e) Representative photos of individuals with pure parental phenotypes (b and c) and admixed individuals (d and e). (Online version in colour.)
Hybridization is extensive where the taxa co-occur and hybrids exhibit a diverse combination of the plumage traits of both parental taxa (figure 1d,e). Previous studies of the hybrid zone analysed colour and morphological variation and speculated that the zone was stable and maintained by selection against hybrids [21], which was later confirmed by using linkage disequilibrium between diagnostic genetic markers [22]. Social pairs do not appear to show assortative mating based on coloration [22], although it is unclear whether hybrid males with mixed plumage characters face any fitness costs in defending territories. While coloration is one of the most conspicuous differences between myrtle and Audubon's warblers, the two forms also differ in their vocalizations, ecology, migratory behaviour and genetic signatures [20,23–26].
The hybrid zone between these two warblers provides an ideal system to investigate the genetic architecture of plumage differences and to perform admixture mapping—the association of genetic variants with phenotypic traits across hybrid offspring. Much of the power of admixture mapping depends on sampling individuals across the full spectrum of ancestry. The distribution of hybrid phenotypes in the myrtle–Audubon's hybrid zone is close to the uniform distribution suggested to be ideal for admixture mapping [22,27]. Admixture mapping also depends on the structure of linkage disequilibrium in hybrids. If too many generations have passed since the admixture event, then blocks of the genome derived from either parent species will be broken down by recombination, and extremely high marker densities will be required to detect associations with phenotype. On the other hand, too few generations of post-admixture recombination will leave large blocks of linkage disequilibrium, making fine-scale mapping difficult. With little or no assortative mating, such that hybrids are continuously produced, and with moderate selection against hybrid combinations [22], the myrtle–Audubon's hybrid zone presents an intermediate level of linkage disequilibrium appropriate for association mapping with a moderate single-nucleotide polymorphism (SNP) density.
Here, we sampled hundreds of individuals from across the myrtle–Audubon's hybrid zone and quantified phenotypic variation across five plumage patches: throat colour, eye spot, eye line, auricular and wing pattern (figure 1). We used genotyping-by-sequencing to survey genomic variation, and tested for associations between SNP genotypes and plumage phenotypes, while controlling for background levels of hybrid ancestry. Our goal was to quantify the genomic substrate that leads to phenotypic divergence between these warblers, in this case by identifying regions of the genome significantly associated with plumage traits and possible candidate genes within these regions. Several recent anonymous genome scans have found differentiation peaks between closely related avian taxa disproportionately populated with pigmentation genes [18,28,29]. Yet, these studies have been focused on taxon comparisons with exceptionally low levels of genome-wide divergence. Therefore, the Audubon's and myrtle warbler system—with a larger number and moderate size of differentiation peaks [25]—provides an important complement to these recent studies and allows us to quantify the proportion of FST peaks (clusters of highly differentiated SNPs) that are also significantly associated with different plumage traits between more highly differentiated taxa.
With these data, we also aimed to test several more specific predictions based on previous genetic studies of avian plumage and pigmentation. First, given that sex chromosomes have been identified in previous research as often being associated with avian plumage traits [30–32], we predicted the Z chromosome might play a disproportionately large role in plumage differences between myrtle and Audubon's warblers. Second, we tested whether distinct plumage traits had associations that co-localized in specific genomic regions, due to either pleiotropy or close linkage of multiple genes (i.e. supergenes; [33,34]). We first tested whether genetic associations for different melanin-based feather patches co-localized, and then asked whether traits that differ in the pigment molecule (i.e. carotenoids versus melanin) had overlapping or distinct association patterns.
2. Material and methods
(a). Sampling methods and plumage scoring
We captured 203 male Audubon's and myrtle warblers on their breeding territories using song playback and mist nets. From each bird, we took a small blood sample from the brachial vein and stored it in Queen's lysis buffer [35]. We focused our sampling on birds within the myrtle–Audubon's hybrid zone (n = 151), but also included some individuals from outside the myrtle–Audubon's hybrid zone (n = 38 myrtle warblers and n = 14 Audubon's warblers). We included individuals that were sampled from across five widely geographically separate transects surveyed for [22], with the number of individuals per transect ranging from 13 (Kananaskis, Alberta) to 49 (Pine Pass, British Columbia). We scored plumage characters as described in [22], relying on the original plumage criteria as outlined by [21]. We focused on the five characters (figure 1) that were identified by [21] as the most repeatable and showed the most between-group differences. We did not include the extent of white on the tail, which has been used as a diagnosable character, as it also shows large age-related differences in addition to subspecies differences [21].
We chose to genotype individuals that exhibited high within-individual variance across the five plumage traits (i.e. focusing on admixed individuals that are Audubon's-like for some colour traits and myrtle-like for others). We did this to maximize our power to detect loci associated with each trait separately—individuals that are intermediate for all colour traits are less informative for the genetic basis of each trait. This is conceptually similar to quantitative trait locus studies that focus on offspring in the high-variance individuals in the F2 generation, as opposed to F1 offspring.
