Abstract
The fossil record in tropical Africa suggests that dry conditions during the Ice Ages caused expansion of savannahs and contraction of the rainforest. Forest refugia have been proposed to be located in areas of Central Africa that currently harbour high rates of endemic species. However, to what extent the forest was fragmented remains unknown. Nuclear microsatellites and plastid sequences of 732 trees of two species occurring in the same habitat—mature lowland evergreen rainforests—but with remarkably different dispersal capacities—animal versus gravity—were analysed. Geographical information system tools revealed intraspecific lineages partially congruent across the two species, suggesting common past barriers to gene flow in Central Africa. According to approximate Bayesian computation, the intraspecific genetic clusters diverged during the Pleistocene (less than 2 Ma), so that intraspecific differentiation is the appropriate scale to test the aridification effect of the Ice Ages on tree populations. Demographic tests revealed clear genetic signals of population expansion in both taxa, possibly following bottleneck events after forest fragmentation, with stronger evidence of expansion after the Penultimate rather than after the Last Glacial Maximum. The differential dispersal capacity may have modulated the particular response of each species to climate change, as revealed by the stronger evidence of expansion found in the animal-dispersed species than in the gravity-dispersed one.
Keywords: approximate Bayesian computation, Atlantic Central Africa, coalescent simulations, comparative phylogeography, nuclear microsatellites, rainforest trees
1. Introduction
Given their temporal proximity, the Ice Ages are considered historical events with the largest influence on species distributions. Their effect is particularly well documented in the Northern Hemisphere, where temperate and Arctic-Alpine organisms were pushed southwards by the ice sheets [1,2]. In the tropics, with more complex environments and older biogeographical histories, the effect of the Ice Ages is difficult to trace. In tropical Africa, the Ice Ages were dominated by dry and cold conditions, especially from 1.05 Myr BP [3]. During these dry periods, the fossil pollen record exhibits a sharp reduction in tree species in favour of grass species [4–6], suggesting that the savannah expanded and the rainforests withdrew to isolated refugia [4]. However, there is still controversy regarding to what extent the African rainforest was fragmented [4,7], as the geographical and temporal coverage of the fossil record is far from complete. In addition, only a few palynological records are older than the Last Glacial Maximum (LGM, 24–12 kyr BP) [5], so that the effect of previous glacial arid periods on the vegetation cannot be assessed.
Given the lack of information in the fossil record, studies have defined the refugia where the rainforest survived during the Ice Ages merely based on species distributions. Ice-age refugia have thus been proposed to be located in areas that currently exhibit high species richness and endemism (Maley's forest refugia, figure 1) [4]. In this context, genetic studies of the rainforest tree populations may be useful to detect forest fragmentation [8,9] because long generation times of trees might preserve genetic signatures of population decline/expansion over long timescales. This approach requires extensive intraspecific sampling of rapidly evolving genetic markers in order to discriminate between alternative evolutionary models.
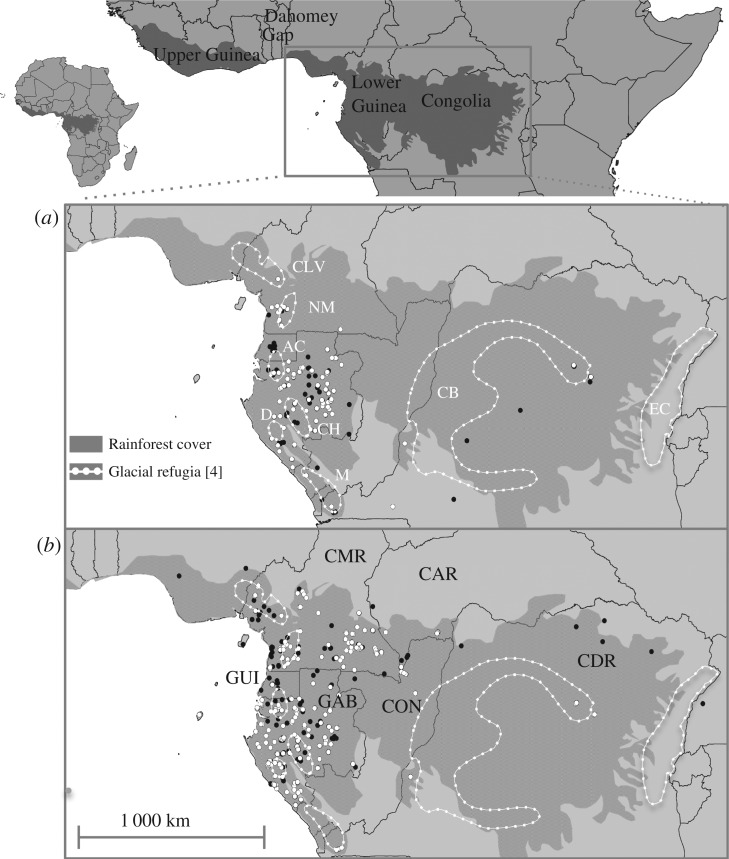
Figure 1.
Distribution of (a) S. zenkeri and (b) G. suaveolens. White dots represent samples in this study; black dots represent all remaining locations based on herbarium vouchers (BR, BRLU, WAG, LBV, MO, Index Herbariorum, http://seetgum.nybg.org/ih/) and tree inventories. Both species are widespread in the Congo Basin, the low number of occurrences just reflecting limited sampling effort by the scientific community. Forest refugia during the Last Glacial Maximum, modified from Maley (a): CVL, Cameroonian Volcanic Line; NM, Ngovayan Massif; AC, Alen-Cristal; D, Doudou; CH, Chaillu; M, Mayombé; CB, Congo Basin; EC, Eastern Congo. Countries (b): CMR, Cameroon; CAR, Central African Republic; GUI, Equatorial Guinea; GAB, Gabon; CON, Republic of the Congo; CDR, Congo Democratic Republic.
The response to forest fragmentation might differ in species adapted to different habitat types [10]. Studies on natural populations of African trees focus almost exclusively on timber trees [11–14], which are light-demanding and thus occur in (previously) disturbed rainforests. Therefore, the study of shade-tolerant species is very relevant, as they might be more sensitive to reduction in canopy cover. To date, only one study provides a timescale of population divergence in a species characteristic of mature African rainforests (0.27–13.47 Myr BP) [13]. The effect of the Ice Ages might have also been modulated by the response of each particular plant species [10]. Important traits to cope with environmental changes like dispersal capacity may trigger dissimilar responses to forest fragmentation. It is thus important to compare the responses of species with contrasting life-history traits.
