Abstract
Maintaining stable intracellular pH (pHi) is essential for homeostasis, and requires the ability to both sense pH changes that may result from internal and external sources, and to regulate downstream compensatory pH pathways. Here we identified the cAMP-producing enzyme soluble adenylyl cyclase (sAC) as the first molecular pH sensor in corals. sAC protein was detected throughout coral tissues, including those involved in symbiosis and calcification. Application of a sAC-specific inhibitor caused significant and reversible pHi acidosis in isolated coral cells under both dark and light conditions, indicating sAC is essential for sensing and regulating pHi perturbations caused by respiration and photosynthesis. Furthermore, pHi regulation during external acidification was also dependent on sAC activity. Thus, sAC is a sensor and regulator of pH disturbances from both metabolic and external origin in corals. Since sAC is present in all coral cell types, and the cAMP pathway can regulate virtually every aspect of cell physiology through post-translational modifications of proteins, sAC is likely to trigger multiple homeostatic mechanisms in response to pH disturbances. This is also the first evidence that sAC modulates pHi in any non-mammalian animal. Since corals are basal metazoans, our results indicate this function is evolutionarily conserved across animals.
Keywords: soluble adenylyl cyclase, acid/base, Symbiodinium, CO2, cAMP, symbiosis
1. Introduction
Regulation of intracellular pH (pHi) is critical for the physiological function of all cells and involves a combination of passive (i.e. buffering by proteins, phosphates and 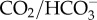 ) and active (i.e. transport of acid/base equivalents energized by ATPases) compensatory mechanisms. Reef-building corals regularly experience pH stress as a result of their own respiration, calcification and photosynthesis, as well as from external sources including the metabolism of reef-associated organisms, tides, water flow, upwelling, and ocean acidification (reviewed in [1]). These diverse pH stressors take place over a wide temporal scale ranging from seconds (e.g. rapid metabolic processes), to hours (e.g. diel photosynthesis and calcification), to months (e.g. seasonal temperature and light variability, upwelling), to years (e.g. ocean acidification). In order to regulate pHi in response to these dynamic disturbances, corals must be able to first sense changes in pH, and to then initiate the appropriate compensatory pathways. The mechanisms of pH sensing in corals have yet to be described, yet they are fundamental for understanding basic coral biology and the capacity of corals to adapt to long-term shifts in seawater pH driven by climate change.
) and active (i.e. transport of acid/base equivalents energized by ATPases) compensatory mechanisms. Reef-building corals regularly experience pH stress as a result of their own respiration, calcification and photosynthesis, as well as from external sources including the metabolism of reef-associated organisms, tides, water flow, upwelling, and ocean acidification (reviewed in [1]). These diverse pH stressors take place over a wide temporal scale ranging from seconds (e.g. rapid metabolic processes), to hours (e.g. diel photosynthesis and calcification), to months (e.g. seasonal temperature and light variability, upwelling), to years (e.g. ocean acidification). In order to regulate pHi in response to these dynamic disturbances, corals must be able to first sense changes in pH, and to then initiate the appropriate compensatory pathways. The mechanisms of pH sensing in corals have yet to be described, yet they are fundamental for understanding basic coral biology and the capacity of corals to adapt to long-term shifts in seawater pH driven by climate change.
Corals have two tissue layers, the ectodermis and gastrodermis, which fold over each other to form the gastrovascular cavity, also known as the coelenteron (figure 1). There is large carbon chemistry and pH variability among extra- and intracellular coral compartments [1,7]. The oral ectodermis is in contact with seawater, which has typical pH values of between 7.9 and 8.3 [3,8]. The coral's symbiotic dinoflagellate algae (Symbiodinium) reside inside the symbiosome of gastrodermal cells, which is around pH 4 [9]. In contrast, the calcifying cells in the calicoblastic epithelium lay on top of alkaline extracellular pockets of fluid, called the sub-calicoblastic medium (SCM), for which pH values of between 8.2 and 10 have been reported [4–6,8]. The coelenteron has its own distinctive pH as well, with values of between 6.6 and 8.5 [3,4,8]. While some of the reported pH variability in these coral microenvironments is likely explained by methodological and species-specific differences, an additional internal source of both temporal and spatial pH variability is Symbiodinium photosynthesis. Photosynthesis induces cytosol alkalinization of Symbiodinium-hosting cells by approximately 0.4 pH units [10–12], while gastrodermal cells lacking Symbiodinium remain at a constant pH across light and dark conditions [11,12]. Meanwhile, the pH of the adjacent coelenteron increases by approximately 0.7 pH units in the light [3,8], presumably due to photosynthesis-driven consumption of CO2. However, there is a sharp drop in pH of the coelenteron immediately above the aboral gastrodermis [4], likely due to removal of H+ from the SCM as a result of calcification. Photosynthesis also induces alkalinization of the SCM, which increases by 0.3–1.0 pH units from dark to light [5,8]. However, the pHi of calicoblastic cells themselves remains stable [5].
Figure 1.
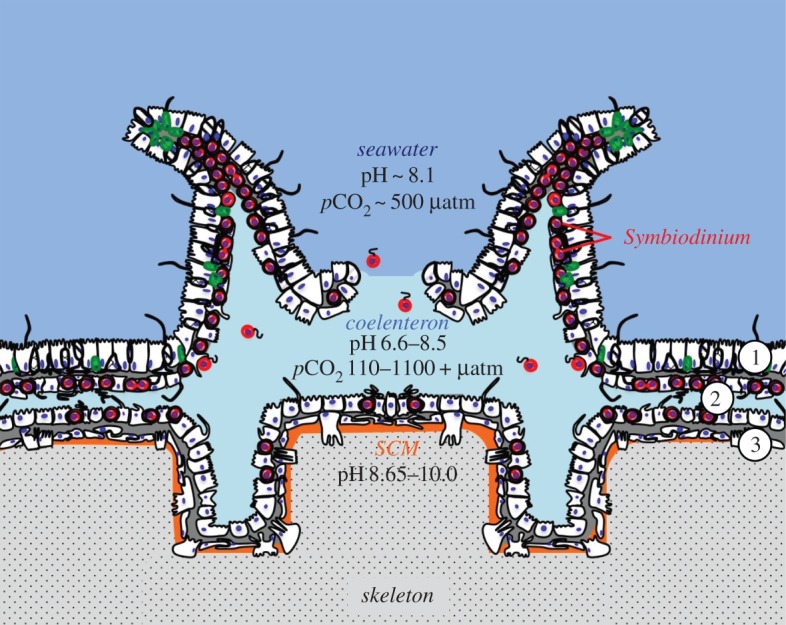
The tissue structure and pH microenvironments of a coral polyp. (Modified from [1]; references for pH: seawater [2], coelenteron [3,4], sub-calicoblastic medium (SCM) [4–6].) Circled numbers: 1 = oral epidermis; 2 = gastrodermis; 3 = calicodermis. (Online version in colour.)
