Abstract
Many fundamental concepts in evolutionary biology were discovered using non-human study systems. Humans are poorly suited to key study designs used to advance this field, and are subject to cultural, technological, and medical influences often considered to restrict the pertinence of human studies to other species and general contexts. Whether studies using current and recent human populations provide insights that have broader biological relevance in evolutionary biology is, therefore, frequently questioned. We first surveyed researchers in evolutionary biology and related fields on their opinions regarding whether studies on contemporary humans can advance evolutionary biology. Almost all 442 participants agreed that humans still evolve, but fewer agreed that this occurs through natural selection. Most agreed that human studies made valuable contributions to evolutionary biology, although those less exposed to human studies expressed more negative views. With a series of examples, we discuss strengths and limitations of evolutionary studies on contemporary humans. These show that human studies provide fundamental insights into evolutionary processes, improve understanding of the biology of many other species, and will make valuable contributions to evolutionary biology in the future.
Keywords: demography, ecology, menopause, phenotypic plasticity, quantitative genetics
1. Introduction
Mendel's studies of peas, Tinbergen's observation of gulls, Morgan's experiments with fruit flies, and Hamilton's consideration of insects are just some examples of organisms being used to make major contributions to the field of evolutionary biology. Recently, an increasing diversity of study organisms have advanced the field owing to factors such as long-term field studies [1] and advancing genetic methods. It is well accepted that many discoveries within evolutionary biology made using such study organisms are applicable to humans. However, the inverse question, whether fundamental concepts in evolutionary biology can be discovered in studies of contemporary or recent human populations and applied to other species, is less clear and sometimes considered with caution or reluctance, even among editors of broad-scope biological journals. For example, as recently as 2015, the editorial policy of Proceedings B specifically stated that studies on humans were only considered ‘if they had clear relevance to fundamental biological principles and processes, or to other groups of organisms’. The frequency of such views among evolutionary biologists, however, remains currently unknown, as do the associated causes for scepticism.
We first investigated to what extent and why, the research community of evolutionary biologists believes that studies on contemporary humans can provide valuable contributions to evolutionary biology. However, even if the respondents believe human studies are useful, it does not necessarily mean they are useful. Hence, in the second section of this article, we discuss the value of studies on contemporary humans through examples drawn from diverse disciplines including demography, genetics, evolutionary medicine, and life-history evolution, to demonstrate that studies on contemporary humans have provided insights into fundamental evolutionary biology, and continue to spark research in a variety of organisms.
2. The extent and causes of scepticism towards human research in evolutionary biology
To investigate the opinion of the evolutionary biology research community with respect to the ability of studies on contemporary human populations to advance the field, we conducted an online survey in April 2017 (survey details and response data available via the Dryad data repository, details below). Researchers were contacted directly, through their institutions, online bulletins, and social media. They were asked about career stage, research area, history of exposure to research on humans, and a series of questions regarding their opinion on the relevance of human studies.
We received 442 responses from 35 countries, with most researchers based in the USA (19.2%), the UK (14.5%), and France (10.6%). The majority (72%) identified their research area as ‘Ecology or Evolution’, followed by ‘Molecular Biology and Genetics’ (8.4%), ‘Psychology or Evolutionary Psychology’ (6.3%), Anthropology (3.6%), and Demography (1.8%). Most were currently affiliated with a research institution (90%). The respondents differed in their educational or research experience, with 45% being or having been principle investigators (PIs) (lecturers, or full, associate, or emeritus professors), followed by postdocs (25%) and PhD students (18%). Hence, the majority of respondents were active researchers with a broad range of expertise and experience in evolutionary biology or ecology.
With seven questions (Q7–Q13), we aimed to gauge attitudes towards human studies, and their perceived general relevance to evolutionary and behavioural ecology. Respondents were asked to rate statements from strongly disagree to strongly agree on a scale of 1–5 accordingly. We also invited comments on the suitability of human studies for evolutionary biology and behavioural ecology (Q14), and general remarks (Q15). While some responses were not always straightforward to interpret, we list some examples here (table 1).
Table 1.
Summary of advantages and disadvantages of human studies to evolutionary and behavioural ecology mentioned and explained by respondents to our survey (Question14: “I believe human studies are/are not suitable for evolutionary and behavioural ecology because…”). The full dataset with all comments is available online.
| advantages | disadvantages | |
|---|---|---|
| features unique to humans |
|
|
| exposure to environmental variation |
|
|
| methodology |
|
|
| applicability |
|
|
Major differences in perspectives on human research may arise depending on the respondent's personal experience with human research. Approximately one-third had performed research using human data themselves (‘direct-exposure’; 35%), while a similar portion had departmental colleagues performing such research (‘peer-exposure’; 31%) or neither type of experience (‘no-exposure’; 34%). That over a third of respondents were conducting or had conducted research on human data is probably larger than expected based on the expertise in the discipline at large. This possible overrepresentation may have arisen because these researchers were either more willing to answer our survey or because our survey was more likely to reach them. We, therefore, investigated whether levels of exposure to human studies influenced the answer to each question using ordinal logistic regression with the ‘ordinal’ package [2] in R (v. 3.3.3). The dependent variable was an ordinate variable of agreement (1–5). We included the exposure of respondents to human studies as a three-level categorical factor, academic position as a two-level factor (‘with-PhD’ (postdoc, lecturer, or professor); ‘without-PhD’ (undergraduate, masters, or PhD student); excluding ‘others’). All results are detailed in electronic supplementary material, table S1.