This study attempts to detect putative genetic signals of the fragmentation of populations (expansions and contractions) in two tropical trees characteristic of mature lowland rainforests: Greenwayodendron suaveolens subsp. suaveolens (Annonaceae) and Scorodophloeus zenkeri (Detaroideae). Both species are widespread in the forests of Central Africa (Lower Guinea and Congolia, figure 1a). Greenwayodendron suaveolens subsp. suaveolens is a medium-sized tree of the upper understory or canopy. Scorodophloeus zenkeri Harms is a large tree of the higher canopy strata [15]. Despite their similar ecologies, the two species exhibit very different dispersal capacities. The berries of G. suaveolens [16] are reported to be dispersed through endozoochory [17–19]. By contrast, S. zenkeri exhibits a characteristic aggregated distribution in the field [20] that suggests limited seed dispersal, and the fruit morphology clearly suggests ballistic dispersal of the seeds through the explosion of the pods (R Piñeiro 2017, personal observation), typical of this family [21,22].
In order to evaluate if the tree populations were fragmented during the dry and cold periods of the Pleistocene, we use nuclear microsatellites and plastid sequences to estimate intraspecific divergence and test demographic models to detect potential genetic signals of population contraction and expansion. In a situation of no forest fragmentation during the Ice Ages, low intraspecific divergence (or significant pre-Pleistocene divergence, greater than 2 Myr BP) and stable constant population sizes through time are expected. Under a scenario of forest fragmentation, high intraspecific divergence during the Pleistocene (less than 2 Myr BP) and signals of population contractions and expansions are expected. Therefore, we test whether the tree populations underwent decline, expansion, bottleneck, or remained stable and evaluate differences between the two species in the light of their different dispersal capabilities. Specifically, we test whether expansion events of the tree populations occurred after the end of the following arid periods: aridification of the climate and expansion of Bantu farmers during the Holocene approximately 3 kyr BP [23], after the LGM approximately 10 kyr BP, or after the Penultimate Glacial Maximum, PGM, approximately 130 kyr BP. Finally, we evaluate if the distribution of the genetic clusters and endemic alleles fits the fragmentation of the rainforest postulated by Maley [4] if we were expecting each forest refuge to host a different cluster.
2. Methods
(a). Study area and species taxonomy
The rainforests of Central and Western Africa are divided into two blocks by a savannah corridor, the ‘Dahomey Gap’ (figure 1). The Western African block corresponds to the Upper Guinea domain, whereas most authors subdivide the Central African block into two domains based on endemism and species composition: Lower Guinea in the West, the richest of the three domains, and Congolia within the Congo Basin [24]. In Lower Guinea, putative forest refugia during the ice-age periods were postulated by Maley [4] in the Cameroonian Volcanic Line, Alen-Cristal, Chaillu, Doudou, and Mayombé (figure 1).
In phylogeographic studies, it is important to make sure that the sequences obtained represent the biogeographic history of the target species alone. Scorodophloeus zenkeri is the only species of the genus occurring in Central Africa (with two other species in East Africa). Therefore, chances for misidentification or hybridization with close relatives are negligible. Within G. suaveolens subsp. suaveolens, a widespread variety (var. suaveolens), in Lower Guinea and Congo (figure 1b), and a narrowly distributed variety (var. gabonica), restricted to Lower Guinea, are currently recognized. Recent DNA evidence [20,25] and a new taxonomic revision of the genus (BJ Lissambou et al., 2017, personal comments) clearly indicate that they should be considered as different species, reproductively isolated. Therefore, only the var. suaveolens, from here on referred to as G. suaveolens, has been used for this comparative phylogeographic study.
(b). Sampling and DNA isolation
Silica-dried cambium or leaves from 240 individuals of S. zenkeri and 450 individuals of G. suaveolens were collected between 2006 and 2012 (figure 1; electronic supplementary material, table S1). Our sampling virtually covers the whole known distribution of the two species in Lower Guinea, along with a few populations occurring in the less accessible rainforests of the Congo Basin, where both species are known to be widespread, but the exact distributions are unknown. Total DNA was extracted using NucleoSpin plant kit (Macherey-Nagel), DNeasy 96 Plant Kit (Qiagen), or cetyl trimethylammonium bromide (CTAB).
(c). Nuclear microsatellites and plastid DNA datasets
Ten and eight nuclear microsatellites (nSSR) were amplified for S. zenkeri and G. suaveolens, in two multiplex reactions per species [26,27], fluorescent labelled with universal sequences, genotyped on an ABI 3730 (Applied Biosystems, Lennik, The Netherlands), and scored with Genemapper 3.7 (Applied Biosystems). Diploid profiles were obtained. When no trace was observed for a locus, it was coded as a null allele if alleles were clearly visible in other loci of the same PCR, and as missing when the other loci showed low quality. A total of 452 individuals previously sequenced for the intergenic spacer trnC-petN1 by Dauby et al. [25] were re-analysed with the software TCS 1.21 [28] in order to explore congruence with nSSR (electronic supplementary material, table S1). Poppr [29] was used to plot the number of different genotypes against the number of loci. To find all different genotypes of G. suaveolens and S. zenkeri six and seven loci are needed, respectively. This indicates that the markers used have enough power to infer the genetic structure of our study species (electronic supplementary material, figure S1).
(d). Number of genetic clusters in the nSSR datasets
Divergent genetic lineages within each species were estimated with STRUCTURE 2.3.4 5 [30] using K = 1 to K = 10 genetic clusters, admixture and independent allele frequency models, 106 generation runs, 105 generation burn-ins, and 10 replicate runs at each K. In order to choose the optimal K, the following statistics were plotted (electronic supplementary material, figure S2): Ln(P), log-likelihood of the data; deltaK (to detect the highest probability increment between consecutive K); similarity coefficient (stability of replicate runs at each K) and clusteredness (to what extent an individual is assigned to a single versus several genetic clusters). Clustering was identical when null allele bias was accounted for using the recessive allele option. The STRUCTURE results were similar subsampling a single sample per square kilometre, showing that our sampling did not cause spurious clusters resulting from isolation by distance.