(b). Sequencing library construction
We used a restriction-site-associated DNA sequencing method [36,37] and detailed library construction details can be found in [25]. Briefly, we performed PCR for each sample separately: each reaction was prepared on ice and included: 0.5 units of PhusionTaq (New England Biolabs), 5 µl of 5× Phusion Buffer, 0.5 µl of 10 µM dNTPs, 0.125 µl of forward and reverse GBS primers (200 µM; see [36] for sequences), 18 µl of UltraPure water and 1 µl of the cleaned DNA fragments from the digestion–ligation reaction. For the PCR, we used the following thermocycling profile: 98°C for 30 s followed by 20 cycles at 98°C for 10 s, 65°C for 30 s and 72°C for 30 s. This was followed by an extension at 72°C for 5 min. We then fluorometrically quantified the product and added equimolar amounts of each sample to a pool. Pooled samples were run on agarose, and 300–400 bp size fragments were excised using a Qiagen gel extraction kit. This size range of each pool was confirmed using a high sensitivity Bioanalyzer chip (Agilent Technologies) and quantified using qPCR. The final libraries were paired-end sequenced with 100 bp read length, across three lanes of an Illumina HiSeq 2000 with 96 individuals per lane.
(c). Data analysis and single-nucleotide polymorphism calling
We demultiplexed sequencing reads using the barcode-splitting program ‘Sabre’ (https://github.com/najoshi/sabre). We allowed for one mismatch in the barcode + enzyme cut-site sequence. We then used AdapterRemoval (v. 1.5.4; [38]) to merge overlapping paired reads. We used BOWTIE2 (v. 2.2.8; [39]) to map each of the individual reads to the myrtle warbler genome [18]. For this mapping, we used the ‘very sensitive local’ set of alignment pre-sets. For SNP discovery and variant calling, we used the UnifiedGenotyper in GATK [40] and followed [41] for GATK ‘best practices' as a guideline. We removed possible variants that had ‘quality by depth’ (QD) of less than 2 and ‘mapping quality’ (MQ) of less than 30. We applied additional filters using the program VCFtools [42]. First, we coded genotypes with a Phred-scaled quality lower than 20 as missing data, which corresponds to a genotyping accuracy of at least 99%. We retained only bi-allelic loci and excluded loci with more than 80% missing data and a minor allele frequency of less than 1%.
(d). Phenotype–genotype analysis
We used the genome-wide efficient mixed model association algorithm (GEMMA; [43]) to associate variation at SNPs with the five plumage traits. GEMMA requires complete or imputed SNP data. Therefore, we first used BEAGLE (v. 4.1; [44]) to impute missing data for those sites with missing data. We retained pre-imputed loci with high levels of missing data to increase marker density, at the cost of genotype accuracy at sites with high levels of imputed genotypes. We were, however, concerned that imputed data at missing sites could influence our results, so we also conducted the GEMMA analysis with a subset of SNPs that had high estimated correlations between imputed and true genotypes at a given SNP (i.e. AR2 > 0.8; electronic supplementary material, figures S1 and S2) and report this analysis separately. AR2 (or allele R2) is the estimated correlation between imputed and (unknown) true genotypes. The goal with this additional analysis was to focus on the SNPs imputed with high certainty to compare differences with this analysis with those from the full dataset.
We transformed our data into binary PLINK BED format [45] using PGDSpider (2.1.0.3; [46]). The analysis in GEMMA estimates a relatedness matrix, which we inspected to confirm that the between-taxon allopatric comparisons showed much lower levels of relatedness than sympatric comparisons did. We used the Wald test to identify significant associations between SNP genotypes and the plumage traits and report the p-values using five univariate linear mixed models. GEMMA uses the relatedness information to account for population stratification and sample structure (in this case related to overall background hybrid ancestry). We used two conservative α-values to judge statistical significance of associations (α = 0.0001 and −log10(α) = 4; or α = 0.000001 and −log10(α) = 6; we focused our interpretation on the more stringent, smaller value of α). To visualize the resulting associations, we used a modified version of the ‘qqman’ package in R to generate Manhattan plots [47]. To quantify co-localization of patterns of divergence with candidate plumage regions, we estimated FST [48] by comparing a subset of the allopatric samples using VCFtools [42].
(e). Functional characterization
To provide insight into the genes associated with plumage differences, we used annotation information at several levels. First, we identified genes containing or located close to SNPs significantly associated with the five plumage characters. We relied on the annotation of the myrtle warbler genome [18], which used gene annotation data from the zebra finch genome [49]. We identified all of the features marked as genes in the annotation file that contained at least one SNP associated with a plumage trait, with a significance cut-off of −log10(p) > 6. We also did the same search, but with a 20 kb buffer around each significant SNP. We used this buffer to highlight potential genes that had SNP associations, but did not fall directly within a coding region.
We compared the genes within and near plumage-associated SNPs with known genes involved in pigmentation pathways. Over three dozen melanogenesis genes have been identified [3,7,50] and we searched through the associated genes for these candidates. By contrast, there are only three known gene families for carotenoid processing [4,5]: β-carotene oxygenases, scavenger receptors and cytochrome P450s. We were particularly interested scavenger receptor genes, which are involved in selective transport and/or uptake of carotenoids [17]. These characteristics are relevant in myrtle and Audubon's warblers as both taxa express carotenoid-based plumage characters, but in different plumage patches, as is expected for carotenoid selectivity. Finally, across all the annotated genes within 20 kb of a significant SNP, we tested whether there was significant evidence of enrichment of gene ontology terms (GO terms) of genes identified with this analysis (http://www.ark-genomics.org/tools/GOfinch; [51]).
3. Results and discussion
Much recent progress has been made identifying putative genes involved in generating coloration differences between related species [4,5,14,28,29,52]. In birds, coloration differences are derived primarily from two molecules—melanins and carotenoids—that have different biochemical processing [4,5]. To date, we have a much better understanding of the genes involved in melanin-based pigmentation, although several candidate carotenoid processing genes have been recently identified [4,5,15–17]. Here, we used admixture mapping in a natural hybrid zone and identified several genomic regions associated with five plumage traits that differ between myrtle and Audubon's warblers. We take advantage of the fact that these taxa differ in both melanic and carotenoid-based plumage characters and report genes within the associated regions for each (electronic supplementary material, table S1).