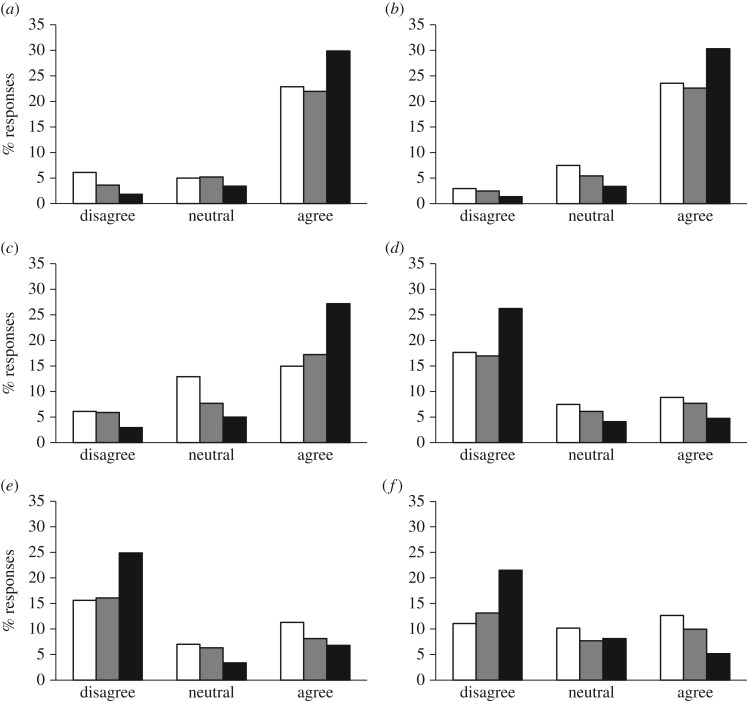
We first gauged the extent to which researchers agreed that humans were subject to evolution—a starting point if we are to use human data to study general evolutionary processes. Almost all respondents (95.7%) agreed that humans are continuing to evolve. However, a smaller proportion (74.9%) agreed that humans still evolve by natural selection, while 11.5% disagreed (figure 1a). There were differences in these frequencies among researchers with direct-, peer, and no-exposure to human research. We report here results as odd ratio (OR) (±asymmetric s.e.), with OR = 1, meaning there is no effect and, for example, OR = 1.25, meaning there is a 25% difference between groups. Compared to researchers with no-exposure, those with direct-exposure were more likely to agree that humans are still evolving (OR = 1.75 [1.27–2.42], p = 0.082), and that humans are evolving under natural selection (OR = 1.97 [1.54–2.50], p = 0.005). However, no differences were detected between peer-exposure and no-exposure respondents (humans are evolving: OR = 0.71 [0.53–0.94], p = 0.220; humans are evolving under natural selection: OR = 1.08 [0.86–1.37], p = 0.720). Academic position of respondents influenced agreement with the statement that humans are still evolving by natural selection: participants with a PhD were far more likely to agree than non-PhD participants (OR = 2.75 [2.23–3.39], p < 0.001). Thus, in summary, most respondents agreed that humans are still evolving, with the majority agreeing on a role for natural selection in that process, but this varied among levels of relevant education and experience. Importantly, this finding is in line with an increasing number of studies showing that, despite large decreases in mortality and reproduction over the last centuries, and in contrast to some beliefs [4,5], contemporary human populations are subject to selection [6]. In other words, genetic variation still influences survival to adulthood, reproduction, and fitness, fuelling continued evolution by natural selection in current human populations [7].
Figure 1.
Responses (n = 442) of evolutionary biologists to a survey concerning the relevance of human studies for advancing the evolutionary research field in general (questions 8–13, [3]). Responses sorted by exposure to human research; direct-exposure through own work (black bars); peer-exposure through departmental colleagues (grey bars), and no-exposure through either route. Respondents were asked to agree on a scale from 1 (strongly disagree) to 5 (strongly agree), with the following statements: (a) humans are still evolving by natural selection; (b) human studies make a valuable contribution to evolutionary and behavioural ecology; (c) results from human studies can be applicable to other species; (d) that humans no longer live in their natural environment limits the suitability of human studies for evolutionary and behavioural ecology; (e) cultural and social influences limit the suitability of human studies for evolutionary and behavioural ecology; (f) the limited scope for experiments prevents humans from being a suitable study organism for evolutionary and behavioural ecology. The original response scale 1–5 was simplified here for illustrative purposes only, with 1 and 2 pooled as ‘disagree’, 3 representing ‘neutral’, and 4 and 5 pooled as ‘agree’.
We then gauged opinion on whether human studies are valuable for evolutionary biology and behavioural ecology. About 76.5% of respondents agreed so, and only 7.0% disagreed (figure 1b). Most respondents (59.3%) agreed that results from human studies are applicable to other species, while 15.2% disagreed (figure 1c). Participants exposed to human research agreed 8% more than no-exposure participants with statements that human studies contribute to evolutionary biology and behavioural ecology (figure 1b, OR = 3.05 [2.39–3.90], p < 0.001) and 18% more that they can be applicable to other species (figure 1c, OR = 3.74 [2.98–4.71], p < 0.001). While we did not have clear expectation on rates, these percentages may seem somewhat low. However, they should not be compared to 100% but rather to the rates of agreement that results from non-human systems are generally applicable, which remain, to the best of our knowledge, unknown. The academic position of respondents did not influence the probability of agreement with the statement that human studies contribute to evolutionary biology and behavioural ecology (OR = 0.94 [0.76–1.15], p = 0.750) or that they can be applicable to other species (OR = 1.17 [0.96–1.44], p = 0.416). Hence, most respondents agreed that research on humans can be relevant for evolutionary biology and behavioural ecology in general, though agreement was more common in those with direct-exposure to human research.
We further investigated which issues could diminish the relevance of human studies to evolutionary biology and behavioural ecology. Our survey directly gauged the opinion for three possible causes: (i) the lack of a ‘natural’ environment (figure 1d), (ii) the presence of cultural influences (figure 1e), and (iii) the limited possibility to perform experiments (figure 1f). Among all respondents, 21.3%, 26.5%, and 28.1%, respectively, agreed that these factors posed issues regarding the broader relevance of human studies. Agreement with these statements depended on exposure to human research, with no-exposure respondents agreeing with these statements more than the peer-exposure and direct-exposure groups (26.0%, 33.3%, and 37.3%, respectively; figure 1d–f). Participants exposed to human research disagreed more than no-exposure participants with statements that the suitability of human research in evolutionary and behavioural ecology is limited because: humans no longer live in their natural environment (OR = 0.37 [0.29–0.46], p < 0.001), because of social and cultural influences (OR = 0.32 [0.25–0.40], p < 0.001), or the limited scope for experiments (OR = 0.30 [0.24–0.38], p < 0.001). Almost half (44.8%) of the respondents considered that at least one of the three issues (absence of natural environment, social and cultural influences, and limited experimental possibilities) limited the relevance of human research for evolutionary or behavioural ecology in general, but this was most pronounced within the no-exposure group (55.3%).
Both the modern human environment and cultural influences were considered problematic and restrict the applicability of human studies to broader contexts; a concern also raised in the comments (table 1). For example: ‘Humans contribute to the evolution of other species but medicine and fertility treatments mean that natural selection can't be very effective in humans, at least in developed countries. Too many people survive and reproduce that would not have contributed to the gene pool without interventions.’ It is worth mentioning here that any influences that humans have on the environment could be considered non-natural. Hence, the concept of a natural environment is somewhat troublesome and could have been interpreted differently among our respondents. However, understanding the evolutionary consequences of human-mediated environmental change is itself a growing field of study. Furthermore, some survey participants saw the human cultural environment as a lesser obstacle, drawing parallels to other organisms that modify their environment: ‘The distinction between “natural” and “human-modified” environments is disappearing. We would also not consider a beaver pond an unnatural environment so why consider our built habitats not natural? As we evolve we change our habitat generating new selective factors.’