The ability of cells to maintain acid/base homeostasis in the face of these dynamic pH processes requires sensory mechanisms to rapidly detect these changes and initiate homeostatic responses. The recently discovered enzyme soluble adenylyl cyclase (sAC) is directly stimulated by bicarbonate ions 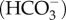 to produce the secondary messenger molecule cyclic AMP (cAMP) [13–15]. Furthermore, due to the rapid and reversible hydration of CO2 into H+ and
to produce the secondary messenger molecule cyclic AMP (cAMP) [13–15]. Furthermore, due to the rapid and reversible hydration of CO2 into H+ and 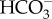 in the presence of carbonic anhydrases (CA), sAC can act not only as a
in the presence of carbonic anhydrases (CA), sAC can act not only as a 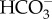 sensor, but also as a sensor of pH and pCO2 (reviewed in [15,16]). Because cAMP can modulate the activity of target proteins through post-translational modifications mediated by protein kinase A (PKA), exchange protein activated by cAMP (EPAC), and cyclic nucleotide gated channels [15,16], sAC has the potential to mediate multiple homeostatic responses in response to acid–base stress. The high conservation of sAC across domains of life suggests that its function as an acid/base sensor is evolutionarily conserved [17,18]. We have previously identified sAC orthologues in genomic databases from the corals Acropora digitifera and Pocillopora damicornis, and reported
sensor, but also as a sensor of pH and pCO2 (reviewed in [15,16]). Because cAMP can modulate the activity of target proteins through post-translational modifications mediated by protein kinase A (PKA), exchange protein activated by cAMP (EPAC), and cyclic nucleotide gated channels [15,16], sAC has the potential to mediate multiple homeostatic responses in response to acid–base stress. The high conservation of sAC across domains of life suggests that its function as an acid/base sensor is evolutionarily conserved [17,18]. We have previously identified sAC orthologues in genomic databases from the corals Acropora digitifera and Pocillopora damicornis, and reported 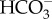 -stimulated cAMP production in A. yongei and P. damicornis tissue homogenates (half-maximal activity or EC50 of approximately 10 mM
-stimulated cAMP production in A. yongei and P. damicornis tissue homogenates (half-maximal activity or EC50 of approximately 10 mM 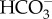 ) that was inhibited by the sAC-specific inhibitor KH7, which are hallmarks of sAC activity [19]. In this study, we cloned several sAC cDNA sequences, characterized sAC protein expression in coral tissues, and examined its role in sensing and counteracting pHi fluctuations derived from both internal and external sources in live coral cells. Our results indicate sAC is a crucial pH sensor and homeostatic regulator in corals, a novel basic mechanism that contributes to our understanding of coral responses to metabolic and environmental acid–base stress.
) that was inhibited by the sAC-specific inhibitor KH7, which are hallmarks of sAC activity [19]. In this study, we cloned several sAC cDNA sequences, characterized sAC protein expression in coral tissues, and examined its role in sensing and counteracting pHi fluctuations derived from both internal and external sources in live coral cells. Our results indicate sAC is a crucial pH sensor and homeostatic regulator in corals, a novel basic mechanism that contributes to our understanding of coral responses to metabolic and environmental acid–base stress.
2. Material and methods
(a). Coral culture
Colonies of P. damicornis at the Hawaii Institute of Marine Biology were maintained in flow through seawater at ambient temperature (24.3 ± 0.8°C) with partially shaded ambient light (approx. 50%). P. damicornis colonies at the Scripps Institution of Oceanography were maintained in flow through seawater (heated to 25°C) with a 12 : 12 h light:dark cycle (approx. 200 µmol photons m−2 s−1).
(b). RT-PCR
To prepare cDNA for RT-PCR, an approximately 2 cm coral fragment was flash frozen in liquid nitrogen and crushed using a chilled mortar and pestle into a fine powder. The powder was homogenized in TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), and total RNA was extracted following the manufacturer's suggested protocol with a few modifications, including an overnight precipitation of the RNA at −80°C and an additional wash step performed with 75% ethanol. Total RNA was heated at 37°C for 2 h and run on an agarose gel to assess RNA quality. Total RNA was cleaned and concentrated using an RNEasy Plus Mini kit (Qiagen, Hilden, Germany). cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) with random hexamers or Oligo(dT) primers according to the manufacturer's protocol. The resulting cDNA was used as a template for RT-PCR. Both full-length sAC sequences were obtained following two rounds of PCR using Phusion High Fidelity taq polymerase (New England Biolabs, Ipswich, MA, USA) and NucleoSpin gel purification (Macherey-Nagel, Düren, Germany). In the second ‘nested’ round of PCR, primers included oligonucleotide overhangs for In-Fusion Cloning (Clontech, Mountain View, CA, USA) into a PCR2.1 vector (Invitrogen).
(c). 5′/3′ RACE PCR
mRNA was isolated from total RNA using a Poly(A)Purist MAG kit (Applied Biosystems, Foster City, CA, USA) and used to synthesize cDNA for 5′ and 3′ Rapid Amplification of cDNA Ends (RACE) PCR (Invitrogen). Following two rounds of PCR using Platinum taq polymerase (Invitrogen), RACE PCR products were TOPO-TA cloned into a PCR2.1 vector (Invitrogen).
The primer sets and conditions used in the first and second (nested) rounds of RT-PCR and RACE PCR are indicated in electronic supplementary material, tables S1 and S2. All cloned PCR products were sequenced by Retrogen, Inc. (San Diego, CA, USA) using a combination of custom, internal primers and the vector primers M13(-20)F and M13R. Phylogenetic analysis was performed using the software package Phylogent.fr with 500 bootstraps (http://www.phylogeny.fr/) [20]; protein domain analyses was performed using PROSITE (http://prosite.expasy.org/) [21] and InterPro (http://www.ebi.ac.uk/interpro/) [22].
(d). Antibodies
Custom rabbit polyclonal antibodies were developed using a peptide antigen and were affinity purified (GenScript USA, Inc.). The peptide antigen sequence, LPGDKHEDDPARAL, was selected from a region of the catalytic domain of the predicted sAC protein sequence from the A digitifera genome [23]. Specificity of the antibodies for P. damicornis sAC was tested by Western blotting on recombinant protein and coral tissue homogenates, and by immunohistochemistry (IHC) on coral histological sections.