(e). Estimation of population genetics parameters
For each STRUCTURE nSSR genetic cluster, the following multi-locus parameters were computed with SPAGeDi 1.4 [31]: rarefied allelic richness (AR), expected heterozygosity (He), and effective number of alleles (ENa). Pairwise genetic differentiation (FST) and pairwise Nei's distances Ds between genetic clusters were visualized in an neighbour-joining tree with PHYLIP 3.6 [32]. nSSR alleles restricted to each genetic cluster were identified and mapped to assess their distribution with respect to putative refugia.
(f). Interpolation with geographical information system tools
In order to compare the spatial location and degree of genetic differentiation of the intraspecific nSSR lineages, we used the R function GDisPal [33]. Adjacent individuals were connected in space, inter-individual ‘genetic’ distances were calculated, and a continuous surface of genetic differentiation over space was produced using a 50 × 50 km2 grid. The inter-individual ‘genetic’ distance was equal to (i) zero between two individuals of the same genetic cluster and (ii) to the FST between genetic clusters for pairs of individuals belonging to different clusters.
(g). Population demography and approximate Bayesian computation analyses
The demographic history of each nSSR genetic cluster was assessed with BOTTLENECK 1.2.02 [34] with 1 000 simulations under the stepwise mutation (SM), and the two-phase mutation (TPM) models for microsatellites. This method compares AR and heterozygosity, which respond differently to demographic events. Heterozygosity-excess may indicate loss of rare alleles during bottlenecks, whereas heterozygosity-deficiency may characterize expansions. The magnitude of deviation was estimated with T2 = (Hobserved−Hexpected)/s.d.
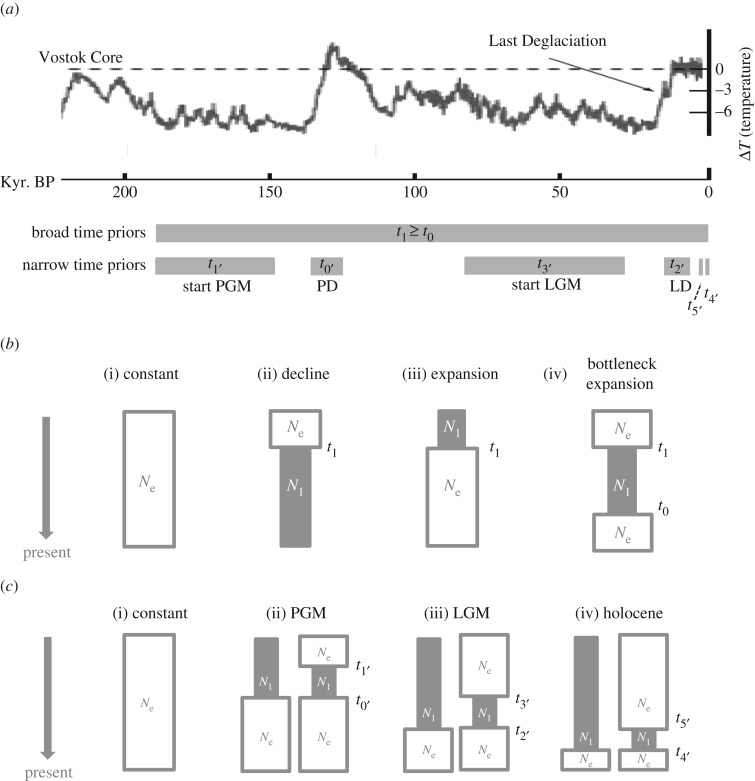
The demography was also investigated with DIYABC 1.0.4.46beta [35]. nSSR datasets were simulated under different demographic scenarios (coalescent simulations) and compared to the real datasets using summary population genetics statistics (approximate Bayesian computation, ABC) in order to estimate the most likely scenario. First, we tested whether the intraspecific genetic clusters diverged during the Pleistocene or earlier (GENETIC CLUSTER DIVERGENCE). Second, the demographic history of each genetic cluster was investigated (figure 2). Using broad time priors, between the beginning of the PGM and the present (190 kyr–0 year BP), we tested four scenarios for each genetic cluster: constant population size, decline, expansion, bottleneck followed by expansion (BROAD TIME MODELS; figure 2b). Once the most likely scenario was identified (see Results), temporal priors were narrowed down in order to test among three specific timescales of bottlenecks: during the Late Holocene climatic pejoration approximately 3 kyr BP, after the LGM approximately 10 kyr BP, or after the PGM approximately 130 kyr BP (NARROW TIME MODELS; figure 2c). The fact that DIYABC cannot implement migration between populations limits the scenario complexity that can be tested (see electronic supplementary material, table S2 for details on the models).
Figure 2.
Demographic models tested against our nSSR datasets with DIYABC. (a) Time priors: priors (in years) in perspective of the Earth climate reconstruction from the Vostok Ice Core in the Antarctic [36]. (b) Broad time models: (i) constant population size, (ii) population decline, (iii) population expansion, and (iv) population bottleneck followed by expansion (time priors in generations for all events: t0 = t1 = 1 900–0). (c) Narrow time models: (i) constant population size, (ii) bottleneck during the PGM and/or subsequent expansion, (iii) bottleneck during the Last Glacial Maximum, LGM, and/or subsequent expansion and (iv) bottleneck during the climatic pejoration of the recent Holocene, and/or subsequent expansion (time priors in generations: beginning PGM bottleneck = t1′ = 1 400–1 900, beginning PD = t0′ = 1 200–1 300, beginning LGM bottleneck = t3′ = 200–700, beginning LD = t2′ = 100–120, beginning Holocene bottleneck = t5′ = 20–40, beginning Holocene expansion = t4′ = 10–30). Effective population sizes were Ne = 1 000–50 000 and N1 = 10–500. PGM, Penultimate Glacial Maximum; LGM, Last Glacial Maximum; PD, Penultimate Deglaciation; LD, Last Deglaciation.