(a). Genomic distribution of divergence and plumage associations
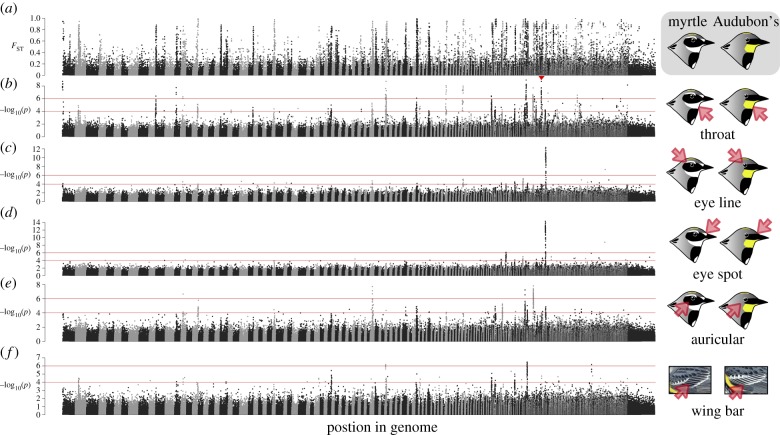
Admixture mapping revealed genomic regions that are strongly associated with plumage traits that differ between Audubon's and myrtle warblers. We tested genotype–phenotype associations with 393 755 or 154 683 SNPs, depending on the stringency for imputation uncertainty of our genomic data (table 1). We found imputation uncertainty has little influence on the interpretation of plumage-associated SNPs: those loci that have strong associations with phenotypic characters also have high correlations between known and imputed genotypes (electronic supplementary material, figure S1), and therefore are retained under this more stringent filter. However, under the most conservative false discovery rate within this reduced dataset, only eye line and eye spot have significant SNP associations (electronic supplementary material, figure S2). We focus our interpretation and further analysis on significant SNP associations with the larger dataset with the more stringent critical value (α = 0.000001 or –log10(α) = 6; table 1), which is a balance between a more inclusive dataset but a more conservative false discovery rate. In this case, under a null of no true associations, we expected to observe less than 1 significantly associated SNP for each plumage trait.
Table 1.
The number of SNPs significantly associated with the five plumage characters. Results derive from the linear mixed model output from GEMMA. The number of SNPs expected (under the null model of no association) to show significant associations for the large dataset (393 755 SNPs) at α = 0.000001 (−log10(α) = 6) and α = 0.0001 (−log10(α) = 4) are less than 1 and 39, respectively. For the smaller dataset (154 683 SNPs), with AR2 = 0.8 (i.e. markers with higher imputation confidence), the expectation is less than 1 and 16, respectively. Numbers in bold indicate that the number of SNPs found exceed the number expected under the null. Throat colour had the largest number of significantly associated genomic regions, suggesting a more polygenic genetic architecture for this carotenoid-based trait than for the four melanin-based traits.
| trait | # SNPs in full dataset, α = 0.0001 (0.000001) | # SNPs in smaller dataset with AR2 > 0.8, α = 0.0001 (0.000001) | scaffolds containing a plumage-associated SNP | genes containing a plumage-associated SNP | genes within 20 kb of a plumage-associated SNP | scaffolds with genes within 20 kb of a plumage-associated SNP |
|---|---|---|---|---|---|---|
| throat | 523 (100) | 56 (0) | 17 | 18 | 81 | 11 |
| eye line | 98 (46) | 60 (38) | 2 | 10 | 19 | 2 |
| eye spot | 113 (49) | 55 (41) | 3 | 11 | 23 | 3 |
| auricular | 228 (37) | 18 (0) | 4 | 10 | 34 | 4 |
| wing bar | 125 (10) | 10 (0) | 3 | 0 | 7 | 2 |
| expected | 39 (<1) | 16 (<1) |
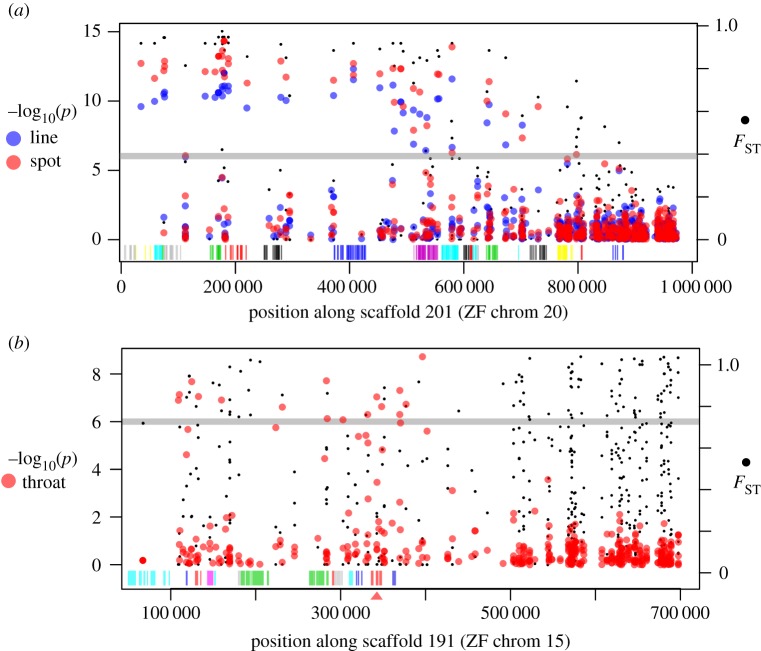
For each of the plumage traits, we found many significant SNP associations and many more than expected by chance (table 1). These associations were clustered in discrete regions (i.e. at least three plumage-associated SNPs on the same scaffold) and these clusters were distributed across the genome (figure 1 and table 1; electronic supplementary material, table S1). For three of the traits—throat colour, auricular colour and wing bar pattern—we found multiple association peaks across several chromosomes (figure 2b,e and f; electronic supplementary material, table S1). Throat coloration—a carotenoid-based trait that differs between the subspecies—had the largest number of associations of all the characters we quantified, with over a dozen regions showing significant SNP–phenotype links. By contrast, eye spot and eye line—two melanin-based traits—each show a single region of very highly significant associations (figure 2c,d). The associated loci are the same between eye line and eye spot and map to the zebra finch chromosome 20 in the interval from 0.061 to 0.937 Mb (figure 3a). These two characters have the highest correlation among the five traits (R2 = 0.53), consistent with a common molecular basis. Two other genomic regions were significantly associated with more than one trait: one association peak for both throat and auricular colour (mapping to finch chromosome 19; electronic supplementary material, table S1), and one association peak for both throat colour and wing bar (mapping to finch chromosome 6; electronic supplementary material, table S1). These results suggest an independent genetic basis for the majority of the plumage traits analysed in our study, with pleiotropy or supergenes playing a lesser role. By contrast, for eye line and eye spot, only a single genomic region was significantly associated with these two characters, and more detailed analyses will be needed to determine whether they are controlled by one gene or multiple tightly linked genes.