The major perceived limitation of human studies was the limited scope for experiments: ‘Generation time too long, replicated experiments impossible, correlation not causation’. Even when attitudes towards human studies were positive, many respondents called for more rigorous approaches: ‘I think human studies can make a valuable contribution, as long as the very substantial conceptual difficulties that such studies present are taken into account. Human studies are problematic when their assumptions are not acknowledged, and when their findings are interpreted in a superficial way’. Further, several respondents raised ethical concerns preventing human experiments and a human-biased interpretation of results. On the positive side, many respondents viewed the diversity of environments that humans live in as an advantage, not a problem and many acknowledged that the large amount of high-quality data available on humans is particularly valuable.
3. Contributions of human studies to evolutionary biology: from genes to populations
Our survey identified numerous strengths and weaknesses in human studies (table 1). We discuss these in the following sections, drawing on examples from topics in which studies in humans have played a key role in advancing the field: demography, quantitative genetics, evolutionary medicine, and life-history evolution. These examples illustrate contributions of different kinds: methodological developments, big data, or new hypotheses. We note though that these are just a small sample of the many areas, such as the role of telomeres in life history [8,9], the evolution of animal personalities [10], and social networks [11], in which human studies have made valuable contributions to fundamental understanding in evolutionary biology, or have even initiated entire fields of research.
(a). Demography
Demography is the study of vital rates: survival and fertility. Demography originated when local priests started tracking births, deaths and, in some cases, migrations of those in their community as early as the seventeenth century [12]. In the late seventeenth century, two members of the British Royal Society, John Graunt and Edmond Halley, began to explore the composition of human populations using such data, counting the number of individuals alive at a given time point and for a given age class. In doing so, they created some of the first known life tables, a method still widely used [13–15], and thereby founded the field of demography in Western society [12].
Using the wealth of data available [16] demographers continue to characterize ever-changing human populations [17–19], but the impact of demography extends much further than humans. At the genetic level, survival and fertility drive the propagation of genes, hence demography captures the two major components of fitness more broadly recognized in evolutionary biology [20]. For example, matrix population and integral projection models can both be used to link demography and quantitative genetics [21,22]. Demography provides further key insights into the biology of ageing: as yet, there is no evidence for an intrinsic limit to human lifespan [23], but the predictive value of individual biomarkers of lifespan declines with increasing age [24]. At the individual and species level, evolutionary biologists use demographic approaches to highlight diversity and identify the factors shaping variation in survival and fecundity [25].
At first sight, human demography may seem very different from that of short-lived species, which may experience regular population crashes and genetic bottlenecks [26,27]. However, a major reason for the multidisciplinary relevance of human demography arises because many demographic concepts and tools were first developed in humans. Classic examples include the description of population collapses following growth [28], which even inspired Darwin when formulating his theory of natural selection [28], logistic population growth [29], and exponential increases in mortality rate with age [30]. These pioneering works were subsequently used, for example, to model the dynamics of tumour growth [31,32], and to describe population-level phenomena of many non-human species [33], such as density dependence [34]. A more recent concept resulting from human work is that of individual-level variation in mortality risk [35], a phenomenon present in many species [36]. In demography it has improved estimates of population-level parameters such as life-expectancy and demographic rates [35] and, in ecology and evolution, it has been used to quantify ageing rates [37,38], developmental plasticity, and behavioural syndromes [39]. Other examples of knowledge transfer from human to non-human systems include the occurrence of late-life mortality plateaus [40] and the convergence of mortality rates of high-and low-quality cohorts [41], both described in humans long before in any other species [42–44]. Many concepts originating from human demographic research have, therefore, provided key insights that are applicable far beyond humans.
(b). Heritability, adaptation, and quantitative genetics
It was data within large demographic records on humans that allowed Fisher to examine the causes of trait variability [45], laying important foundations for the field of quantitative genetics [46]. Fisher's research was initially based on the calculation of heritability: the proportion of phenotypic variation in a trait attributable to additive genetic effects [47]. Despite the complex genetics underlying trait inheritance [48], heritability remains a useful tool in evolutionary biology [49]. For example, because fitness can have genetic variation, heritability quantifies potential rates of adaptation [50]. Heritability is important, not only to studies of fundamental concepts within evolutionary biology, but also to issues of immediate wider concern to human populations such as the potential long-term impacts of environmental change [51], pesticide and antibiotic resistance, and the evolution of disease. Heritability, clearly, is a fundamental concept within evolutionary biology which originates from ideas initially formulated and tested in humans.
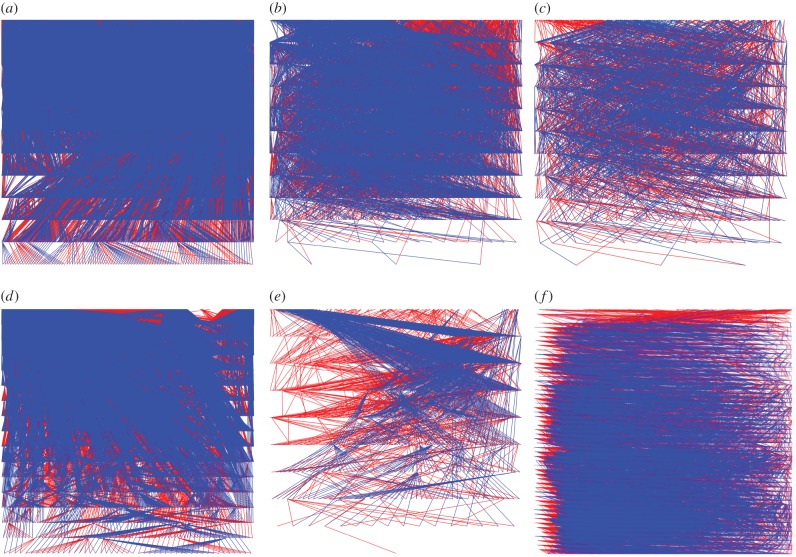
Despite the utility of heritability, recent work has highlighted the limited, and potentially misleading, insight that heritability provides in terms of modelling evolution and using the breeder’s equation [52–54]. For example, heritability does not necessarily reflect the ability of populations to evolve in an adaptive way, perhaps better captured in the more recent concept of evolvability [55]. Quantitative geneticists are, therefore, increasingly pursuing alternative approaches when studying evolution and the response to selection [56–58]. Such goals often require large quantities of data and, despite statistical approaches that can make use of complex pedigree relationships in large datasets [59], this limits the potential for such studies in natural populations. Human-based research can, however, make a valuable contribution in this area owing to the vast multi-generational datasets available [60], which are comparable to highly regarded datasets on natural populations of other species (figure 2), and have previously been used to estimate heritability [7,61] and other quantitative genetic parameters (e.g. G-matrices [58] and intersexual genetic correlations [62]).
Figure 2.