Time parameters were estimated in generations and converted into years assuming a generation time of 100 years [37]. The prior microsatellite mutation rate per generation across loci was log [10−5 to 10−3]. One million genetic datasets were simulated per scenario. The posterior probabilities of each scenario and demographic parameter were estimated using local logistic regression [35]. Bayes factors (BF) were calculated [38] to measure support for the most likely model. Five hundred datasets (pseudo-observed) were simulated based on known parameter values (true values). Confidence for a scenario was the number of ‘pseudo-observed’ datasets drawn from this scenario and correctly assigned to it. Confidence in parameter estimates was calculated as mean bias (difference between the estimated and the ‘true value’ divided by the ‘true value’) and factor 2 (proportion of estimated values falling 50–200% of the ‘true value’). The estimates of the NARROW TIME MODELS were similar using generation times of 200 years (250 000 simulations per scenario).
3. Results
(a). Inferred nSSR genetic clusters and plastid lineages
Five and four intraspecific genetic clusters were inferred for S. zenkeri and G. suaveolens, respectively. For S. zenkeri, the most likely partitions were K = 2 and K = 5 (electronic supplementary material, figure S2a). Clusteredness and similarity coefficients were stable for K = 2–5 and unstable from K = 6 and higher. For G. suaveolens, the most likely partitions were at K = 2 and K = 4 (electronic supplementary material, figure S2b), clusteredness was stable across K, and highest similarity occurred at K = 2 and K = 4. Congruence of the most likely genetic clusters with the geography was better at K = 5 for S. zenkeri and K = 4 for G. suaveolens than at K = 2 (electronic supplementary material, figure S2a,b; table 1). In demographic model testing, it is important to avoid genetic substructure that can confound the demographic signal within each genetic cluster, which additionally justifies the selection of K = 5 and K = 4 over K = 2 [39]. Multi-locus diversity parameters for each genetic cluster are shown in table 1.
Table 1.
Population genetics parameters for each nSSR genetic cluster of S. zenkeri (10 loci, 240 individuals), and G. suaveolens (8 loci, 450 individuals) in Lower Guinea. N, sample size; AR, rarefied allele richness (K = 44, K = 14); He, expected heterozygosity corrected for sample size; Fi,s inbreeding coefficient; NAe, number of effective alleles; Coor., averaged geographical coordinates for each genetic cluster.
| S. zenkeri | N | AR | He | NAe | Coor. | G. suaveolens | N | AR | He | NAe | Coor. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Northwest | 24 | 6.32 | 0.53 | 2.83 | 10.79/2.4 | Northwest | 57 | 1.7 | 0.69 | 7.1 | 10.73/2.92 |
| West | 51 | 6.68 | 0.64 | 3.46 | 10.81/0.11 | Southwest | 62 | 1.82 | 0.82 | 7.83 | 10.37/−0.17 |
| Southwest | 51 | 6.5 | 0.57 | 3 | 11.47/−1.96 | Northeast | 142 | 1.85 | 0.85 | 7.31 | 13.32/2.02 |
| East | 85 | 6.67 | 0.63 | 3.89 | 13.43/1.99 | Southeast | 189 | 1.76 | 0.76 | 5.35 | 12.37/−0.59 |
| Congoa | 29 | — | — |
aDiversity parameters in the Congo region are not provided given that sampling consists of only two locations.
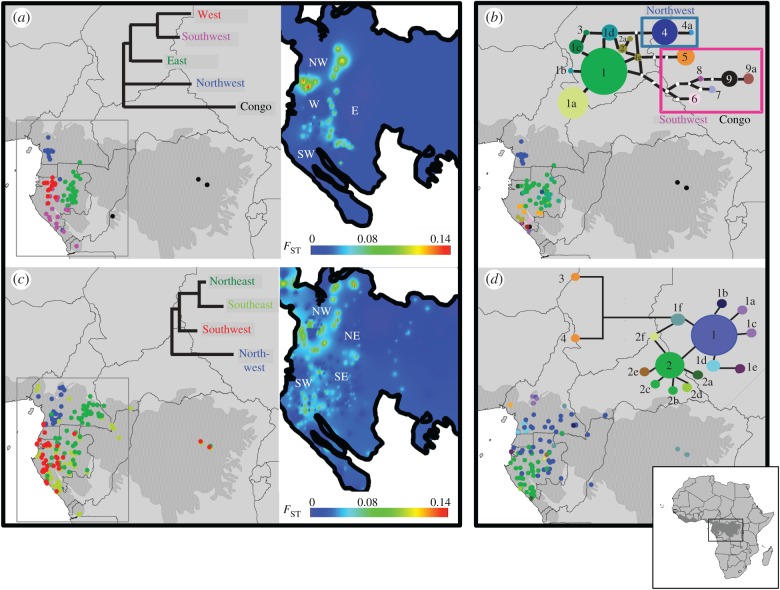
For S. zenkeri, pairwise FST differentiation among the Lower Guinean genetic clusters (figure 3a; electronic supplementary material, table S3) revealed that the ‘Northwest’ cluster was the most differentiated from all other populations (FST > 0.13), followed by the ‘East’, and the ‘West’ and ‘Southwest’ clusters. Ds distance, less affected by genetic drift, revealed comparable results. A fifth genetic cluster was detected in the Congo Basin (Congo), but samples came from only three locations. For the plastid data (figure 3c), 17 haplotypes were retrieved in 192 individuals. The differentiation of the ‘Northwest’ genetic cluster was supported by two divergent haplotypes (4, 4a). By contrast, the remaining genetic clusters were not supported.
Figure 3.
(a,c) Geographical distribution and genetic distances among nSSR Bayesian genetic clusters of (a) S. zenkeri (10 loci, 240 individuals) and (c) G. suaveolens (8 loci, 450 individuals). Only non-admixed individuals, i.e. showing more than 70% ancestry in the Bayesian clustering, are represented. Neighbour-joining trees were built from pairwise FST among nSSR genetic clusters. Heat maps represent interpolation surfaces of FST implemented with SPADS. (b,d) Geographical distribution and genealogy of trnC-petN1R for (b) S. zenkeri (17 haplotypes, 192 individuals) and (d) G. suaveolens (16 haplotypes, 260 individuals).
For G. suaveolens, the ‘Northwest’ genetic cluster was also the most differentiated (FST > 0.12), followed by the ‘Southwest’ cluster, while the two eastern clusters were closely related (FST = 0.04). The genotypes from the Congo Basin were assigned to several genetic clusters (figure 3b; electronic supplementary material, table S3). Except for the finding of remarkably divergent haplotypes north of the Cameroonian Volcanic Line, not paralleled by nSSR data, our plastid marker displayed little phylogeographic resolution in G. suaveolens (figure 3d) with two largely distributed haplotypes (1, 2) and three exclusive to the ‘Northwest’ genetic cluster (1a, 1c, 1d).