Figure 2.
Manhattan plots of genomic differentiation and plumage associations. (a) FST between allopatric myrtle and Audubon's warblers at 393 755 SNPs across the genome with scaffolds ordered by size and neighbouring scaffolds distinguished by alternating grey or black coloration. (b–f) Phenotype–genotype associations for the five plumage characters. The triangle in (b) shows the cluster of loci that aligns to the zebra finch chromosome 15. This region includes SCARF2, a strong candidate gene for selective carotenoid transport. (Online version in colour.)
Figure 3.
Patterns of divergence and associations. (a) Phenotype–genotype associations for eye line (blue points) and eye spot (red points) for a region aligning to the zebra finch chromosome 20. Associations between these two traits are highly correlated with each other as well as patterns of divergence as measured by FST (filled black points). Strings of coding regions for genes are shown by the vertical bars, with different genes coloured arbitrarily, and different colours between adjacent genes. (b) Association with throat coloration (red points) in a region aligning to the zebra finch chromosome 15, in this case only associated with part of the FST peak. This region includes SCARF2 (exons shown in vertical red bars at 350 kb highlighted by the red triangle), a strong candidate gene for selective carotenoid transport. (Online version in colour.)
Counter to our original prediction, we found only two of the 29 plumage–genotype association clusters mapped to the Z chromosome. Given that the plumage traits differing between myrtle and Audubon's warblers are most strongly expressed in males in the breeding season, and that sex chromosomes have been identified in previous research as often being involved in avian plumage traits [18,30–32], we expected the Z chromosome to account for more of the plumage differences between myrtle and Audubon's warblers. Recent anonymous genome scans of phenotypically distinct birds that have also investigated pigmentation have identified both a large proportion of differentiation peaks on the Z chromosome and many that include pigmentation genes [18,28,52]. For example, among Vermivora warblers and Sporophila seedeaters, two of six (33%) and 10 of 25 divergent regions (40%) were associated with the Z chromosome, respectively [18,28]. This contrasts to the pattern between myrtle and Audubon's warblers, which have few divergent regions on the Z chromosome [25] and, of those, only two from the current study that show a plumage association. This result suggests that important sexually dimorphic plumage genes are autosomal and might instead be activated by sex hormones—possibly testosterone-dependent—rather than being located on the Z chromosome.
To quantify the genomic substrate that leads to phenotypic divergence between these warblers, we calculated the proportion of scaffolds with clusters of high-FST markers that also contain SNPs significantly associated with plumage traits. There were 90 scaffolds that had three or more SNPs above the 99% FST quantile, which is approximately three times the number of scaffolds that had significant plumage associations (table 1). This finding is consistent with most of the regions differing between the taxa being related to non-plumage differences between the taxa.
(b). Candidate genes and functional interpretations
Gene ontology enrichment analysis using GOFinch, based on 115 genes located within 20 kb of a plumage-associated SNP, returned five significant GO terms at a false discovery rate of 0.05. The most significantly enriched GO term was keratin filament (Fisher's exact test uncorrected p = 5.8 × 10−5). Keratin genes are highly clustered in avian genomes, and one of these clusters on chromosome 22 is associated with throat colour. Other significantly enriched GO terms (uncorrected p-values between 0.0001 and 0.0006) include LIM domain binding, regulation of innate immune response, regulation of cell growth and structural molecule activity. Myrtle warbler genes located near plumage-associated SNPs, and their corresponding zebra finch Ensembl genes, are reported in the electronic supplementary material, table S1.
Given the moderately large regions of divergence and multiple genes within each of these regions, pinpointing causal genes will take additional fine mapping, functional annotation and RNA expression data. However, one gene appears to be a strong candidate for its involvement in the carotenoid processing pathway. Within the cluster of SNPs associated with throat colour and mapping to the finch chromosome 15 is the Scavenger Receptor Class F Member 2 (SCARF2) gene (figure 3b). Of the three classes of genes that have been previously been associated with carotenoid transport, scavenger receptors have been implicated across several taxonomic groups, although this gene member has not be implicated previously [5,53,54]. Carotenoids are not water-soluble and therefore require special transport by lipoproteins through plasma to tissues for deposition. Scavenger receptors at target tissues are thought to recognize these lipoproteins and facilitate the selective movement of carotenoids into the target cells [5]. Most recently, Toomey et al. [17] identified a related gene (SCARB1) as likely involved with the uptake of carotenoids into the feathers of domesticated canaries.