The cDNA encoding for the first 499 amino acids of pdsACFL1, which include the catalytic domains, was cloned and recombinantly expressed in E. coli. Western blots indicated that recombinant pdsAC was specifically recognized by our anti-coral sAC antibodies (figure 2b and electronic supplementary material, figure S2). Controls using pre-immune serum and antibodies pre-incubated with excess antigen peptide showed no signal, indicating that the anti-coral sAC antibodies specifically recognize recombinant pdsAC (electronic supplementary material, figure S2).
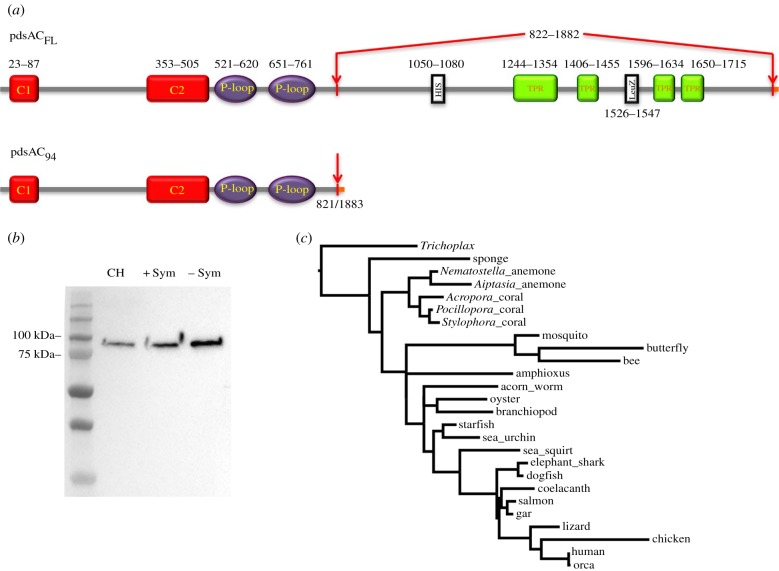
Figure 2.
Protein structure, expression, and phylogenetic analysis of sAC from the coral Pocillopora damicornis. (a) Predicted functional domains of pdsACFL and pdsAC94. C1, C2: catalytic domains, P-loop: P-loop motifs; HIS: histidine-rich region; TPR: tertratricopeptide-like helical domains; LeuZ: leucine zipper pattern. pdsAC94 is identical to pdsACFL until amino acid 821. A potential alternative splicing event results in the loss of amino acids corresponding to HIS, TPRs, and LeuZ and change of open reading frame that also changes the C-terminus (indicated in orange in the online version). (b) Anti-coral sAC antibodies were used in a western blot against P. damicornis protein preparations, yielding a single approximately 90 kDa band; far left lane shows protein molecular weight marker (kDa). (c) Phylogenetic tree of soluble adenylyl cyclase from diverse organisms. Human (Homo sapiens) NP_060887.2; chicken (Gallus gallus) XM_015280795.1; lizard (Anolis carolinensis) XP_008117859.2; elephant shark (Callorhinchus milii) XP_007888388.1; dogfish shark (Squalus acanthias) ACA52542; coelocanth (Latimeria chalumnae) XP_014348984.1; salmon (Salmo salar) XP_014007204.1; gar (Lepisosteus oculatus) XP_015214518.1; sea squirt (Ciona intestinalis) XP_002121952.1; amphioxus (Branchiostoma floridae) XP_002214797.1; sea urchin (Strongylocentrotus purpuratus) NP_001020380.1; oyster (Crassostrea gigas) XM_011451882.2; mosquito (Culex quinquefasciatus) XP_001842661.1; acorn worm (Saccoglossus kowalevskii) XP_006812110.1; branchiopod (Lingula anatina) XP_013401312.1; Pocillopora coral (Pocillopora damicronis) KX910691; Acropora coral (Acropora digitifera) XP_015758344.1; Stylophora coral (Stlylophora pistillata) SpisGene23569; Nematostella anemone (Nematostella vectensis) XP_001623318.1, Aiptasia Anemone (Aiptasia spp.), sponge (Amphimedon queenslandica) XM_019993565.1; Trichoplax (Trichoplax adhaerens) XP_002117857.1. (Online version in colour.)
(e). Protein expression and localization
P. damicornis tissue was homogenized in S22 buffer supplemented with a protease inhibitor mixture (Sigma-Aldrich, St Louis, MO, USA) and phosphatase inhibitor (PhosStop, Roche Applied Biosciences, Indianapolis, IN, USA) as previously described [9]. Protein concentration was determined using the Bradford method with a BSA standard. Western blots were carried out as described in [24], using 5.5 ng ml−1 of the anti-sAC antibodies (1 : 10 000 from stock). Corals were prepared for IHC as in [25] and pdsAC was immunolabelled using 0.11 µg ml−1 anti-sAC antibodies (1 : 500 from stock). The specificity of anti-pdsAC antibodies for immunohistochemistry is shown in electronic supplementary material, figure S3.
(f). Coral cell preparation
Coral cells were isolated from colonies of P. damicornis as previously described [12]. Isolated cell suspensions were loaded with 10 µM SNARF-1AM and 0.1% DMSO in 0.22 µm filtered seawater (FSW) for 30 min in the dark [12]. Cells were pelleted by centrifugation to remove the dye and resuspended in the corresponding treatment; cells were incubated with the treatment for 15 min prior to imaging. All treatments were prepared immediately before use and consisted of FSW supplemented with either 50 µM KH7, 0.5% DMSO (control), 100 µM DCMU, or 50 µM KH7 and 100 µM DCMU. For external pH (pHe) experiments, FSW (pH 8.1) was adjusted to pH 7.4 using HCL immediately before use, and supplemented with either 50 µM KH7 or 0.5% DMSO. In the KH7 washout experiment, cells were treated with 50 µM KH7 for 15 min and imaged, after which the KH7-containing medium was gently removed with a pipette and replaced with 0.5% DMSO in FSW. During light treatments, cells were continuously exposed to 400 µmol photons m−2 s−1 of white light. Light intensity was determined using a Licor LI-1400. These are the same light conditions previously used for isolated P. damicornis cells from this same location [10]. Each experiment was conducted on cells isolated from at least five different coral fragments (45–65 total cells analysed per time point per treatment).