(b). Geographical distribution of genetic discontinuities and private alleles
Putting aside the Congo Basin, with poor sampling, in Lower Guinea, the genetic differentiation surfaces of S. zenkeri and G. suaveolens revealed remarkable differentiation of the northwestern populations in both species (‘Northwest’ genetic clusters; figure 3a,b), spanning putative forest refugia in Ngovayan Massif and the Cameroonian Volcanic Line (figure 1a), and showing lower diversity than the remaining clusters (table 1). Secondary phylogeographic breaks appear to be West/East in both species, although with different geographical boundaries in each one. The western clusters span several forest refugia proposed by Maley. In S. zenkeri genetic clusters, ‘Northwest’, ‘West’, and ‘Southwest’ overlap with the refugia in Ngovayan, Alen-Cristal, and Chaillu-Doudou-Mayombé, respectively. In G. suaveolens, the cluster ‘Southwest’ spans all southern refugia. By contrast, the origin the eastern genetic clusters do not overlap with Maley's [4] refugia. Large rivers, like the Ogooué in Gabon or the Sanaga in Cameroon, do not coincide with the boundaries between genetic clusters.
The geographical overlap of intraspecific genetic clusters was different in the two species: the heat maps show discrete borders in S. zenkeri (figure 3a) and overlapping distributions in G. suaveolens, especially between the eastern genetic clusters (figure 3b). However, the centroids (average coordinates) of each gene cluster were separated by 2–3 latitude or longitude degrees (table 1).
Each genetic cluster carried endemic nSSR alleles, 7.69–12.64% of the total number of alleles in S. zenkeri, and from 7.91 to 12.98% in G. suaveolens (electronic supplementary material, table S4). However, no significant correlation between Maley's refugia and the abundance of endemic alleles was detected (electronic supplementary material, figure S3). In S. zenkeri, the cumulated frequency of endemic alleles, averaged over loci, was 2.3% within refugia and 1.8% outside refugia (χ2 test: p = 0.4). In G. suaveolens, 1.79% of the endemic alleles occurred within refugia and 1.06% outside (χ2 test: p = 0.034). This effect is mostly due to the high rate of endemic alleles and higher proportion of samples within a refuge in the Northwest genetic cluster and the correlation disappears if we consider each cluster separately.
(c). Demographic changes and scenario testing
The GENETIC CLUSTER DIVERGENCE analyses strongly supported the model of Pleistocene divergence (less than 2 Myr BP) for both species (BF > 8.4), discarding older splits in the Pliocene or Miocene (table 2a). According to the posterior parameter distributions under the Pleistocene divergence model, the timing of divergence between pairs of Lower Guinean genetic clusters ranged from 615 to 1 090 kyr BP in S. zenkeri and from 428 to 1 340 kyr BP in G. suaveolens (electronic supplementary material, tables S2 and S5). Hence, divergence occurred well before the LGM, although the very wide 95% credible intervals of these estimates (e.g. 83–1 230 kyr between the closest G. suaveolens genetic clusters) does not allow for highlighting of a particular Pleistocene period.
Table 2.
Demographic analyses of nSSR data of S. zenkeri and G. suaveolens implemented with DIYABC and BOTTLENECK (figure 2). (a) Genetic cluster divergence, divergence of intraspecific genetic clusters: Pleistocene divergence (less than 2 Myr BP), Pliocene divergence (2–4 Myr BP), and Mio/Pliocene divergence (4–6 Myr BP). (b) Bottleneck, heterozygosity-excess tests (probability of constant population size) of each nSSR genetic cluster using a Wilcoxon's signed-rank test. The magnitude of deviation was estimated with the parameter T2 = (Hobserved−Hexpected)/s.d. under the TPM and SMM is indicated in parentheses. (c) Broad time models, for each intraspecific genetic cluster four demographic models were tested between the beginning of the PGM and the present (PGM, 190 kyr–0 yr BP): constant population size, decline, expansion, and bottleneck followed by expansion; (d) Narrow time models, constant population size, bottleneck during the PGM (190–120 kyr BP), and/or subsequent expansion; bottleneck during the Last Glacial Maximum (LGM, 70–10 kyr BP) and/or subsequent expansion; and bottleneck during the climatic pejoration of the recent Holocene (4–2 kyr BP) and/or subsequent expansion (less than 2 kyr BP). Posterior probability of the most likely scenarios is highlighted in bold. BF against the alternative scenarios in parentheses. Conf., confidence in scenario choice under the most likely scenario; abs (absolute), the probability of the least likely scenario is 0; PGM, Penultimate Glacial Maximum; LGM, Ultimate Glacial Maximum.