Myrtle warblers do not have the conspicuous yellow throat feathers expressed by Audubon's warblers and instead are characterized by white throat feathers. HPLC data are consistent with the absence of carotenoids in myrtle warbler throat feathers (electronic supplementary material, table S2). By contrast, in Audubon's warblers, there are high levels of carotenoids, with high amounts of lutein (electronic supplementary material, table S2), as has been found with previous studies of the carotenoid-rich yellow feathers of yellow warblers (Setophaga petechia) [55]. However, both myrtle and Audubon's warblers express yellow feathers in some other patches across their body, including their crown, breast and rump feathers. This suggests selectivity for the lipid transport of carotenoids into the throat feathers of Audubon's, with the myrtle warblers possibly blocking carotenoid incorporation in the developing feathers at these sites. This pattern is consistent with the presumed selective activity of scavenger receptors identified in other taxa—and possibly SCARF2 in this case—although additional expression profiles of developing feathers will be necessary to confirm this finding.
While the genomic regions associated with the eye line and eye spot characters did not contain any previously annotated pigmentation genes, the strong associations for these two traits along a single scaffold suggest that this region—and the genes within it—deserve additional scrutiny. Genotypic variation at the SNPs showing the strongest associations with these characters are consistent with a pattern of additive inheritance (electronic supplementary material, figure S3). While caution must be taken when interpreting these raw genotype–phenotype associations—unlike the GEMMA analysis, they do not control for background hybrid ancestry—the additive patterns are suggestive. If true, this contrasts with the recessive inheritance pattern of black throat colour in golden-winged warblers, which was predicted to be a recessive Mendelian trait [56] and recently confirmed with genomic data [18]. Other melanin-based traits in birds with known genetic basis are consistent with dominant [57] or partially dominant inheritance [14].
It is in fact notable how few previously annotated melanogenesis genes we recovered in our association study, particularly when contrasted with other genomic comparisons of related avian taxa that differ in pigmentation patterns [18,28,29]. For instance, several studies have consistently identified coding changes as well as putative regulatory differences in the gene agouti signalling protein (ASIP) between birds with melanic and non-melanic plumage patches [1,14,18,28]. Our assay identified 72 SNPs within 50 kb of the ASIP coding region, but none were highly differentiated between myrtle and Audubon's warblers (average FST in this region = 0.02, lower than the genome-wide average of 0.045). In the comparisons of other related avian taxa, melanic differences involved either relatively substantial pattern differences (i.e. a black versus plain throat; [18,28]) or completely melanic morphs [14]. By contrast, the melanic patches that differ between myrtle and Audubon's warblers are small and confined mostly to the face and wing. Therefore, genes within the associated genomic regions for these melanin-based traits between myrtle and Audubon's warblers may include feather-tract specific regulatory factors or involve selective transport or deposition of melanin.
(c). Heterogeneous differentiation and warbler speciation
How are these significant phenotype–genotype associations related to the evolution of reproductive isolation in this species complex and in the radiation of wood warblers more generally? Recent studies of closely related species pairs have revealed that genetic differentiation is highly heterogeneous between different portions of the genome across a diversity of taxa. Moreover, there is compelling evidence that selection has contributed to regions that exhibit elevated differentiation in several cases (e.g. [25,58,59]). However, questions remain regarding the connection between these two observations: are differentiation peaks related to the evolution of reproductive isolation?
The phenotypic associations we report here between myrtle and Audubon's warblers fall within a subset of the characterized divergence peaks between these two taxa. This subset of peaks with plumage associations can therefore be interpreted as genetic locations associated with clear phenotypic changes during speciation. However, it is not clear whether or how much these loci and their associated phenotypes contribute to reproductive isolation. Previous research on Audubon's and myrtle warblers shows that the two taxa have moderate levels of reproductive isolation [22]. This is likely due to post-mating selection against hybrids, because no assortative mating based on plumage was observed in the hybrid zone, based on observations of social pairs [22]. However, it is still possible that there is some amount of cryptic assortative mating via extra-pair mating, and also possible that plumage differences influence male–male interactions.
The low fraction of all high-FST clusters that are associated with plumage suggests that characteristics other than plumage could play important roles in reproductive isolation (e.g. migration, genetic incompatibilities, song). It is therefore possible that plumage diverged through selection in allopatry, but plays only a weak role in generating reproductive isolation. More generally, the patterns in the Audubon's and myrtle warbler system are notable in that they are intermediate when compared with several recent studies that found few, small genomic regions of divergence that disproportionately include pigmentation genes [18,28,29] and others that have found very large regions of divergence—over half the length of several chromosomes in some cases—that encompass thousands of genes [52,60]. Understanding the evolutionary processes that give rise to the genomic differences among these and other systems is an active area of research, and these data from Audubon's and myrtle warblers provide a more complete representation for future comparative analyses.