(g). Confocal imaging
Coral cells were visualized by confocal microscopy using a Zeiss LSM 710 confocal microscope. Cells were maintained at 25°C using a temperature-controlled stage throughout the experiments. SNARF1 was excited using a 561 nm laser at 10% power, and fluorescence emission of SNARF1 was recorded at 585 (λ1) and 640 (λ2) ± 10 nm, and chlorophyll at 680 ± 10 nm. These excitation and emission wavelengths were optimized to maximize SNARF excitation while minimizing excitation and emission overlap with endogenous chlorophyll (Symbiodinium) and green fluorescence protein (P. damicornis; electronic supplementary material, figure S8). At least eight coral cells containing Symbiodinium and eight cells without Symbiodinium were imaged for each treatment at each time point. The selected Symbiodinium-free cells were typically spherical with approximately 10 µm diameter, so most likely they were gastrodermal and oral ectodermal cells as calcifying cells are much smaller (see a representative image of coral cells with and without Symbiodinium in electronic supplementary material, figure S4).
The relationship between the ratio (R) of SNARF1 fluorescence at 585 and 640 nm and pHi was calibrated in vivo from pH 6–8.5 (electronic supplementary material, figure S9) as previously described [12]. Calibrations were conducted approximately weekly under light or dark conditions according to the subsequent experiment.
Image analysis was conducted on a region of interest (ROI) drawn in an area of the cytoplasm of each coral cell that did not overlap with Symbiodinium chlorophyll fluorescence. SNARF1 fluorescence intensity (F) within the ROI was recorded at 585 nm (λ1) and 640 nm (λ2). An ROI was also drawn in the surrounding medium, and background fluorescence was subtracted from each channel before calculating R (Fλ1/Fλ1). The equation pH = pKA − log [R − RB/RA − R × FB(λ2)/FA(λ2)] was used to calculate pHi from R, where B denotes the basic endpoint (pH 8.5) and A denotes the acidic endpoint (pH 6) of the calibration [12].
3. Results and discussion
(a). High complexity of the P. damicornis sAC gene(s)
Using RT-PCR and RACE-PCR, we cloned two complete sAC cDNA sequences (accession numbers KX910691 and KY853034) (electronic supplementary material, figure S1). KX910691 contains the longest open reading frame (ORF) and encodes for a 212.8 kDa protein, which we named P. damicornis sAC full length 1 (pdsACFL1). KY853034 has a different start codon compared to KX910691, likely due to an alternatively spliced exon. As a result, the encoded protein (pdsACFL2) is six amino acids shorter than pdsACFL1 and differs in the first 17 amino acids, resulting in a shorter 212.4 kDa protein. However, the remaining 1868 C-terminal amino acids of pdsACFL1 and pdsACFL2 are identical to each other. This region contains all the features essential for sAC enzymatic activity and regulation: the two N-terminal catalytic domains (C1 and C2) essential for 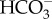 -stimulated cAMP production, and the two P-loop motifs, leucine zipper pattern, and four tetratricopeptide (TPR)-like helical domains previously identified in sAC sequences from vertebrate animals ([13] reviewed in [16,26]) and sea urchins [27] (figure 2a). Both pdsACFL proteins contain a histidine-rich region not previously described in any other sAC protein (figure 2a). Interestingly, both pdsACFL proteins lack the autoinhibitory nine amino acid region immediately downstream of C2 that is present in sAC from humans, rats and mice [28], which might explain the high cAMP-producing activity reported in coral tissues [19].
-stimulated cAMP production, and the two P-loop motifs, leucine zipper pattern, and four tetratricopeptide (TPR)-like helical domains previously identified in sAC sequences from vertebrate animals ([13] reviewed in [16,26]) and sea urchins [27] (figure 2a). Both pdsACFL proteins contain a histidine-rich region not previously described in any other sAC protein (figure 2a). Interestingly, both pdsACFL proteins lack the autoinhibitory nine amino acid region immediately downstream of C2 that is present in sAC from humans, rats and mice [28], which might explain the high cAMP-producing activity reported in coral tissues [19].
In addition to pdsACFL1 and pdsACFL2, we cloned two other partial pdsAC cDNA sequences, each with a unique 5′ untranslated region (UTR) (KY853039 and KY853041), as well as a cDNA sequence that lacks 3181 base pairs encoding for amino acids 822–1882 of pdsACFL (KY853037). The multiple pdsAC 5′ UTRs suggest the presence of alternative promoters, while the shorter mRNA sequences indicate alternative splicing; both of these features have previously been reported for mammalian sAC [29,30]. The predicted size of the putative alternatively spliced pdsAC protein is approximately 94 kDa (pdsAC94) (figure 2a). This shorter pdsAC variant contains C1, C2 and the P-loop motifs, but lacks the leucine zipper pattern, the TPR-like domains, and the histidine-rich region (figure 2a). The physiological implications of the pdsAC genetic complexity are currently unknown, but could be related to differential expression in specific cell types and sub-cellular compartments, association with different proteins or DNA, or the response to specific metabolic or environmental conditions.
Phylogenetic analysis of sAC's catalytic domains indicated that pdsAC was most similar to the predicted sAC proteins from Stylophora pistillata (another coral from the Pocilloporidae family), followed by A. digitifera, and all three coral sequences grouped together with predicted sAC sequences from Nematostella and Aiptasia anemones (figure 2c). BLAST searches in nucleotide databases revealed sAC homologues in several other coral species including Porites lutea, P. asteroides, Orbicella faveolata and Galaxea fascicularis. However, sAC sequences from those four coral species are partial, possibly due to limitations of bioinformatics programs to process multiple gene splice variants.
(b). pdsAC is expressed throughout coral tissues
Western blot analyses revealed pdsAC94 was the predominant sAC protein expressed in coral tissues (figure 2b; see electronic supplementary material, figures S2–S3 for antibody validation). Immunohistochemical analyses confirmed pdsAC was abundantly expressed in cells throughout P. damicornis tissues, including seawater-facing cells in the oral ectodermis, symbiont-containing and symbiont-free cells in the oral and aboral gastrodermis that line the coelenteron, and skeleton-associated desmocytes and calicoblastic cells (figure 3 and electronic supplementary material, figure S3).