| (a) Genetic cluster divergence |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| less than 2 Myr BP | 2–4 Myr BP | 4–6 Myr BP | conf. | |||||||||
| Scorodophloeus zenkeri | 0.97 | 0.03 (32.9) | 0 (abs) | 0.83 | ||||||||
| Greenwayodendron suaveolens | 0.88 | 0.1 (8.4) | 0.02 (45.4) | 0.81 | ||||||||
| (b) bottleneck |
(c) broad time models |
(d) narrow time models |
||||||||||
| TP model | SM model | constant | decline | expansion | bottleneck | conf. | constant | PGM | LGM | Holocene | conf. | |
| Scorodophloeus zenkeri Northwest | 0.02 (−2.84) | 0 (−4.8) | 0 (571.9) | 0 (abs) | 0.43 (1.3) | 0.57 | 0.51 | 0.01 (116.04) | 0.99 | 0 (205.5) | 0 (2465.8) | 0.66 |
| Scorodophloeus zenkeri West | 0.23 (−1.17) | 0.01 (−3.66) | 0.29 (2.4) | 0 (2264) | 0.03 (20.7) | 0.68 | 0.46 | 0.07 (13.92) | 0.92 | 0.01 (74) | 0 (203.8) | 0.81 |
| Scorodophloeus zenkeri Southwest | 0 (−5.43) | 0 (−14.4) | 0 (1644.5) | 0 (abs) | 0.66 | 0.34 (1.9) | 0.9 | 0 (8211) | 0.82 | 0.18 (4.59) | 0 (abs) | 0.78 |
| Scorodophloeus zenkeri East | 0.38 (−1.81) | 0 (−7.33) | 0.25 (1.7) | 0 (4261) | 0.33 (1.3) | 0.43 | 0.46 | 0.14 (4.37) | 0.59 | 0.27 (2.2) | 0 (179.6) | 0.7 |
| Greenwayodendron suaveolens Northwest | 0.02 (−12.2) | 0.02 (−26.19) | 0.01 (108.9) | 0 (abs) | 0 (abs) | 0.99 | 0.44 | 0 (1110.11) | 1 | 0 (abs) | 0 (abs) | 0.7 |
| Greenwayodendron suaveolens Southwest | 0.02 (−5.41) | 0.01 (−11.33) | 0.08 (10) | 0 (abs) | 0.79 | 0.13 (6) | 0.89 | 0.07 (12.57) | 0.93 | 0 (abs) | 0 (abs) | 0.78 |
| Greenwayodendron suaveolens Northeast | 0.03 (−2.35) | 0 (−9.23) | 0.1 (7.4) | 0 (abs) | 0.73 | 0.17 (4.4) | 0.89 | 0.01 (78.37) | 0.99 | 0 (abs) | 0 (abs) | 0.75 |
| Greenwayodendron suaveolens Southeast | 0.02 (−8.72) | 0 (−18.24) | 0.01 (85.7) | 0 (abs) | 0.32 (2.1) | 0.67 | 0.41 | 0.01 (157.7) | 0.99 | 0 (3311.67) | 0 (abs) | 0.67 |
BOTTLENECK rejected constant population sizes for all genetic clusters under the SMM, and for all but two clusters in S. zenkeri, ‘West’ and ‘Southeast’, under the TPM model (table 2b). Negative values of T2 (heterozygozity-deficiency) suggested recent population expansions
The BROAD TIME MODELS revealed, for all genetic clusters of the two species, highest posterior probabilities for scenarios involving demographic expansions, either alone or following bottlenecks (table 2c). The mean population expansion times were estimated to be 65.4–98.8 kyr BP for S. zenkeri and 100–154 kyr BP for G. suaveolens (electronic supplementary material, tables S2 and S5). In line with BOTTLENECK, evidence against constant population was positive for all four G. suaveolens genetic clusters and for clusters ‘Northwest’ and ‘Southwest’ of S. zenkeri (BF > 7.4), and non-significant for the other two clusters of S. zenkeri (BF = 2.4 and 1.7, respectively). BF showed very strong evidence (BF > 2 264) against scenarios of population declines in all cases (table 2d). Our data showed limited ability to distinguish between the two most likely (and similar) models involving demographic expansions (expansion alone versus preceded by a bottleneck, BF ≤ 1.9, table 2d).
In the NARROW TIME MODELS, the most likely scenarios were population expansions after the PGM (table 2d). Evidence against LGM or Holocene scenarios was strong in all cases (BF > 4.37) except for the LGM model of the genetic cluster ‘East’ of S. zenkeri (BF = 2.2).
4. Discussion
The tropical trees S. zenkeri and G. suaveolens show significant intraspecific genetic discontinuities in rainforests of Central Africa. Our results also reveal a congruent signal of genetic differentiation between the northern (‘Northwest’ genetic clusters) and the southern populations in both species, based on rapidly evolving nuclear microsatellites but also on more conserved plastid DNA. This finding is in line with North/South discontinuities reported in six trees and two herbaceous species in Lower Guinea [8,9,40]. Congruent genetic discontinuities in unrelated species suggest a common driver for the differentiation of the genetic clusters, and therefore, support a scenario of past fragmentation of the rainforest. The intraspecific genetic clusters exhibit not only different allele frequencies but also significant proportions of private alleles, which further suggests isolation in forest fragments. This genetic break does not coincide with discontinuities of the species' distributions or main geographical barriers, such as large rivers.
The divergence of the intraspecific genetic clusters during the Pleistocene (less than 2 Myr BP) in both taxa suggests that the intraspecific level is the right scale to study the effect of the Ice Ages in our study species. The divergence times between genetic clusters, although imprecise, largely preceded the LGM. This suggests that subsequent Ice Ages may have contributed to the genetic differentiation of the genetic clusters and subsequent expansions during the interglacial periods were not enough to admix them. Consistent with a history of fragmentation and re-colonization of the rainforest, ABC tests also revealed clear signals of population expansions in G. suaveolens and in the northernmost and southernmost genetic clusters of S. zenkeri. Stronger evidence of expansion was found in G. suaveolens, dispersed by mammals and birds, as revealed by the current geographical overlap of genetic clusters. The sharp genetic discontinuities over space observed in S. zenkeri along with the lower signal of expansion in two of the genetic clusters suit the expectations of limited dispersal capacity for this species [10].
Demographic tests found strong support for population expansions after the PGM (130 kyr BP onwards) in both species and low support for more recent expansions after the LGM (10 kyr BP onwards) or in the Holocene (2 kyr BP onwards). Consistently, independent analyses using broad time priors also discarded recent rainforest expansion, with estimated mean expansion times at 99–65 kyr BP for S. zenkeri and 154–100 kyr BP for G. suaveolens. The exact dates should be taken with caution as we are testing simplified models but the finding of pre-LGM divergence times retrieved for unrelated species with similar habitat requirements add credibility to our estimates. An indication that the tropics were likely drier during the PGM than during the LGM is shown by stratigraphic studies on lakes in East Africa [41] and by the relative effect of both episodes on West African vegetation in at least two palynological cores [3]. It is also worth noting that while the shade-tolerant species studied here display signatures of population expansion after the PGM, the light-demanding legume tree Erythrophleum suaveolens shows evidence of population decline starting approximately 125–50 kyr BP [13]. The signatures of past expansion or decline may thus reflect global shift in forest vegetation types, and not only changes in forest cover.
Although our data support a scenario of fragmentation of the rainforest during the Ice Ages, the spatial distribution of the intraspecific genetic clusters cannot be fully explained by Maley's [4] refugia, at least if we were expecting each forest refuge to host a different cluster. The northern and western genetic clusters span several forest refugia, while the eastern clusters do not overlap any refugia (figure 1). In addition, similar percentages of private alleles were detected both inside and outside Maley's refugia. This pattern might be explained by the existence of three or four large forest refugia, instead of the six refugia postulated by Maley, or by the presence of microrefugia outside the main forest fragments, such as gallery forests. Alternatively, the forest fragments may have experienced different waves of contractions and expansions through glacial and interglacial cycles with changing levels of connectivity between them over time [4].