Pedigrees from a human population and several large datasets of wild non-human populations. (a) Human, Finnish Human Population Data; (b) Soay Sheep, St Kilda Soay Sheep Project (data provided by Alastair Wilson); (c) Seychelles Warbler, Seychelles Warbler Project (figure provided by Hannah Dugdale and David Richardson); (d) Collared Flycatcher, Gotland (data provided by Simon Evans); (e) Bighorn Sheep, Alpine Ungulate Research Group (figure provided by David Coltman); (f) Great Tit, Wytham Tit Project. (Figure provided by Josh Firth.)
Furthermore, extensive records on ecological parameters such as crop yields, disease dynamics, and climate are available for many human populations, offering quite unique depths of information and opportunity. Many natural populations of non-human species now face dramatically changing environments, the consequences of which remain poorly understood. Combining records of human populations with environmental data, there is potential to draw parallels on the general evolutionary and ecological consequences of rapid environmental change, improving understanding of how changing environments affect quantitative genetic parameters and the potential to adapt to environmental change. For example, such data allow evolutionary biologists to address how rapid environmental change affects genetic (co)variance, and the response to selection in different conditions [58]. There is a pressing need for a greater general understanding of how changing environments affect the genetic basis of adaptation [63], which can be aided by contributions from human studies.
One issue highlighted by our survey is the influence of heritable non-genetic information, such as cultural influences. Identifying and quantifying such effects remains a major challenge [7], but one not limited to humans. Culture in non-human societies was acknowledged only relatively recently [64,65], but studies in non-human systems will benefit by drawing parallels with human studies. Furthermore, while gene–culture coevolution in humans is an established field [66], research on the impact of culture at the genetic level has only begun relatively recently in non-human systems [67,68], and the insights and methods from human studies will be relevant within this emerging field.
Finally, research on human genetics and genomics has produced a wealth of data over the last two decades, allowing humans to be among the species at the forefront of evolutionary quantitative genetics. For example, using genome-wide association studies (GWAS), researchers can identify the underlying alleles involved in trait variation. Until now, however, researchers have only been able to identify alleles which explain a fraction of the heritability in a given trait, and a considerable part of trait heritability remains unexplained, the so-called ‘missing heritability’ [69]. Although first identified in humans, the same observation has since been made in a number of other species [70]. This has led to the suggestion that most adaptive change comes through relatively modest changes accumulating over gene networks, a so-called ‘omnigenic’ model [71,72]. Clearly, human studies have made important contributions to the field of quantitative genetics and, owing to the sheer amount of data and techniques available, continue to do so while offering the opportunity to address critical issues of broader interest.
(c). Human health and the predictive adaptive response
A substantial body of work has accumulated on phenotypic plasticity which considers the long-term effects of developmental conditions in humans. Early work on humans showed positive associations between early- and late-life cohort mortality [73,74] and between early-life conditions and adult lifespan [75]. Such associations have subsequently been found across a range of species [76,77]. The consistent pattern among these studies is that individuals exposed to better developmental conditions outperform those that experienced challenging conditions in terms of health, lifespan, and reproduction, referred to as the ‘silver-spoon’ effect [78]. These concepts are important for evolutionary biologists because such long-term effects can drive individual differences in fitness [79], hence affecting eco-evolutionary dynamics.
Despite considerable support for the silver-spoon hypothesis across taxa, human studies have also revealed some cases where cohorts exposed to good conditions show poorer adult health. For example, the negative association between birthweight and the risk of developing type 2 diabetes and heart disease is most profound for cohorts with better developmental conditions [80]. Examples like this triggered the hypothesis that individuals exposed to poor environmental conditions go through physiological changes that better prepare them to cope with similar challenges in the future, referred to as ‘predictive adaptive response’ (PAR).
Since its original formulation in humans, the PAR hypothesis has sparked further developments [81,82] and research in other species. The silver-spoon and PAR hypotheses offer contrasting predictions, and a major line of research has formed to disentangle these from each other in humans [83] and other species [84]. Human studies though remain correlative, which can be an issue, for example, when there are mortality biases [85]. Disentangling the silver-spoon and PAR hypotheses without confounding variables requires experiments, which are only feasible in non-human systems. A meta-analysis, pooling most experimental studies to date, showed some evidence for PAR, but the effect was weak [84]. This weak signal may arise either because PAR effects are infrequent or because the benefit that can be gained from physiological adjustments is small. Studies in different species are crucial in disentangling these two possibilities.
(d). Senescence and the evolution of post-reproductive lifespan
Senescence is the decline in organismal functioning, survival, and/or reproduction with increasing age [86]. As recently as the 1950s, senescence was considered a uniquely human trait that did not occur in nature. Hence our evolutionary theories on the ageing conundrum (why does senescence occur despite natural selection?) are based on human observations. It was much later that descriptions of senescence began to emerge for other species. Now, a vast body of evidence shows that senescence occurs in a wide variety of taxa [25,87]. This diversity is now a major strength in the study of evolutionary processes and physiological pathways involved in senescence. The study of the evolution of senescence is an example which was motivated by human studies, while the subsequent broadening to other study organisms helps us better understand the process in humans.
An interesting ageing case is post-reproductive lifespan (PRLS)—lifespan beyond the age at reproductive cessation. Until the mid-1980s, evolutionary biologists paid little attention to the evolution of this specific aspect of human life-history and ageing pattern, because it was initially viewed as a uniquely human trait [88,89], and was mostly studied by scientists working exclusively on humans. PRLS also represents an evolutionary conundrum. After all, what fitness benefit could the cessation of reproduction give? The first adaptive theories to explain PRLS were rooted both in the fact that late-life reproduction is associated with increased mortality risk in mothers, and that social individuals might increase their fitness by helping relatives [86,90]. Indeed, (grand)mothers increase (grand)offspring survival [91] or reproduction in humans [92]. More recently, PRLS is increasingly being studied in non-human systems [93–97], with some species showing extents of PRLS comparable with those found in humans [96].
A major focus within evolutionary biology is the identification of the common features of species exhibiting PRLS, which may help to explain how this pattern evolves. These features include sociality and cooperation [98] and some eco-demographic variables. For example, age-specific patterns of relatedness among females in social groups can affect kin benefits, reproductive costs, and the evolution of PRLS in humans and cetaceans [99]. This hypothesis has now received support from studies on historical humans [100] and comparative analysis of mammals [101]. Research to identify ecological conditions and life-history traits that favour the evolution of PRLS, an insight mostly gained from non-human studies, is flourishing both in humans and non-human systems.
While initially observed in humans, PRLS occurs in a wide variety of species. Based on human observation, various mutually non-exclusive adaptive explanations for PRLS have been proposed. However, the origin of PRLS is difficult to infer solely from human studies, and further theoretical developments and empirical tests of the adaptive benefits of PRLS require non-human studies. Indeed, current theoretical and empirical knowledge, from human and non-human studies, suggests that the evolution of PRLS is heterogeneous, the result of several selective pressures (natural, kin, and sexual selection), and is explained by both adaptive and non-adaptive processes [101]. However, as a long-lived species with a long PRLS, complex sociality and the most dissected physiological knowledge of reproductive cessation of any species, humans remain essential to furthering our understanding of the evolution of PRLS, in general.