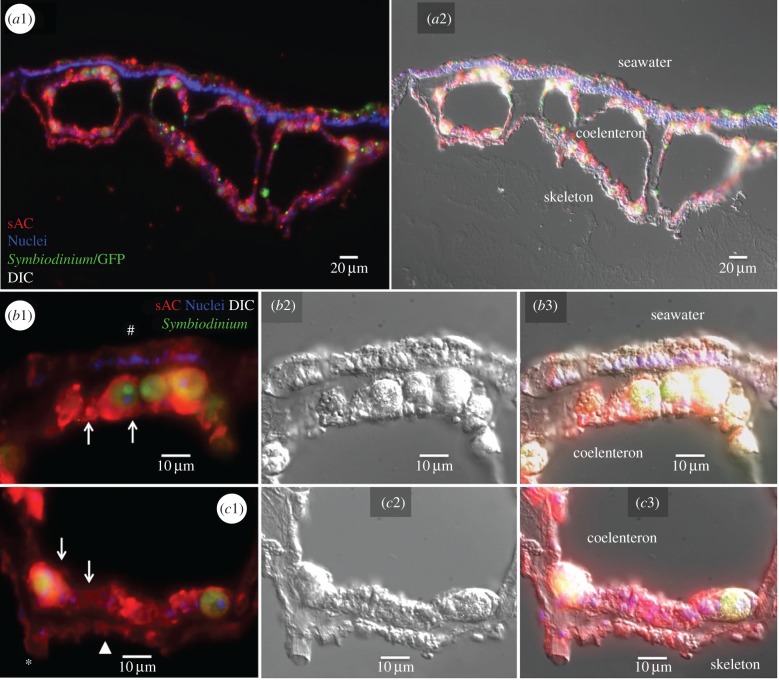
Figure 3.
Immunolocalization of sAC in P. damicornis. (a1) General view of a coenosarc showing widespread pdsAC localization throughout coral tissues. pdsAC (red); nuclei (blue); coral GFP and Symbiodinium chlorophyll autofluorescence (green). a2 shows an overlaid differential interference contrast (DIC) image. (b1) Detail of oral tissue showing abundant pdsAC expression in gastrodermal cells (arrows) as well as in ectodermal cells (hash sign); b2 shows the corresponding DIC image and b3 shows the overlaid image. (c1) Detail of aboral tissue showing abundant pdsAC expression in gastrodermal cells (arrows), calicoblastic cells (arrowhead) and desmocytes (asterisk); c2 shows the corresponding DIC image and c3 shows the overlaid image.
(c). pdsAC is a pH sensor in coral cells
The widespread expression of pdsAC throughout P. damicornis cells suggests regulatory roles in multiple coral physiological functions, potentially including pHi regulation, photosynthesis and calcification. However, the intrinsic relationship between those three functions [9,11,12,31] prevents studying the roles of pdsAC by looking at the effect of sAC pharmacological inhibitors in coral colonies. Instead, we studied the potential role of pdsAC in pHi regulation in isolated P. damicornis cells exposed to various acid–base conditions.
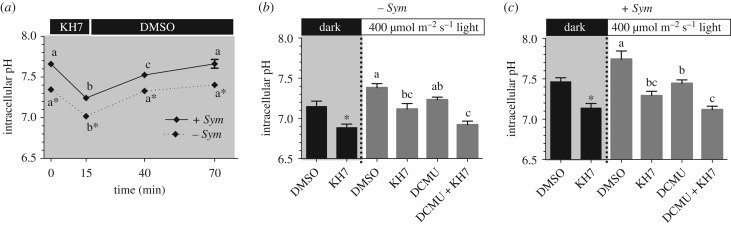
The small molecule KH7 is a specific inhibitor of sAC from mammals [32], sharks [33], sea urchins [34], and also corals [19]. In a first set of experiments, exposure to KH7 for 15-min in the dark resulted in significant pHi acidification in both symbiont-free (from 7.35 ± 0.02 to 7.02 ± 0.02) and symbiont-containing (from 7.66 ± 0.03 to 7.24 ± 0.04) cells (figure 4a and electronic supplementary material, figure S5a). Exposure to KH7 for up to 40 min did not result in any further changes in pHi, and the effect of KH7 on both cell types was completely reversed after washout (figure 4a and electronic supplementary material, figure S5a). These results established that 15 min of exposure to KH7 is sufficient to induce a maximum effect on pHi, and that it is not toxic to coral cells. Furthermore, a potential KH7-induced mitochondrial uncoupling that was reported in some cultured transgenic mammalian cell lines [35] was ruled out in coral cells (electronic supplementary material, figure S7). Thus, these results indicate pdsAC is essential for coral pHi regulation. Since sAC is an evolutionarily conserved acid–base sensor, we conclude pdsAC senses the acidifying effect of cellular respiration and modulates the activity of pHi regulatory proteins to maintain pHi homeostasis. Thus, when pdsAC is inhibited, H+ builds up in the cytoplasm and acidifies pHi. The specific pHi regulatory proteins under pdsAC control are unknown; however, likely candidates include Na+/H+-exchangers, V-H+-ATPases and Na+/K+-ATPases. Evidence for the presence of all of these proteins has been reported in anthozoans [9,24,36], and their activities are regulated by sAC in cells from vertebrate animals [33,37–40].
Figure 4.
Role of pdsAC in intracellular pH regulation in response to metabolic acidification. (a) Intracellular pH of coral cells with (+Sym) or without Symbiodinium (−Sym) following exposure to 50 µM KH7 for 15 min, followed by washout with 0.5% DMSO in FSW in the dark. Letters indicate statistically significant groupings (48–62 cells from five different corals per time point, N = 5; two-way RM ANOVA: p < 0.0001; Tukey's post hoc test). Intracellular pH of coral cells (b) without Symbiodinium (−Sym) or (c) with Symbiodinium (+Sym) following exposure to 0.5% DMSO (control), 50 µM KH7, 100 µM DCMU, or a combination of 50 µM KH7 and 100 µM DCMU under either dark (shaded) or light (400 µmol m−2 s−1) conditions. Asterisks denote statistically significant differences in the dark experiments (45–53 cells from five different corals, N = 5; paired t-test, p < 0.05). Letters indicate statistically significant groupings in the light experiments (47–67 cells from five different corals per time point, N = 5; ANOVA: p < 0.0001; Tukey's post hoc test). Error bars indicate s.e.m., where not visible they fall within the symbol of the data point. The shaded boxes indicate dark conditions.