5. Conclusion
Based on our extensive sampling, a history of rainforest fragmentation in western Central Africa during cold and dry periods of the Pleistocene seems the most likely explanation for the significant, partially congruent, intraspecific genetic discontinuities detected in two shade-tolerant trees. The clear signatures of expansion of tree populations during the Pleistocene also support this scenario. The differential dispersal capacity may have modulated the response of each species to climate change, with more efficient colonization in the animal-dispersed species than in the gravity-dispersed one. Importantly, the main Central African rivers do not seem to have acted as barriers to gene flow, in contrast with what is documented in murid rodents [42], Mandrills [43], gorillas [44], and chimpanzees [44]. Our time estimates point at a stronger fragmentation during the PGM, when palaeoenvironmental studies report a drier climate in tropical Africa than during the LGM. If we assume that climate cycles were the main driver of the genetic signatures recorded, we can conclude that the genetic discontinuities have built up over multiple glacial cycles, so that interglacial periods were not long enough to erase differentiation between populations through their admixture. In addition, microrefugia might have contributed to forest re-colonization.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgement
We thank Gonzalo Nieto Feliner, Manuel Pimentel Pereira, and five anonymous referees for additional comments on our manuscript.
Data accessibility
Microsatellite data and plastid sequences: DRYAD entries (http://dx.doi.org/10.5061/dryad.5h5t6) [45].
Authors' contributions
O.J.H. and R.P. designed the study, interpreted the results, and wrote the article. E.K., R.P., and G.D. did the laboratory work. R.P. performed the analyses.
Competing interests
We declare we have no competing interests.
Funding
R.P. was a postdoctoral fellow financed by the Belgian Fund for Scientific Research, FRS-FNRS (project FRFC 2.4577.10). G.D. and O.J.H. are, respectively, Postdoctoral Researcher and Senior Research Associate of FRS-FNRS. E.K. was funded through project T.0163.13 of FRS-FNRS. The study was also financed by the AFRIFORD project financed by BELSPO (Belgian Science Policy).
References
- 1.Hewitt G. 2000. The genetic legacy of the quaternary ice ages. Nature 405, 907 ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 2.Stewart JR, Lister AM, Barnes I, Dalén L. 2010. Refugia revisited: individualistic responses of species in space and time. Proc. R. Soc. B 277, 661–671. ( 10.1098/rspb.2009.1272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dupont L. 2011. Orbital scale vegetation change in Africa. Quat. Sci. Rev. 30, 3589–3602. ( 10.1016/j.quascirev.2011.09.019) [DOI] [Google Scholar]
- 4.Maley J. 1996. The African rain forest–main characteristics of changes in vegetation and climate from the upper cretaceous to the quaternary. Proc. R. Soc. Lond. B 104, 31–73. ( 10.1017/S0269727000006114) [DOI] [Google Scholar]
- 5.Morley RJ. 2000. Origin and evolution of tropical rain forests. Chichester, UK: John Wiley and Sons. [Google Scholar]
- 6.Miller C, Gossling W. 2014. Quaternary forest associations in lowland tropical West Africa. Quat. Sci. Rev. 84, 7–25. ( 10.1016/j.quascirev.2013.10.027) [DOI] [Google Scholar]
- 7.Anhuf D, et al. 2006. Paleo-environmental change in Amazonian and African rainforest during the LGM. Palaeogeogr. Palaeoclimatol. Palaeoecol. 239, 510–527. ( 10.1016/j.palaeo.2006.01.017) [DOI] [Google Scholar]
- 8.Heuertz M, Duminil J, Dauby G, Savolainen V, Hardy OJ. 2014. Comparative phylogeography in rainforest trees from Lower Guinea, Africa. PLoS ONE 9, e84307 ( 10.1371/journal.pone.0084307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy OJ, et al. 2013. Comparative phylogeography of African rain forest trees: a review of genetic signatures of vegetation history in the Guineo-Congolian region. C.R. Geosci. 345, 284–296. ( 10.1016/j.crte.2013.05.001) [DOI] [Google Scholar]
- 10.Hamrick JL, Murawski DA, Nason JD. 1993. The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio 107, 281–297. [Google Scholar]
- 11.Born C, Kjellberg F, Chevallier M-H, Vignes H, Dikangadissi J-T, Sanguié J, Wickings EJ, Hossaert-McKey M. 2008. Colonization processes and the maintenance of genetic diversity: insights from a pioneer rainforest tree, Aucoumea klaineana. Proc. R. Soc. B 275, 2171–2179. ( 10.1098/rspb.2008.0446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daïnou K, Bizoux JP, Doucet JL, Mahy G, Hardy OJ, Heuertz M. 2010. Forest refugia revisited: nSSRs and cpDNA sequences support historical isolation in a wide-spread African tree with high colonization capacity, Milicia excelsa (Moraceae). Mol. Ecol. 19, 4462–4477. ( 10.1111/j.1365-294X.2010.04831.x) [DOI] [PubMed] [Google Scholar]
- 13.Duminil J, Mona S, Mardulyn P, Doumenge C, Walmacq F, Doucet JL, Hardly OJ. 2015. Late Pleistocene molecular dating of past population fragmentation and demographic changes in African rain forest tree species supports the forest refuge hypothesis. J. Biogeogr. 42, 1443–1454. ( 10.1111/jbi.12510) [DOI] [Google Scholar]
- 14.Newbery DM, Praz CJ, Van der Burgt XM, Norghauer JM, Chuyong GB. 2010. Recruitment dynamics of the grove-dominant tree Microberlinia bisulcata in African rain forest: extending the light response versus adult longevity trade-off concept. Plant Ecol. 206, 151–172. [Google Scholar]
- 15.Lewis GP, Schrire B, Mackinder B, Lock M. 2005. Legumes of the world. Kew Richmond, UK: Royal Botanic Gardens. [Google Scholar]
- 16.Le Thomas A, Aubréville A. 1969. Annonaceae. Flore du Gabon. Paris, France: Muséum National d'Histoire Naturelle. [Google Scholar]