4. Conclusion
Several pioneering studies on human populations have provided key insights, concepts, and tools to evolutionary biology (table 2). These studies focused on topics such as genetics, life-history evolution, and demography, all discussed in our article, as well as topics not covered here. The majority of researchers within evolutionary biology and associated fields acknowledged that human studies can provide insights applicable to non-human systems, with those more exposed to human studies expressing more positive views. The main issues identified with human studies are the lack of experimental manipulations, and the non-natural and cultural environment. Indeed, these issues can alter the course of evolution. However, these limitations also apply to other species with a slower pace of life, such as elephants, cetaceans, primates, seabirds, turtles, and some fish species. Human studies can inspire research in such systems, for example, through the ground-breaking work of gene–culture coevolution together with the detailed knowledge of genetic dynamics of human populations. Carefully designed experiments are essential to exclude effects of confounding factors and to confirm the causality of the patterns seen. Predictive adaptive responses illustrate that a fundamental concept can be formulated based on non-experimental human studies, after which they can be investigated experimentally in a wide variety of species. The historical monitoring of births and deaths in various human populations has given rise to demography, from which methods and concepts are now applied in various disciplines and many species alike. Detailed knowledge of family relationships made it possible to study heritability and genetic variation, and to study the potential benefits of grand-mothering and PRLS, concepts relevant to other species and to evolutionary biology in general. We have demonstrated here that human studies have made a host of valuable contributions to the understanding of fundamental concepts within evolutionary biology, and highlighted some of the many ways in which they are highly suited to making a continued contribution applicable across a variety of disciplines and sparking research in many other species.
Table 2.
Examples of evolutionary concepts and methods first discovered in humans and subsequently applied to non-human species. Note that, for more recent examples, the time between the human and the non-human references decreases, indicative of increased rates of interdisciplinary communication.
| example | references |
|
|---|---|---|
| human | non-human | |
| demography | ||
| life tables | Graunt [102] | Deevey [13] |
| mortality equation | Gompertz [30] | Kimball [103] |
| logistic equation | Verhulst [29] | Allee [34]a |
| individual heterogeneity | Vaupel et al. [35] | Harcombe [104] |
| genetics | ||
| missing heritability | Maher [69] | Brachi et al. [70] |
| gene–culture coevolution | Durham [105]a | Whitehead [67] |
| predictive adaptive response | Gluckman & Hanson [106] | Taborsky [107]b |
| post-reproductive lifespan | Williams [86] | Cohen [95] |
aAnd references therein.
bUsing a full-factorial experimental design.
Supplementary Material
Acknowledgement
We thank Erik Postma, Sarah Brosnan, Simon Verhulst, Jean-Michel Gaillard, and Gert Stulp for discussions.
Data accessibility
Survey data are available at: http://dx.doi.org/10.5061/dryad.513b9 [3].
Authors' contributions
All the authors were involved in the planning the survey, analysis of the data, and writing of the manuscript.
Competing interests
We have no competing interests.
Funding
Funding was provided by the Academy of Finland and by the European Research Council to V.L.
References
- 1.Clutton-Brock T, Sheldon BC. 2010. Individuals and populations: the role of long-term, individual-based studies of animals in ecology and evolutionary biology. Trends Ecol. Evol. 25, 562–573. ( 10.1016/j.tree.2010.08.002) [DOI] [PubMed] [Google Scholar]
- 2.Christensen RHB. 2015. Ordinal: regression models for ordinal data. https://cran.r-project.org/web/packages/ordinal/index.html.
- 3.Briga M, Griffin RM, Berger V, Pettay JE, Lummaa V. 2017. Data from: what have humans done for evolutionary biology? Contributions from genes to populations Dryad Digital Repository. ( 10.5061/dryad.513b9) [DOI] [PMC free article] [PubMed]
- 4.Tait L. 1869. Art. XII-has the law of natural selection by survival of the fittest failed in the case of man? Dublin Q. J. Med. Sci. 47, 102–113. ( 10.1007/BF02944468) [DOI] [Google Scholar]
- 5.Meikle J. 2013. Sir David Attenborough warns against large families and predicts things will only get worse. The Guardian. http://www.theguardian.com/tv-and-radio/2013/sep/10/david-attenborough-human-evolution-stopped. [Google Scholar]
- 6.Scranton K, Lummaa V, Stearns SC. 2016. The importance of the timescale of the fitness metric for estimates of selection on phenotypic traits during a period of demographic change. Ecol. Lett. 19, 854–861. ( 10.1111/ele.12619) [DOI] [PubMed] [Google Scholar]
- 7.Stearns SC, Byars SG, Govindaraju DR, Ewbank D. 2010. Measuring selection in contemporary human populations. Nat. Rev. Genet. 11, 611–622. ( 10.1038/nrg2831) [DOI] [PubMed] [Google Scholar]
- 8.Blackburn EH. 1991. Structure and function of telomeres. Nature 350, 569–573. ( 10.1038/350569a0) [DOI] [PubMed] [Google Scholar]
- 9.Nussey DH, et al. 2014. Measuring telomere length and telomere dynamics in evolutionary biology and ecology. Methods Ecol. Evol. 5, 299–310. ( 10.1111/2041-210X.12161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf M, Weissing FJ. 2012. Animal personalities: consequences for ecology and evolution. Trends Ecol. Evol. 27, 452–461. ( 10.1016/j.tree.2012.05.001) [DOI] [PubMed] [Google Scholar]
- 11.Kurvers RHJM, Krause J, Croft DP, Wilson ADM, Wolf M. 2014. The evolutionary and ecological consequences of animal social networks: emerging issues. Trends Ecol. Evol. 29, 326–335. ( 10.1016/j.tree.2014.04.002) [DOI] [PubMed] [Google Scholar]
- 12.Greenwood M. 1938. The first life table. Notes Rec. R. Soc. Lond. 1, 70–72. ( 10.1098/rsnr.1938.0017) [DOI] [Google Scholar]
- 13.Deevey ESJ. 1947. Life tables for natural populations of animals. Q. Rev. Biol. 22, 283–314. ( 10.1086/395888) [DOI] [PubMed] [Google Scholar]
- 14.Cox D. 1972. Regression models and life-tables. J. R. Stat. Soc. B 34, 187–220. [Google Scholar]
- 15.Colchero F, Jones OR, Rebke M. 2012. BaSTA: an R package for Bayesian estimation of age-specific survival from incomplete mark-recapture/recovery data with covariates. Methods Ecol. Evol. 3, 466–470. ( 10.1111/j.2041-210X.2012.00186.x) [DOI] [Google Scholar]
- 16.Human mortality Database. In press. University of California, Berkeley (USA) and Max Planck Institute for Demographic Research (Germany). See www.mortality.org or www.humanmortality.de.