(d). Genotype–phenotype associations with reduced representation sequencing
Recently, there has been debate regarding whether reduced representation genome sequencing approaches can identify SNPs at high enough densities to perform the kinds of association studies presented here [61]. For example, a reduced representation genome scan of Vermivora warblers failed to detect highly divergent genomic regions, which were only discovered after whole-genome sequencing [18]. Using simulations, Kardos et al. [62] showed that tens of thousands of SNP markers may not be adequate to detect genotype–phenotype associations in natural populations, and recommended whole-genome sequencing of extreme phenotypes. Lowry et al. [61] argued that most reduced-representation sequencing studies genotype far too few SNPs to find significant associations, given the extent of linkage disequilibrium. However, when linkage disequilibrium is high, such as in hybrid zones (e.g. [52]) and in supergenes (e.g. [63]), reduced representation approaches are clearly useful in identifying genotype–phenotype associations. The strong results from our study of genotype–phenotype associations in the myrtle–Audubon's warbler hybrid zone bode well for other studies of similar cases of natural admixture. It is important to note, however, that reduced representation sequencing necessarily assays variation at only a subset of the overall genome. Indeed, there are likely additional genomic regions that differ between myrtle and Audubon's warblers, and a subset of these may be linked to other plumage gene candidates. Therefore, whole-genome re-sequencing of hybrids will be an important next step to test whether and how many of these additional candidates exist in the genome.
(e). Conclusions
Here, we have identified genomic associations for a variety of plumage coloration traits, derived from various pigment molecules requiring different biochemical processing. In particular, we identified an additional candidate for carotenoid processing (SCARF2), as well as a single genomic region that is highly associated with melanic variation in two traits, eye line and eye spot. Other plumage traits (auricular colour, and wing bar pattern) show associations with additional independent genomic regions, although each have associations that are less statistically significant. With the exception of eye line and eye spot, the characters mostly differ in the associated loci, and only a small fraction of the genomic regions of high differentiation between these warblers are associated with plumage. These results indicate a strong genetic basis for plumage, but that those plumage differences explain only a small amount of overall differentiation and that their role in reproductive isolation remains to be determined. More generally, genomic studies of natural systems have made substantial progress in identifying the genes associated with coloration phenotypes, and set the stage for important comparative analyses across this and other rapid radiations.
Supplementary Material
Supplementary Material
Acknowledgements
Field sampling was conducted under permits from Canadian Wildlife Service, Parks Canada, Alberta Community Development, and the Animal Care Committees of University of British Columbia and University of Calgary. Alana Demko, Allison Patterson, Olga Lansdorp and Michael Moretti assisted with fieldwork, and Miguel Alcaide provided advice and guidance regarding the molecular analysis. We thank two anonymous reviewers for their input on an earlier version of this manuscript. Erin Morrison completed the HPLC analysis of the warbler throat feathers and provided valuable input on its interpretation.
Ethics
This research adhered to the University of British Columbia's Animal Care Committee guidelines for the use of vertebrate animals in research (protocol no. A09-0131). Field permits were provided by the Canadian Wildlife Service, Prairie and Northern Region office (AB Scientific Permit 11-AB-SC023) and Alberta Parks (11-107 and 15-162).
Data accessibility
Genotype data and phenotype information can be found in the Dryad Digital Repository: (http://dx.doi.org/10.5061/dryad.24s5j; [64]).
Authors' contributions
D.P.L.T., A.B. and D.E.I. conceived the idea and design. D.P.L.T. and A.B. collected samples and analysed the data. D.P.L.T. generated the sequence data. D.P.L.T. and A.B. and wrote the original draft of the paper; all authors contributed to writing the final version of the paper.
Competing interests
We declare we have no competing interests.
Funding
Field sampling and molecular analysis was supported by NSERC Discovery Grants (311931-2005, 311931-2010 and 311931-2012) to D.E.I., as well as an Alberta Conservation Association Grant in Biodiversity and a University of Lausanne 450th Anniversary Foundation grant to A.B. A.B. was supported by an NSF Graduate Research Fellowship. D.P.L.T. was supported by an NSERC CGS-D and a Banting postdoctoral fellowship.
References
- 1.Cuthill IC, et al. 2017. The biology of color. Science 357, eaan0221–9. ( 10.1126/science.aan0221) [DOI] [PubMed] [Google Scholar]
- 2.San-Jose LM, Roulin A. 2017. Genomics of coloration in natural animal populations. Phil. Trans. R. Soc. B 372, 20160337 ( 10.1098/rstb.2016.0337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard JK, Uy J, Hauber ME, Hoekstra HE. 2010. Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 26, 231–239. ( 10.1016/j.tig.2010.02.002) [DOI] [PubMed] [Google Scholar]
- 4.Walsh N, Dale J, McGraw KJ, Pointer MA, Mundy NI. 2011. Candidate genes for carotenoid coloration in vertebrates and their expression profiles in the carotenoid-containing plumage and bill of a wild bird. Proc. R. Soc. B 279, 58–66. ( 10.1042/BJ20040554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toews DPL, Hofmeister NR, Taylor SA. 2017. The evolution and genetics of carotenoid processing in animals. Trends Genet. 33, 171–182. ( 10.1016/j.tig.2017.01.002) [DOI] [PubMed] [Google Scholar]