In subsequent experiments, we examined the role of pdsAC in pHi sensing and regulation in isolated coral cells kept in dark or during light exposure following a 15-min incubation with DMSO (control) or KH7. Again, pHi of symbiont-free cells was lower than in symbiont-containing cells (7.15 ± 0.07 versus 7.47 ± 0.05, respectively; figure 4b,c and electronic supplementary material, figure S5b,c). As previously described [10–12], exposure to light caused pHi alkalinization in symbiont-containing cells to 7.70 ± 0.11 (figure 4c and electronic supplementary material, figure S5c); the pHi of symbiont-free cells also alkalinized to 7.36 ± 0.06 (figure 4b and electronic supplementary material, figure S5b). Inhibition of pdsAC by KH7 under exposure to light resulted in significant pHi acidification in both cell types (symbiont-free cells to pHi 7.15 ± 0.07; symbiont-containing cells to pHi 7.31 ± 0.06) (figure 4b,c and electronic supplementary material, figure S5b,c). Thus, pdsAC activity is essential for proper pHi regulation in coral cells both in the dark and when exposed to light. Interestingly, light affected pHi in both symbiont-containing and symbiont-free cells. The discrepancy with a previous study on the same species [10] is probably due to a higher cell density in our experiments, causing photosynthetic activity from symbiont-containing cells to alkalinize the seawater and affect neighbouring symbiont-free cells. We speculate our experimental conditions of higher cell density more likely resemble in vivo conditions, and therefore photosynthetic activity in live corals has the potential to affect the acid–base status of multiple cell types, not only Symbiodinium-hosting gastrodermal cells.
To rule out a potential effect of KH7 on photosynthesis, the highly specific photosynthesis inhibitor DCMU was applied alone and in combination with KH7. As previously reported in P. damicornis cells exposed to light [10], DCMU treatment did not significantly affect pHi of symbiont-free cells (figure 4b and electronic supplementary material, figure S5b), but it resulted in significant acidification in symbiont-containing cells from 7.70 ± 0.11 to 7.44 ± 0.04 (figure 4c and electronic supplementary material, figure S5c). When KH7 was applied in combination with DCMU, additional pHi acidification was induced in symbiont-containing cells to 7.12 ± 0.10 (figure 4c and electronic supplementary material, figure S5c). Thus, the effect of KH7 on pHi regulation during exposure to light is due to inhibition of pdsAC and not to an off-target effect on photosynthesis.
Overall, these results demonstrate pdsAC is necessary for coral pHi sensing and downstream pHi regulation both under dark and light conditions, a mechanism that prevents coral pHi acidification by constant metabolic CO2/H+ production. Future research will investigate potential downstream physiological roles in specific coral cell subtypes such as coordination of host pHi regulation and symbiont photosynthesis, and ion and acid/base transport for calcification in calcifying cells.
(d). sAC activity regulates pHi in response to extracellular acidification
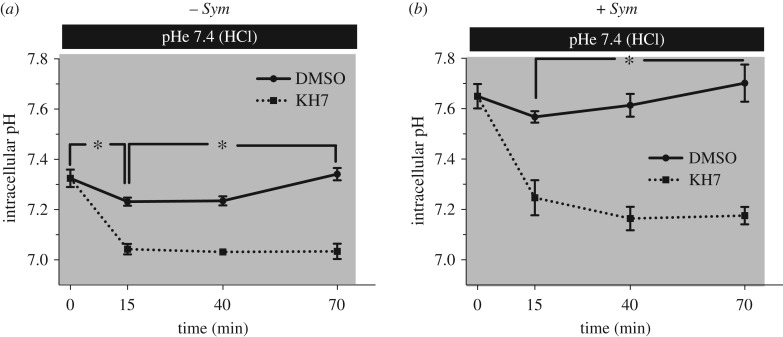
Finally, we explored the hypothesis that sAC plays a role in sensing acidification of the coelenteron caused by H+ transport by the aboral gastrodermis [4]. pHe acidification was induced by exposing isolated coral cells to seawater that was acidified from pH 8.1 to pH 7.4 with HCl, and pHi was followed over time. After 15 min of exposure to external acidification, the pHi of control symbiont-free cells acidified from 7.33 ± 0.03 to 7.23 ± 0.02; pHi recovered to control values by t = 70 min (pHi = 7.34 ± 0.02) (figure 5a and electronic supplementary material, figure S6a). Inhibition of pdsAC by KH7 in symbiont-free cells exposed to external acidification resulted in more pronounced pHi acidification after 15 min (pHi 7.04 ± 0.02). Moreover, those cells failed to recover pHi after 70 min (pHi = 7.03 ± 0.03).
Figure 5.
Role of sAC in intracellular pH regulation in response to external acidification. External acidification was achieved by lowering seawater pH from 8.1 to 7.4 by adding HCl, in the dark (indicated by shaded boxes). (a) Coral cells without Symbiodinium (−Sym), and (b) coral cells with Symbiodinium (+Sym). Asterisks denote statistically significant differences between selected time points (50–59 cells from four different corals per time point, N = 4; RM-ANOVA; Bonferroni's multiple comparison test). Error bars indicate s.e.m.; where not visible they fall within the symbol of the data point.
Exposure to external acidification for 15 min also acidified the pHi of symbiont-containing cells, from 7.65 ± 0.05 to 7.57 ± 0.02 (figure 5b and electronic supplementary material, figure S6b). The more modest effect compared to symbiont-free cells is consistent with the higher buffering capacity of symbiont-containing cells [10,36], which is probably due to the higher buffering capacity of the 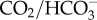 buffer system at higher pH [41] and to H+ transport into the symbiosome [9]. After 70 min of exposure to external acidification, pHi of symbiont-containing cells recovered to 7.70 ± 0.07, which was significantly higher than baseline values (figure 5b and electronic supplementary material, figure S6b). This ‘overshoot’ in pHi is an indication of active H+ secretion and
buffer system at higher pH [41] and to H+ transport into the symbiosome [9]. After 70 min of exposure to external acidification, pHi of symbiont-containing cells recovered to 7.70 ± 0.07, which was significantly higher than baseline values (figure 5b and electronic supplementary material, figure S6b). This ‘overshoot’ in pHi is an indication of active H+ secretion and 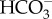 accumulation in response to acidosis [42]; a similar response has previously been reported in P. damicornis symbiont-containing cells [10]. In the presence of KH7, external acidification caused pHi acidification to 7.25 ± 0.07, and, similar to symbiont-free cells, pHi failed to recover after 70 min and even continued to drop to 7.18 ± 0.04. These results indicate that, in addition to sensing and regulating pHi in response to metabolically generated H+ in each cell, pdsAC is essential for sensing and compensating pHi acidification caused by extracellular acidification both in symbiont-containing and symbiont-free cells.
accumulation in response to acidosis [42]; a similar response has previously been reported in P. damicornis symbiont-containing cells [10]. In the presence of KH7, external acidification caused pHi acidification to 7.25 ± 0.07, and, similar to symbiont-free cells, pHi failed to recover after 70 min and even continued to drop to 7.18 ± 0.04. These results indicate that, in addition to sensing and regulating pHi in response to metabolically generated H+ in each cell, pdsAC is essential for sensing and compensating pHi acidification caused by extracellular acidification both in symbiont-containing and symbiont-free cells.