- 17.Vaupel JW, et al. 1998. Biodemographic trajectories of longevity. Science 280, 855–860. ( 10.1126/science.280.5365.855) [DOI] [PubMed] [Google Scholar]
- 18.Vaupel JW. 2010. Biodemography of human ageing. Nature 464, 536–542. ( 10.1038/nature08984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen K, Doblhammer G, Rau R, Vaupel JW. 2009. Ageing populations: the challenges ahead. Lancet 374, 1196–1208. ( 10.1016/S0140-6736(09)61460-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf CJE, Pavard S. 2007. Why evolutionary biologists should be demographers. Trends Ecol. Evol. 22, 205–212. ( 10.1016/j.tree.2006.12.001) [DOI] [PubMed] [Google Scholar]
- 21.Caswell H. 2001. Matrix population models: construction, analysis, and interpretation. 2nd edn Sunderland, MA: Sinauer Associates. [Google Scholar]
- 22.Smallegange IM, Coulson T. 2013. Towards a general, population-level understanding of eco-evolutionary change. Trends Ecol. Evol. 28, 143–148. ( 10.1016/j.tree.2012.07.021) [DOI] [PubMed] [Google Scholar]
- 23.Oeppen J, Vaupel JW. 2002. Broken limits to life expectancy. Science 296, 1029–1031. ( 10.4054/DemRes.2011.24.11) [DOI] [PubMed] [Google Scholar]
- 24.Boonekamp JJ, Simons MJP, Hemerik L, Verhulst S. 2013. Telomere length behaves as biomarker of somatic redundancy rather than biological age. Aging Cell 12, 330–332. ( 10.1111/acel.12050) [DOI] [PubMed] [Google Scholar]
- 25.Jones OR, et al. 2014. Diversity of ageing across the tree of life. Nature 505, 169–173. ( 10.1038/nature12789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coulson T, Catchpole EA, Albon SD, Morgan BJ, Pemberton JM, Clutton-Brock TH, Crawley MJ, Grenfell BT. 2001. Age, sex, density, winter weather, and population crashes in Soay sheep. Science 292, 1528–1531. ( 10.1126/science.292.5521.1528) [DOI] [PubMed] [Google Scholar]
- 27.Ozgul A, Childs DZ, Oli MK, Armitage KB, Blumstein DT, Olson LE, Tuljapurkar S, Coulson T. 2010. Coupled dynamics of body mass and population growth in response to environmental change. Nature 466, 482–485. ( 10.1038/nature09210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malthus TR. 1798. An essay on the principle of population, as it affects the future improvement of society, with remarks on the speculations of Mr Godwin, M. Condoret and other writers. London, UK: Johnson; ( 10.2307/2064821) [DOI] [Google Scholar]
- 29.Verhulst P. 1838. Notice sur la loi que la population suit dans son accroisement. Corresp. mathématique Phys. 10, 113–121. [Google Scholar]
- 30.Gompertz B. 1825. On the nature of the function expressive of the law of human mortality, and on a new mode of determining the value of life contingencies. Phil. Trans. R. Soc. 115, 513–585. ( 10.1098/rstb.2014.0379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atici FM, Şengül S. 2010. Modeling with fractional difference equations. J. Math. Anal. Appl. 369, 1–9. ( 10.1016/j.jmaa.2010.02.009) [DOI] [Google Scholar]
- 32.Norton L. 1988. A Gompertzian model of human breast cancer growth. Cancer Res. 48, 7067–7071. [PubMed] [Google Scholar]
- 33.Winsor CP. 1932. The Gompertz curve as a growth curve. Proc. Natl Acad. Sci. USA 18, 1–8. ( 10.1073/pnas.18.1.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allee WC. 1931. Animal aggregations, a study in general sociobiology. Chicago, IL: University of Chicago Press. [Google Scholar]
- 35.Vaupel JW, Manton K, Stallard E. 1979. The impact of heterogeneity in individual frailty on the dynamics of mortality. Demography 16, 439–454. ( 10.2307/2061224) [DOI] [PubMed] [Google Scholar]
- 36.Hanson MA, Gluckman PD. 2014. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiol. Rev. 94, 1027–1076. ( 10.1152/physrev.00029.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cam E, Link WA, Cooch EG, Monnat J, Danchin E. 2002. Individual covariation in life-history traits: seeing the trees despite the forest. Am. Nat. 159, 96–105. ( 10.1086/324126) [DOI] [PubMed] [Google Scholar]
- 38.van de Pol M, Verhulst S. 2006. Age-dependent traits: a new statistical model to separate within- and between- individual effects. Am. Nat. 167, 766–773. [DOI] [PubMed] [Google Scholar]
- 39.van de Pol M, Wright J. 2009. A simple method for distinguishing within- versus between-subject effects using mixed models. Anim. Behav. 77, 753–758. ( 10.1016/j.anbehav.2008.11.006) [DOI] [Google Scholar]
- 40.Brownlee J. 1919. Notes on the biology of a life-table. J. R. Stat. Soc. 82, 34–77. ( 10.2307/2340732) [DOI] [Google Scholar]
- 41.Strehler B, Mildvan A. 1960. General theory of mortality and aging. Science 132, 14–21. ( 10.1126/science.132.3418.14) [DOI] [PubMed] [Google Scholar]
- 42.Carey JR, Liedo P, Orozco D, Vaupel JW. 1992. Slowing of mortality rates at older ages in large medfly cohorts. Science 258, 457–461. ( 10.1126/science.1411540) [DOI] [PubMed] [Google Scholar]
- 43.Bronikowski AM, Morgan TJ, Garland T, Carter PA. 2006. The evolution of aging and age-related physical decline in mice selectively bred for high voluntary exercise. Evolution 60, 1494–1508. ( 10.1111/j.0014-3820.2006.tb01228.x) [DOI] [PubMed] [Google Scholar]
- 44.Briga M, Koetsier E, Boonekamp J, Jimeno B, Verhulst S. 2017. Food availability affects adult survival trajectories depending on early developmental conditions. Proc. R. Soc. B 284, 20162287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisher RA. 1918. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52, 399–433. ( 10.1017/S0080456800012163) [DOI] [Google Scholar]
- 46.Roff DA. 2007. A centennial celebration for quantitative genetics. Evolution 61, 1017–1032. ( 10.1111/j.1558-5646.2007.00100.x) [DOI] [PubMed] [Google Scholar]
- 47.Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics. 4th edn London, UK: Pearson Prentice Hall. [Google Scholar]
- 48.Hansen TF, Pélabon C, Houle D. 2011. Heritability is not evolvability. Evol. Biol. 38, 258–277. ( 10.1007/s11692-011-9127-6) [DOI] [Google Scholar]
- 49.Visscher PM, Hill WG, Wray NR. 2008. Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet. 9, 255–266. ( 10.1038/nrg2322) [DOI] [PubMed] [Google Scholar]