- 17.Whitney KD, Fogiel MK, Lamperti AM, Holbrook KM, Stauffer DJ, Hardesty BD, Parker VT, Smith TB. 1998. Seed dispersal by Ceratogymna hornbills in the dja reserve, cameroon. J. Trop. Ecol. 14, 351–371. ( 10.1017/S0266467498000273) [DOI] [Google Scholar]
- 18.Feer F. 1995. Morphology of fruits dispersed by African forest elephants. Afr. J. Ecol. 33, 279–284. [Google Scholar]
- 19.Gautier-Hion A, Michaloud G. 1989. Are figs always keystone resources for tropical frugivorous vertebrates? A test in gabon. Ecology 70, 1826–1833. ( 10.2307/1938115) [DOI] [Google Scholar]
- 20.Dauby G. 2012. Structuration spatiale de la diversité intra- et interspecifique en Afrique centrale. PhD dissertation, Université Libre de Bruxelles, Brussels. [Google Scholar]
- 21.Rietkerk M, Ketner P, De Wilde J. 1996. Caesalpinioideae and the study of forest refuges in central Africa. In The biodiversity of African plants (eds van der Maesen LJG. et al.), pp. 618–623. Berlin, Germany: Springer. [Google Scholar]
- 22.Norghauer JM, Newbery DM. 2015. Tree size and fecundity influence ballistic seed dispersal of two dominant mast-fruiting species in a tropical rain forest. Forest Ecol. Manag. 338, 100–113. ( 10.1016/j.foreco.2014.11.005) [DOI] [Google Scholar]
- 23.Bonnefille R. 2007. Rain forest responses to past climate changes in tropical Africa. Tropical rain forest responses to climate change, pp. 117–170. Chichester, England: Praxis Publishing. [Google Scholar]
- 24.White F. 1983. The vegetation of Africa. Switzerland: Unesco. [Google Scholar]
- 25.Dauby G, Duminil J, Heuertz M, Koffi KG, Stévart T, Hardy OJ. 2014. Congruent phylogeographic patterns of eight tree species in Atlantic Central Africa provide insights on the past dynamics of forest cover. Mol. Ecol. 23, 2299–2312. ( 10.1111/mec.12724) [DOI] [PubMed] [Google Scholar]
- 26.Piñeiro R, Micheneau C, Dauby G, Hardy OJ. 2013. Isolation of nuclear microsatellite loci in the African tree Scorodophloeus zenkeri (Fabaceae). Conserv. Genet. Res. 5, 219–221. ( 10.1007/s12686-012-9773-8) [DOI] [Google Scholar]
- 27.Piñeiro R, Micheneau C, Dauby G, Hardy OJ. 2016. Isolation, characterisation and cross-species amplification of nuclear microsatellites in the African tree genus Greenwayodendron (annonaceae). J. Trop. For. Sci. 28, 121. [Google Scholar]
- 28.Clement M, Posada D, Crandall KA. 2009. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. ( 10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 29.Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2, e281 ( 10.7717/peerj.281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy OJ, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620. ( 10.1046/j.1471-8286.2002.00305.x) [DOI] [Google Scholar]
- 32.Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package), version 3.6. Distributed by the author. [Google Scholar]
- 33.Dellicour S, Mardulyn P. 2013. spads 1.0: a toolbox to perform spatial analyses on DNA sequence data sets. Mol. Ecol. Res. 14, 647–651. ( 10.1111/1755-0998.12200) [DOI] [PubMed] [Google Scholar]
- 34.Cornuet JM, Luikart G. 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144, 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornuet J-M, Santos F, Beaumont MA, Robert CP, Marin J-M, Balding DJ, Guillemaud T, Estoup A. 2008. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24, 2713–2719. ( 10.1093/bioinformatics/btn514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Augustin L, et al. 2004. Eight glacial cycles from an Antarctic ice core. Nature 429, 623–628. ( 10.1038/nature02599) [DOI] [PubMed] [Google Scholar]
- 37.Baker TR, et al. 2014. Fast demographic traits promote high diversification rates of Amazonian trees. Ecol. Lett. 17, 527–536. ( 10.1111/ele.12252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kass RE, Raftery AE. 1995. Bayes factors. J. Am. Stat. Assoc. 90, 773–795. [Google Scholar]
- 39.Broquet T, Angelone S, Jaquiery J, Joly P, Lena JP, Lengagne T, Plenet S, Luquet E, Perrin N. 2010. Genetic bottlenecks driven by population disconnection. Conserv. Biol. 24, 1596–1605. ( 10.1111/j.1523-1739.2010.01556.x) [DOI] [PubMed] [Google Scholar]
- 40.Ley AC, Dauby G, Köhler J, Wypior C, Röser M, Hardy OJ. 2014. Comparative phylogeography of eight herbs and lianas (Marantaceae) in central African rainforests. Front. Genet. 5, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moernaut J, Verschuren D, Charlet F, Kristen I, Fagot M, De Batist M. 2010. The seismic-stratigraphic record of lake-level fluctuations in Lake Challa: hydrological stability and change in equatorial East Africa over the last 140kyr. Earth Planet. Sci. Lett. 290, 214–223. ( 10.1016/j.epsl.2009.12.023) [DOI] [Google Scholar]
- 42.Anthony NM, et al. 2007. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc. Natl Acad. Sci. USA 104, 20 432–20 436. ( 10.1073/pnas.0704816105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telfer PT, et al. 2003. Molecular evidence for deep phylogenetic divergence in Mandrillus sphinx. Mol. Ecol. 12, 2019–2024. ( 10.1046/j.1365-294X.2003.01877.x) [DOI] [PubMed] [Google Scholar]
- 44.Hey J. 2010. The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Mol. Biol. Evol. 27, 921–933. ( 10.1093/molbev/msp298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piñeiro R, Dauby G, Kaymak E, Hardy OJ.2017. Data from: Pleistocene population expansions of shade-tolerant trees indicate fragmentation of the African rainforest during the Ice Ages. Dryad Digital Repository. ( ) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Piñeiro R, Dauby G, Kaymak E, Hardy OJ.2017. Data from: Pleistocene population expansions of shade-tolerant trees indicate fragmentation of the African rainforest during the Ice Ages. Dryad Digital Repository. ( ) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Microsatellite data and plastid sequences: DRYAD entries (http://dx.doi.org/10.5061/dryad.5h5t6) [45].