- 6.Mallarino R, Henegar C, Mirasierra M, Manceau M, Schradin C, Vallejo M, Beronja S, Barsh GS, Hoekstra HE. 2016. Developmental mechanisms of stripe patterns in rodents. Nature 539, 518–523. ( 10.1038/nature20109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313, 101–104. ( 10.2307/3846602) [DOI] [PubMed] [Google Scholar]
- 8.Nachman MW, Hoekstra HE, D'Agostino SL. 2003. The genetic basis of adaptive melanism in pocket mice. Proc. Natl Acad. Sci. USA 100, 5268–5273. ( 10.2307/3139701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linnen CR, Poh YP, Peterson BK, Barrett RDH, Larson JG, Jensen JD, Hoekstra HE. 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339, 1312–1316. ( 10.1126/science.1233213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho WW, Smith SD. 2016. Molecular evolution of anthocyanin pigmentation genes following losses of flower color. BMC Evol. Biol. 16, 1–10. ( 10.1186/s12862-016-0675-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ligon RA, Simpson RK, Mason NA, Hill GE, McGraw KJ. 2016. Evolutionary innovation and diversification of carotenoid-based pigmentation in finches. Evolution 70, 2839–2852. ( 10.1111/evo.13093) [DOI] [PubMed] [Google Scholar]
- 12.Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. 2009. On the origin and spread of an adaptive allele in deer mice. Science 325, 1095–1098. ( 10.1126/science.1175826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenblum EB, Rompler H, Schoneberg T, Hoekstra HE. 2010. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl Acad. Sci. USA 107, 2113–2117. ( 10.1073/pnas.0911042107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uy JAC, Cooper EA, Cutie S, Concannon MR, Poelstra JW, Moyle RG, Filardi CE. 2016. Mutations in different pigmentation genes are associated with parallel melanism in island flycatchers. Proc. R. Soc. B 283, 20160731 ( 10.1007/s00265-013-1492-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes RJ, et al. 2016. Genetic basis for red coloration in birds. Curr. Biol. 26, 1427–1434. ( 10.1016/j.cub.2016.03.076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mundy NI, et al. 2016. Red carotenoid coloration in the zebra finch is controlled by a cytochrome P450 gene cluster. Curr. Biol. 26, 1435–1440. ( 10.1016/j.cub.2016.04.047) [DOI] [PubMed] [Google Scholar]
- 17.Toomey MB, et al. 2017. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl Acad. Sci. USA 114, 5219–5224. ( 10.1073/pnas.1700751114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toews DPL, Taylor SA, Vallender R, Brelsford A, Butcher BG, Messer PW, Lovette IJ. 2016. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr. Biol. 26, 2313–2318. ( 10.1016/j.cub.2016.06.034) [DOI] [PubMed] [Google Scholar]
- 19.Price T, Gibbs H, de Sousa L, Richman A. 1998. Different timing of the adaptive radiations of North American and Asian warblers. Proc. R. Soc. B 265, 1969–1975. ( 10.1098/rspb.1998.0527) [DOI] [Google Scholar]
- 20.Hunt P, Flaspohler DJ. 1998. Yellow-rumped warbler (Setophaga coronata). In Birds of North America (ed. Rodewald PG.). Ithaca, NY: Cornell Lab of Ornithology. [Google Scholar]
- 21.Hubbard JP. 1969. Relationships and evolution of the Dendrocia coronata complex. The Auk 86, 393–432. [Google Scholar]
- 22.Brelsford A, Irwin DE. 2009. Incipient speciation despite little assortative mating: the yellow-rumped warbler hybrid zone. Evolution 63, 3050–3060. ( 10.1111/j.1558-5646.2009.00777.x) [DOI] [PubMed] [Google Scholar]
- 23.Milá B, Smith T, Wayne R. 2007. Speciation and rapid phenotypic differentiation in the yellow-rumped warbler Dendroica coronata complex. Mol. Ecol. 16, 159–174. ( 10.1111/j.1365-294X.2006.03119.x) [DOI] [PubMed] [Google Scholar]
- 24.Toews DPL, Brelsford A, Irwin DE. 2014. Isotopic variation across the Audubon's–myrtle warbler hybrid zone. J. Evol. Biol. 27, 1179–1191. ( 10.1111/jeb.12392) [DOI] [PubMed] [Google Scholar]
- 25.Toews DPL, Brelsford A, Grossen C, Mila B, Irwin DE. 2016. Genomic variation across the yellow-rumped Warbler species complex. The Auk 133, 698–717. ( 10.1642/AUK-16-61.1) [DOI] [Google Scholar]
- 26.Brelsford A, Milá B, Irwin DE. 2011. Hybrid origin of Audubon's warbler. Mol. Ecol. 20, 2380–2389. ( 10.1111/j.1365-294X.2011.05055.x) [DOI] [PubMed] [Google Scholar]
- 27.Buerkle CA, Lexer C. 2008. Admixture as the basis for genetic mapping. Trends Ecol. Evol. 23, 686–694. ( 10.1016/j.tree.2008.07.008) [DOI] [PubMed] [Google Scholar]
- 28.Campagna L, Repenning M, Silveira LF, Fontana CS, Tubaro PL, Lovette IJ. 2017. Repeated divergent selection on pigmentation genes in a rapid finch radiation. Sci. Adv. 3, e1602404 ( 10.1101/075713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poelstra JW. 2014. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science 344, 1405–1410. ( 10.1126/science.1253823) [DOI] [PubMed] [Google Scholar]
- 30.Saether SA, et al. 2007. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318, 95–97. ( 10.1126/science.1141506) [DOI] [PubMed] [Google Scholar]
- 31.Toews DPL, Campagna L, Taylor SA. 2016. Genomic approaches to understanding population divergence and speciation in birds. The Auk 133, 13–30. ( 10.1642/AUK-15-51.1) [DOI] [Google Scholar]
- 32.Albert AYK, Otto SP. 2005. Sexual selection can resolve sex-linked sexual antagonism. Science 310, 119–121. ( 10.2307/3842876) [DOI] [PubMed] [Google Scholar]