- 50.Fisher RA. 1930. The genetical theory of natural selection. Oxford, UK: Clarendon Press. [Google Scholar]
- 51.Hoffmann A, Sgrò C. 2011. Climate change and evolutionary adaptation. Nature 470, 479–485. ( 10.1038/nature09670) [DOI] [PubMed] [Google Scholar]
- 52.Blows MW. 2007. A tale of two matrices: multivariate approaches in evolutionary biology. J. Evol. Biol. 20, 1–8. ( 10.1111/j.1420-9101.2006.01164.x) [DOI] [PubMed] [Google Scholar]
- 53.Morrissey MB, Kruuk LEB, Wilson AJ. 2010. The danger of applying the breeder's equation in observational studies of natural populations. J. Evol. Biol. 23, 2277–2288. ( 10.1111/j.1420-9101.2010.02084.x) [DOI] [PubMed] [Google Scholar]
- 54.Wilson AJ, Poissant J. 2016. Quantitative genetics in natural populations. In Encylopedia of evolutionary biology (ed Kliman RM.), pp. 361–371. Oxford, UK: Academic Press; ( 10.1016/B978-0-12-800049-6.00046-9361) [DOI] [Google Scholar]
- 55.Houle D. 1992. Comparing evolvability and variability of quantitative traits. Genetics 130, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coulson T, Tuljapurkar S. 2008. The dynamics of a quantitative trait in an age-structured population living in a variable environment. Am. Nat. 172, 599–612. ( 10.1086/591693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozgul A, Tuljapurkar S, Benton TG, Pemberton JM, Clutton-Brock TH, Coulson T. 2009. The dynamics of phenotypic change and the shrinking sheep of St. Kilda. Science 325, 464–467. ( 10.1126/science.1173668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolund E, Hayward A, Pettay JE, Lummaa V. 2015. Effects of the demographic transition on the genetic variances and covariances of human life-history traits. Evolution 69, 747–755. ( 10.1111/evo.12598) [DOI] [PubMed] [Google Scholar]
- 59.Kruuk L. 2004. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. Lond. B 359, 873–890. ( 10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolund E, Hayward A, Lummaa V. 2016. Life history evolution, human. In Encyclopedia of evolutionary biology (ed. Kliman RM.), pp. 328–334. Oxford, UK: Academic Press; ( 10.1016/B978-0-12-800049-6.00097-4) [DOI] [Google Scholar]
- 61.Pettay JE, Kruuk LEB, Jokela J, Lummaa V. 2005. Heritability and genetic constraints of life-history trait evolution in preindustrial humans. Proc. Natl Acad. Sci. USA 102, 2838–2843. ( 10.1073/pnas.0406709102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolund E, Bouwhuis S, Pettay JE, Lummaa V. 2013. Divergent selection on, but no genetic conflict over, female and male timing and rate of reproduction in a human population. Proc. R. Soc. B 280, 20132002 ( 10.1098/rspb.2013.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brooks RC, Garratt MG. 2017. Life history evolution, reproduction, and the origins of sex-dependent aging and longevity. Ann. N. Y. Acad. Sci. 1389, 92–107. ( 10.1111/nyas.13302) [DOI] [PubMed] [Google Scholar]
- 64.Whiten A, Goodall J, McGrew WC, Nishida T, Reynolds V, Sugiyama Y, Tutin CEG, Wrangham RW, Boesch C. 1999. Cultures in chimpanzees. Nature 399, 682–685. ( 10.1038/21415) [DOI] [PubMed] [Google Scholar]
- 65.Rendell L, Whitehead H. 2001. Culture in whales and dolphins. Behav. Brain Sci. 24, 309–324; discussion 324–382 ( 10.1016/B978-0-12-373553-9.00068-7) [DOI] [PubMed] [Google Scholar]
- 66.Laland KN, Odling-Smee J, Myles S. 2010. How culture shaped the human genome: bringing genetics and the human sciences together. Nat. Rev. Genet. 11, 137–148. ( 10.1038/nrg2734) [DOI] [PubMed] [Google Scholar]
- 67.Whitehead H. 1998. Cultural selection and genetic diversity in matrilineal whales. Science 282, 1708–1711. ( 10.1126/science.282.5394.1708) [DOI] [PubMed] [Google Scholar]
- 68.Foote A, et al. 2016. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat. Commun. 7, 11 693 ( 10.1038/ncomms11693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maher B. 2008. Personal genomes: The case of the missing heritability. Nature 456, 18–21. ( 10.1038/456018a) [DOI] [PubMed] [Google Scholar]
- 70.Brachi B, Morris GP, Borevitz JO. 2011. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12, 232 ( 10.1186/gb-2011-12-10-232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rockman MV. 2012. The QTN program and the alleles that matter for evolution: all that's gold does not glitter. Evolution 66, 1–17. ( 10.1111/j.1558-5646.2011.01486.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186. ( 10.1016/j.cell.2017.05.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Derrick VPA. 1927. Observations on (1) Errors of age in the population statistics of England and Wales, and (2) the changes in mortality indicated by the national records. J. Inst. Actuar. 58, 117–159. ( 10.1017/S0020268100031474) [DOI] [Google Scholar]
- 74.Kermack WO, McKendrick AG, McKinlay PL. 1934. Death-rates in Great Britain and Sweden: expression of specific mortality rates as products of two factors, and some consequences thereof. J. Hyg. (Lond). 34, 433–457. ( 10.1017/S0022172400043230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore SE, Cole TJ, Poskitt EME, Sonko BJ, Whitehead RG, McGregor IA, Prentice AM. 1997. Season of birth predicts mortality in rural Gambia. Nature 388, 434 ( 10.1038/41245) [DOI] [PubMed] [Google Scholar]
- 76.Barrett ELB, Hunt J, Moore AJ, Moore PJ. 2009. Separate and combined effects of nutrition during juvenile and sexual development on female life-history trajectories: the thrifty phenotype in a cockroach. Proc. R. Soc. B 276, 3257–3264. ( 10.1098/rspb.2009.0725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lindström J. 1999. Early development and fitness in birds and mammals. Trends Ecol. Evol. 14, 343–348. ( 10.1016/S0169-5347(99)01639-0) [DOI] [PubMed] [Google Scholar]
- 78.Grafen A. 1988. On the uses of data on lifetime reproductive success. In Reproductive success (ed Clutton-Brock TH.), pp. 454–471. Chicago, IL: University of Chicago Press. [Google Scholar]
- 79.Harrison XA, Blount JD, Inger R, Norris DR, Bearhop S. 2011. Carry-over effects as drivers of fitness differences in animals. J. Anim. Ecol. 80, 4–18. ( 10.1111/j.1365-2656.2010.01740.x) [DOI] [PubMed] [Google Scholar]