5. Conclusion
This study lays the foundation for understanding the mechanisms that allow corals to detect and respond to pHi disturbances caused by both metabolic and environmental sources. The dynamic interplay of symbiont photosynthesis, coral calcification and cellular respiration involves the production and consumption of acid–base equivalents at different rates and locations within a coral throughout the diel cycle. Because sAC produces the ubiquitous messenger molecule cAMP in response to acid–base disturbances, it can potentially modulate every aspect of physiology by post-translational modifications of target proteins. Thus, the role of sAC in coral physiology is likely critical for the many essential processes that depend on CO2, pH and 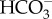 . This type of fundamental mechanistic understanding of coral biology is also essential for predicting how corals will fare in the face of global climate change. Finally, the observation of sAC-dependent pHi sensing in corals, similar to that found in mammalian cells, confirms sAC is an evolutionarily conserved acid–base sensor.
. This type of fundamental mechanistic understanding of coral biology is also essential for predicting how corals will fare in the face of global climate change. Finally, the observation of sAC-dependent pHi sensing in corals, similar to that found in mammalian cells, confirms sAC is an evolutionarily conserved acid–base sensor.
Supplementary Material
Acknowledgements
We would like to thank several people at the University of Hawaii, especially Dr Ruth Gates (U. Hawaii) for providing access to laboratory equipment and support, Dr Ross Cunning and Dr Raphael Ritson-Williams for providing coral colonies (Hawaii DAR Special Activities Permit 2015-17), and Dr Hollie Putnam for coral tank temperature data. We are also grateful to Mr Fernando Nosratpour (Birch Aquarium at Scripps) for providing additional coral colonies at SIO, and Ms Sidney Perez, Dr Cristina Salmerón-Salvador (Tresguerres laboratory at SIO) and Dr Jonathan Matalonga (Sanford Burnham Medical Research Institute, San Diego, CA) for their help with Western blotting and cloning.
Data accessibility
This article has no additional data.
Authors' contributions
Designed the study: K.L.B. and M.T. Performed experiments: K.L.B., M.E.B. and M.T. Analysed data: K.L.B., M.E.B. and M.T. Wrote the paper: K.L.B. and M.T.
Competing interests
The authors declare no competing interests.
Funding
This work was supported by the National Science Foundation (EF-1220641 to M.T., OCE-1226396 to K.L.B., GRFP to M.E.B.) and by an Alfred P. Sloan Research Fellowship (BR2013-103 to M.T.).
References
- 1.Tresguerres M, Barott KL, Barron ME, Deheyn DD, Kline DI, Linsmayer LB. 2017. Cell biology of reef-building corals: ion transport, acid/base regulation, and energy metabolism. In Acid–base balance and nitrogen excretion in invertebrates (eds Weihrauch D, O'Donnell M), pp. 193–218. Springer International Publishing; http://link.springer.com/chapter/10.1007/978-3-319-39617-0_7. (Accessed 25 January 2017.) [Google Scholar]
- 2.Doney SC, Fabry VJ, Feely RA, Kleypas JA. 2009. Ocean acidification: the other CO2 problem. Annu. Rev. Mar. Sci. 1, 169–192. ( 10.1146/annurev.marine.010908.163834) [DOI] [PubMed] [Google Scholar]
- 3.Agostini S, Suzuki Y, Higuchi T, Casareto BE, Yoshinaga K, Nakano Y, Fujimura H. 2011. Biological and chemical characteristics of the coral gastric cavity. Coral Reefs 31, 147–156. ( 10.1007/s00338-011-0831-6) [DOI] [Google Scholar]
- 4.Cai W-J, et al. 2016. Microelectrode characterization of coral daytime interior pH and carbonate chemistry. Nat Commun. 7, 11144 ( 10.1038/ncomms11144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venn A, Tambutté E, Holcomb M, Allemand D, Tambutté S. 2011. Live tissue imaging shows reef corals elevate pH under their calcifying tissue relative to seawater. PLoS ONE 6, e20013 ( 10.1371/journal.pone.0020013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCulloch M, Falter J, Trotter J, Montagna P. 2012. Coral resilience to ocean acidification and global warming through pH up-regulation. Nat. Clim. Change 2, 623–627. ( 10.1038/nclimate1473) [DOI] [Google Scholar]
- 7.Putnam HM, Barott KL, Ainsworth TD, Gates RD. 2017. The vulnerability and resilience of reef-building corals. Curr. Biol. 27, R528–R540. ( 10.1016/j.cub.2017.04.047) [DOI] [PubMed] [Google Scholar]
- 8.Al-Horani FA, Al-Moghrabi SM, De Beer D. 2003. The mechanism of calcification and its relation to photosynthesis and respiration in the scleractinian coral Galaxea fascicularis. Mar. Biol. 142, 419–426. ( 10.1007/s00227-002-0981-8) [DOI] [Google Scholar]
- 9.Barott KL, Venn AA, Perez SO, Tambutté S, Tresguerres M. 2015. Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. Proc. Natl Acad. Sci. USA 112, 607–612. ( 10.1073/pnas.1413483112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibbin EM, Putnam HM, Davy SK, Gates RD. 2014. Intracellular pH and its response to CO2-driven seawater acidification in symbiotic versus non-symbiotic coral cells. J. Exp. Biol. 217, 1963–1969. ( 10.1242/jeb.099549) [DOI] [PubMed] [Google Scholar]
- 11.Laurent J, Tambutté S, Tambutté É, Allemand D, Venn A. 2013. The influence of photosynthesis on host intracellular pH in scleractinian corals. J. Exp. Biol. 216, 1398–1404. ( 10.1242/jeb.082081) [DOI] [PubMed] [Google Scholar]
- 12.Venn AA, Tambutté E, Lotto S, Zoccola D, Allemand D, Tambutté S. 2009. Imaging intracellular pH in a reef coral and symbiotic anemone. Proc. Natl Acad. Sci. USA 106, 16 574–16 579. ( 10.1073/pnas.0902894106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buck J, Sinclair M, Schapal L, Cann M, Levin L. 1999. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc. Natl Acad. Sci. USA 96, 79–84. ( 10.1073/pnas.96.1.79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J. 2000. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science 289, 625–628. ( 10.1126/science.289.5479.625) [DOI] [PubMed] [Google Scholar]