- 33.Küpper C, et al. 2015. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48, 79–83. ( 10.1038/ng.3443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuttle EM, et al. 2016. Divergence and functional degradation of a sex chromosome-like supergene. Curr. Biol. 26, 344–350. ( 10.1016/j.cub.2015.11.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seutin G, White B, Boag P. 1991. Preservation of avian blood and tissue samples for DNA analyses. Can. J. Zool. 69, 82–90. ( 10.1139/z91-013) [DOI] [Google Scholar]
- 36.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, e19379 ( 10.1371/journal.pone.0019379.g006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcaide M, Scordato ESC, Price T, Irwin DE. 2014. Genomic divergence in a ring species complex. Nature 511, 83–85. ( 10.1038/nature13285) [DOI] [PubMed] [Google Scholar]
- 38.Lindgreen S. 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 5, 337 ( 10.1186/1756-0500-5-337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. ( 10.1038/ng.806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van der Auwera GA, et al. 2002. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
- 42.Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27, 2156–2158. ( 10.1093/bioinformatics/btr330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Stephens M. 2014. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 11, 407–409. ( 10.1038/nmeth.2848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097. ( 10.1086/521987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-cased linkage analyses. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lischer HEL, Excoffier L. 2012. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28, 298–299. ( 10.1093/bioinformatics/btr642) [DOI] [PubMed] [Google Scholar]
- 47.Turner SD. 2014. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. bioRxiv 1–2. ( 10.1101/005165) [DOI]
- 48.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. ( 10.2307/2408641) [DOI] [PubMed] [Google Scholar]
- 49.Warren WC, et al. 2010. The genome of a songbird. Nature 464, 757–762. ( 10.1038/nature08819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mundy NI. 2005. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc. R. Soc. B 272, 1633–1640. ( 10.1146/annurev.ecolsys.27.1.543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Watson M. 2009. CORNA: testing gene lists for regulation by microRNAs. Bioinformatics 25, 832–833. ( 10.1093/bioinformatics/btp059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delmore KE, Toews DPL, Germain RR, Owens GL, Irwin DE. 2016. The genetics of seasonal migration and plumage color. Curr. Biol. 26, 2167–2173. ( 10.1016/j.cub.2016.06.015) [DOI] [PubMed] [Google Scholar]
- 53.Kiefer C, Sumser E, Wernet MF, von Lintig J. 2002. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl Acad. Sci. USA 99, 10 581–10 586. ( 10.1073/pnas.162182899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakudoh T, Kuwazaki S, Iizuka T, Narukawa J, Yamamoto K, Uchino K, Sezutsu H, Banno Y, Tsuchida K. 2013. CD36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm Bombyx mori. J. Lipid Res. 54, 482–495. ( 10.1194/jlr.M032771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGraw KJ, Beebee MD, Hill GE, Parker RS. 2003. Lutein-based plumage coloration in songbirds is a consequence of selective pigment incorporation into feathers. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 135, 689–696. ( 10.1016/S1096-4959(03)00164-7) [DOI] [PubMed] [Google Scholar]
- 56.Nichols JT. 1908. Lawrence's and Brewster's warblers and Mendelian inheritance. The Auk 25, 86 ( 10.2307/4070261) [DOI] [Google Scholar]
- 57.Bourgeois YXC, et al. 2017. A novel locus on chromosome 1 underlies the evolution of a melanic plumage polymorphism in a wild songbird. R. Soc. Open Sci. 4, 160805 ( 10.1098/rsos.160805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruegg K, Anderson EC, Boone J, Pouls J, Smith TB. 2014. A role for migration-linked genes and genomic islands in divergence of a songbird. Mol. Ecol. 23, 4757–4769. ( 10.1111/mec.12842) [DOI] [PubMed] [Google Scholar]
- 59.Delmore KE, Hübner S, Kane NC, Schuster R, Andrew RL, Câmara F, Guigó R, Irwin DE. 2015. Genomic analysis of a migratory divide reveals candidate genes for migration and implicates selective sweeps in generating islands of differentiation. Mol. Ecol. 24, 1873–1888. ( 10.1111/mec.13150) [DOI] [PubMed] [Google Scholar]
- 60.Ellegren H, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491, 756–760. ( 10.1038/nature11584) [DOI] [PubMed] [Google Scholar]
- 61.Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, Storfer A. 2016. Breaking RAD: an evaluation of the utility of restriction site associated DNA sequencing for genome scans of adaptation. Mol. Ecol. Res. 17, 142–152. ( 10.1111/1755-0998.12635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kardos M, Husby A, McFarlane SE, Qvarnström A, Ellegren H. 2015. Whole-genome resequencing of extreme phenotypes in collared flycatchers highlights the difficulty of detecting quantitative trait loci in natural populations. Mol. Ecol. Res. 16, 727–741. ( 10.1111/1755-0998.12498) [DOI] [PubMed] [Google Scholar]
- 63.Purcell J, Brelsford A, Wurm Y, Perrin N, Chapuisat M. 2014. Convergent genetic architecture underlies social organization in ants. Curr. Biol. 24, 2728–2732. ( 10.1016/j.cub.2014.09.071) [DOI] [PubMed] [Google Scholar]
- 64.Brelsford A, Toews DPL, Irwin DE. 2017. Data from: Admixture mapping in a hybrid zone reveals loci associated with avian feather coloration Dryad Digital Repository. ( 10.5061/dryad.24s5j) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Brelsford A, Toews DPL, Irwin DE. 2017. Data from: Admixture mapping in a hybrid zone reveals loci associated with avian feather coloration Dryad Digital Repository. ( 10.5061/dryad.24s5j) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Genotype data and phenotype information can be found in the Dryad Digital Repository: (http://dx.doi.org/10.5061/dryad.24s5j; [64]).