- 80.Forsén T, Erikkson J, Tuomilehto J, Reunanen A, Osmond C, Barker D. 2000. The fetal and childhood growth of persons who develop type 2 diabetes. Ann. Intern. Med. 133, 176–182. ( 10.7326/0003-4819-133-3-200008010-00008) [DOI] [PubMed] [Google Scholar]
- 81.Rickard IJ, Lummaa V. 2007. The predictive adaptive response and metabolic syndrome: challenges for the hypothesis. Trends Endocrinol. Metab. 18, 94–99. ( 10.1016/j.tem.2007.02.004) [DOI] [PubMed] [Google Scholar]
- 82.Nettle D, Frankenhuis WE, Rickard IJ. 2013. The evolution of predictive adaptive responses in human life history. Proc. R. Soc. B 280, 20131343 ( 10.1098/rspb.2013.1343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hayward AD, Rickard IJ, Lummaa V. 2013. Influence of early-life nutrition on mortality and reproductive success during a subsequent famine in a preindustrial population. Proc. Natl Acad. Sci. USA 110, 13 886–13 891. ( 10.1073/pnas.1301817110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uller T, Nakagawa S, English S. 2013. Weak evidence for anticipatory parental effects in plants and animals. J. Evol. Biol. 26, 2161–2170. ( 10.1111/jeb.12212) [DOI] [PubMed] [Google Scholar]
- 85.Ronget V, Gaillard J-M, Coulson T, Garratt M, Gueyffier F, Lega J-C, Lemaitre J-F. 2017. Causes and consequences of variation in offspring body mass: meta-analyses in birds and mammals. Biol. Rev. http://onlinelibrary.wiley.com/doi/10.1111/brv.12329/full. [DOI] [PubMed] [Google Scholar]
- 86.Williams GC. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411. ( 10.2307/2406060) [DOI] [Google Scholar]
- 87.Nussey DH, Froy H, Lemaitre JF, Gaillard JM, Austad SN. 2013. Senescence in natural populations of animals: widespread evidence and its implications for bio-gerontology. Ageing Res. Rev. 12, 214–225. ( 10.1016/j.arr.2012.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pavelka MSM, Fedigan LM. 1991. Menopause: a comparative life history perspective. Yearb. Phys. Anthropol. 34, 13–38. ( 10.1016/0378-5122(92)90095-L) [DOI] [Google Scholar]
- 89.Hawkes K, O'Connell JF, Jones NGB, Alvarez H, Charnov EL. 1998. Grandmothering, menopause, and the evolution of human life histories. Proc. Natl Acad. Sci. USA 95, 1336–1339. ( 10.1073/pnas.95.3.1336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hamilton WD. 1964. The genetical evolution of social behaviour I & II. J. Theor. Biol. 7, 1–52. ( 10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 91.Sear R, Mace R. 2008. Who keeps children alive? A review of the effects of kin on child survival. Evol. Hum. Behav. 29, 1–18. ( 10.1016/j.evolhumbehav.2007.10.001) [DOI] [Google Scholar]
- 92.Lahdenperä M, Lummaa V, Helle S, Tremblay M, Russell AF. 2004. Fitness benefits of prolonged postreproductive lifespan in women. Nature 428, 178–181. ( 10.1038/nature02367) [DOI] [PubMed] [Google Scholar]
- 93.Bourke AFG. 2007. Kin selection and the evolutionary theory of aging. Annu. Rev. Ecol. Evol. Syst. 38, 103–128. ( 10.1146/annurev.ecolsys.38.091206.095528) [DOI] [Google Scholar]
- 94.Holmes DJ, Ottinger MA. 2003. Birds as long-lived animal models for the study of aging. Exp. Gerontol. 38, 1365–1375. ( 10.1016/j.exger.2003.10.018) [DOI] [PubMed] [Google Scholar]
- 95.Cohen AA. 2004. Female post-reproductive lifespan: a general mammalian trait. Biol. Rev. 79, 733–750. ( 10.1017/S1464793103006432) [DOI] [PubMed] [Google Scholar]
- 96.Croft DP, Brent LJN, Franks DW, Cant MA. 2015. The evolution of prolonged life after reproduction. Trends Ecol. Evol. 30, 407–416. ( 10.1016/j.tree.2015.04.011) [DOI] [PubMed] [Google Scholar]
- 97.Lahdenperä M, Mar KU, Lummaa V. 2014. Reproductive cessation and post-reproductive lifespan in Asian elephants and pre-industrial humans. Front. Zool. 11, 54 ( 10.1186/s12983-014-0054-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carey JR, Judge DS. 2001. Life span extension in humans is self-reinforcing: a general theory of longevity. Popul. Dev. Rev. 27, 411–436. ( 10.1111/j.1728-4457.2001.00411.x) [DOI] [Google Scholar]
- 99.Johnstone RA, Cant MA. 2010. The evolution of menopause in cetaceans and humans: the role of demography. Proc. R. Soc. B 277, 3765–3771. ( 10.1098/rspb.2010.0988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lahdenperä M, Gillespie DOS, Lummaa V, Russell AF. 2012. Severe intergenerational reproductive conflict and the evolution of menopause. Ecol. Lett. 15, 1283–1290. ( 10.1111/j.1461-0248.2012.01851.x) [DOI] [PubMed] [Google Scholar]
- 101.Nichols HJ, Zecherle L, Arbuckle K. 2016. Patterns of philopatry and longevity contribute to the evolution of post-reproductive lifespan in mammals. Biol. Lett. 12, 20150992 ( 10.1098/rsbl.2015.0992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Graunt J. 1662. Natural and political observations made upon the bills of mortality. London, UK: The Royal Society. [Google Scholar]
- 103.Kimball AW. 1960. Estimation of mortality intensities in animal experiments. Biometrics 16, 505–521. ( 10.2307/2527758) [DOI] [Google Scholar]
- 104.Harcombe PA. 1987. Tree life tables. Bioscience 37, 557–568. ( 10.1525/bio.2009.59.3.13) [DOI] [Google Scholar]
- 105.Durham WH. 1991. Co-evolution: genes, culture and human diversity. Stanford, UK: Stanford University Press. [Google Scholar]
- 106.Gluckman PD, Hanson MA. 2004. Living with the past: evolution, development, and patterns of disease. Science 305, 1733–1736. ( 10.1126/science.1095292) [DOI] [PubMed] [Google Scholar]
- 107.Taborsky B. 2006. The influence of juvenile and adult environments on life-history trajectories. Proc. R. Soc. B 273, 741–750. ( 10.1098/rspb.2005.3347) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Briga M, Griffin RM, Berger V, Pettay JE, Lummaa V. 2017. Data from: what have humans done for evolutionary biology? Contributions from genes to populations Dryad Digital Repository. ( 10.5061/dryad.513b9) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Survey data are available at: http://dx.doi.org/10.5061/dryad.513b9 [3].