- 15.Tresguerres M, Buck J, Levin LR. 2010. Physiological carbon dioxide, bicarbonate, and pH sensing. Pflugers Arch 460, 953–964. ( 10.1007/s00424-010-0865-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tresguerres M, Levin LR, Buck J. 2011. Intracellular cAMP signaling by soluble adenylyl cyclase. Kidney Int. 79, 1277–1288. ( 10.1038/ki.2011.95) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tresguerres M. 2014. sAC from aquatic organisms as a model to study the evolution of acid/base sensing. Biochim. Biophys. Acta 1842, 2629–2635. ( 10.1016/j.bbadis.2014.06.021) [DOI] [PubMed] [Google Scholar]
- 18.Tresguerres M, Barott KL, Barron ME, Roa JN. 2014. Established and potential physiological roles of bicarbonate-sensing soluble adenylyl cyclase (sAC) in aquatic animals. J. Exp. Biol. 217, 663–672. ( 10.1242/jeb.086157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barott KL, Helman Y, Haramaty L, Barron ME, Hess KC, Buck J, Levin LR, Tresguerres M. 2013. High adenylyl cyclase activity and in vivo cAMP fluctuations in corals suggest central physiological role. Sci. Rep. 3, 1737 ( 10.1038/srep01379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Suppl. 2), W465–W469. ( 10.1093/nar/gkn180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41(Database issue), D344–D347. ( 10.1093/nar/gks1067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finn RD, et al. 2017. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45(D1), D190–D199. ( 10.1093/nar/gkw1107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. ( 10.1038/nature10249) [DOI] [PubMed] [Google Scholar]
- 24.Barott KL, Perez SO, Linsmayer LB, Tresguerres M. 2015. Differential localization of ion transporters suggests distinct cellular mechanisms for calcification and photosynthesis between two coral species. Am. J. Physiol. Regul. Integr. Comp. Physiol. 309, R235–R246. ( 10.1152/ajpregu.00052.2015) [DOI] [PubMed] [Google Scholar]
- 25.Barott KL, Tresguerres M. 2015. Immunolocalization of proteins in corals: the V-type H+-ATPase proton pump. Bio-Protoc. 5, e1573 ( 10.21769/BioProtoc.1573) [DOI] [Google Scholar]
- 26.Steegborn C. 2014. Structure, mechanism, and regulation of soluble adenylyl cyclases—similarities and differences to transmembrane adenylyl cyclases. Biochim. Biophys. Acta 1842, 2535–2547. ( 10.1016/j.bbadis.2014.08.012) [DOI] [PubMed] [Google Scholar]
- 27.Nomura M, Beltrán C, Darszon A, Vacquier VD. 2005. A soluble adenylyl cyclase from sea urchin spermatozoa. Gene 353, 231–238. ( 10.1016/j.gene.2005.04.034) [DOI] [PubMed] [Google Scholar]
- 28.Chaloupka JA, Bullock SA, Iourgenko V, Levin LR, Buck J. 2006. Autoinhibitory regulation of soluble adenylyl cyclase. Mol. Reprod. Dev. 73, 361–368. ( 10.1002/mrd.20409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrell J, Ramos L, Tresguerres M, Kamenetsky M, Levin LR, Buck J. 2008. Somatic ‘soluble’ adenylyl cyclase isoforms are unaffected in Sacytm1Lex/Sacytm1Lex ‘knockout’ mice. PLoS ONE 3, e3251 ( 10.1371/journal.pone.0003251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geng W, Wang Z, Zhang J, Reed BY, Pak CYC, Moe OW. 2005. Cloning and characterization of the human soluble adenylyl cyclase. Am. J. Physiol. Cell Physiol. 288, C1305–C1316. ( 10.1152/ajpcell.00584.2004) [DOI] [PubMed] [Google Scholar]
- 31.Venn AA, Tambutté E, Holcomb M, Laurent J, Allemand D, Tambutté S. 2013. Impact of seawater acidification on pH at the tissue–skeleton interface and calcification in reef corals. Proc. Natl Acad. Sci. USA 110, 1634–1639. ( 10.1073/pnas.1216153110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess KC, et al. 2005. The ‘soluble’ adenylyl cyclase in sperm mediates multiple signaling events required for fertilization. Dev. Cell 9, 249–259. ( 10.1016/j.devcel.2005.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tresguerres M, Parks SK, Salazar E, Levin LR, Goss GG, Buck J. 2010. Bicarbonate-sensing soluble adenylyl cyclase is an essential sensor for acid/base homeostasis. Proc. Natl Acad. Sci. USA 107, 442 ( 10.1073/pnas.0911790107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beltrán C, Vacquier VD, Moy G, Chen Y, Buck J, Levin LR, Darszon A. 2007. Particulate and soluble adenylyl cyclases participate in the sperm acrosome reaction. Biochem. Biophys. Res. Commun. 358, 1128–1135. ( 10.1016/j.bbrc.2007.05.061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Benedetto G, Scalzotto E, Mongillo M, Pozzan T. 2013. Mitochondrial Ca2+ uptake induces cyclic AMP generation in the matrix and modulates organelle ATP levels. Cell Metab. 17, 965–975. ( 10.1016/j.cmet.2013.05.003) [DOI] [PubMed] [Google Scholar]
- 36.Laurent J, Venn A, Tambutté É, Ganot P, Allemand D, Tambutté S. 2014. Regulation of intracellular pH in cnidarians: response to acidosis in Anemonia viridis. FEBS J. 281, 683–695. ( 10.1111/febs.12614) [DOI] [PubMed] [Google Scholar]
- 37.Roa JN, Tresguerres M. 2017. Bicarbonate-sensing soluble adenylyl cyclase is present in the cell cytoplasm and nucleus of multiple shark tissues. Physiol. Rep. 5, e13090 ( 10.14814/phy2.13090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, Buck J, Levin LR, Breton S. 2003. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J. Biol. Chem. 278, 49 523–49 529. ( 10.1074/jbc.M309543200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallows KR, et al. 2009. Regulation of epithelial Na+ transport by soluble adenylyl cyclase in kidney collecting duct cells. J. Biol. Chem. 284, 5774–5783. ( 10.1074/jbc.M805501200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Hu J, Bobulescu IA, Quill TA, McLeroy P, Moe OW, Garbers DL. 2007. A sperm-specific Na+/H+ exchanger (sNHE) is critical for expression and in vivo bicarbonate regulation of the soluble adenylyl cyclase (sAC). Proc. Natl Acad. Sci. USA 104, 9325–9330. ( 10.1073/pnas.0611296104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putnam RW, Roos A. 1997. Intracellular pH. In Handbook of physiology (ed. Hoffman JF.), pp. 389–440. http://doi.wiley.com/10.1002/cphy.cp140109. (Accessed 16 May 2017.) [Google Scholar]
- 42.Roos A, Boron WF. 1981. Intracellular pH. Physiol. Rev. 61, 296–434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.