Abstract
Spatial clustering is thought to favour the evolution of cooperation because it puts cooperators in a position to help each other. However, clustering also increases competition. The fate of cooperation may depend on how much cooperators cluster relative to defectors, but these clustering differences have not been the focus of previous models and experiments. By competing siderophore-producing cooperator and defector strains of the opportunistic pathogen Pseudomonas aeruginosa in experimental microhabitats, we found that at the spatial scale of individual interactions, cooperator clustering lowers cooperation, but defector clustering favours cooperation. A theoretical model and individual-based simulations show these counterintuitive effects can arise when competition and cooperation occur at a single resource-determined scale, with population dynamics crucially allowing cooperators and defectors to cluster differently. The results suggest that cooperation relies on the regulation of sufficient defector clustering relative to cooperator clustering, which may be important in bacteria, social amoeba and cancer inhibition.
Keywords: evolution of cooperation, kin competition, Pseudomonas aeruginosa, defector clustering, interaction scale, population dynamics
1. Introduction
The spatial clustering of cooperators is generally thought to favour the evolution of cooperation in the presence of defectors [1–3]. By being close together, cooperators help each other more than potential defectors. At the same time, clustering also increases competition between cooperators [4,5], potentially halting the evolution of cooperation. A variety of mechanisms make clustering generally favourable to cooperation in theory despite increased competition. When the scale of cooperation is smaller than the scale of competition, clustering at the small scale benefits cooperators without a proportional increase in local competition [6]. However, in systems where cooperation and competition is mediated by a single common-pool resource through production and consumption there is no spatial scale separation, prompting us to look for other, more general explanations for how cooperation can evolve with competition. Population dynamics can sometimes render clustering favourable to cooperation, as the availability of unoccupied habitat can allow cooperator clusters to expand [7]. These theoretical studies focused on the effects of life-history parameters rather than clustering itself, but implied that cooperator and defector clustering can develop without perfect correlation to each other when the habitat is not saturated [5,7–9]. It is currently unknown how cooperator clustering and defector clustering affect cooperation separately, and how they interact to determine the evolution of cooperation.
Microbial experiments on the evolution of cooperation are increasingly prominent because they are medically relevant, experimentally tractable and ecologically important. In these experiments, clustering is consistently observed to favour cooperation, but most studies manipulated clustering through various degrees of random mixing [6,10] so that cooperators and defectors clustering are not distinguished. Other studies of cooperation allowed spatial patterns to emerge [11,12], but individual-level locations were not tracked. Individual-level resolution is important, because without exogenous mixing it is unknown at which scale clustering should be measured a priori. The diffusivity of the common-pool resource in fresh media only partly determines the appropriate clustering scale because bacterial biomass and activity (such as production of extracellular polymeric substances (EPS) and resource withholding) can lead to bioclotting and reduced diffusion [13]. Microbial experiments with emergent spatial and population dynamics can be coupled with theory to show whether cooperators and defectors clustering are always correlated, at what spatial scales cooperation and defection occur, and what aspects of clustering favour or impede the evolution of cooperation.
Previously [14], we used a microfluidic device to test whether spatial features that are hundreds of times bigger than a bacterium can affect the relative success of Pseudomonas aeruginosa cooperator and defector phenotypes. By increasing habitat patchiness, or the edge-to-area ratio, clustering is generally expected to be higher, and thus cooperation more favourable. While patchiness was demonstrated to stabilize the coexistence of cooperators and defectors, it did not significantly affect cooperator frequency. However, large variations in cooperator frequency occurred, ranging from defector dominance to cooperator dominance, prompting further investigations into the role of spatial heterogeneities at scales finer than the habitat features.
We hypothesize that cooperators and defectors cluster differently when spatial patterns emerge in an unsaturated habitat, and that cooperator clustering by itself impedes the evolution of cooperation because of increased competition between cooperators. We consider the following cooperative phenomenon in P. aerugionsa. Wild-type cooperators produce siderophores called pyoverdines, which are high affinity iron chelators that diffuse in the extracellular environment. Pyoverdine binds to iron (Fe(III)), is transported into bacteria where iron disassociates and becomes available for bacterial growth and function and then is recycled back into the environment [15]. On the one hand, pyoverdines have traditionally been considered public goods when iron is withheld by other iron chelators [16], such as human apo-transferrin, because secreted (or extracellular) pyoverdines are non-excludable, or freely shared and recycled. On the other hand, pyoverdines are common-pool resources, because once uptaken by a bacterium, they are temporarily unavailable to neighbours [17], resulting in a concurrent competitive effect. We thus consider bioavailable iron, mediated by pyoverdines, as the common-pool resource that forces cooperation and competition to operate at one spatial scale. Pyoverdine production is indiscriminate [18] (no kin recognition) so it is expected that spatial factors predominantly drive its evolution. Loss-of-function mutations leading to defection appears common for the within-host evolution of P. aeruginosa in cystic fibrosis (CF) patients [19], so the experimental evolution between cooperators and defectors has potential medical implications [20].
We competed cooperators and defectors in a microfluidic device, which contains three microhabitat types with spatial features at the 100 µm scale (figure 1). We measured over a range of spatial scales the clustering among cooperators, among defectors, and between cooperators and defectors, and determined their effects on cooperator frequency and population density. We expected to find the strongest clustering effects at scales much smaller than 100 µm despite known spatial features at that scale [14]. These experimental analyses then motivated analytical and simulation models that tease apart the potential causality of clustering scale on the evolution of cooperation. We link our results to existing theoretical frameworks. We end by discussing the contrasting roles of cooperator and defector clustering in bacteria, social amoeba and cancer evolution.
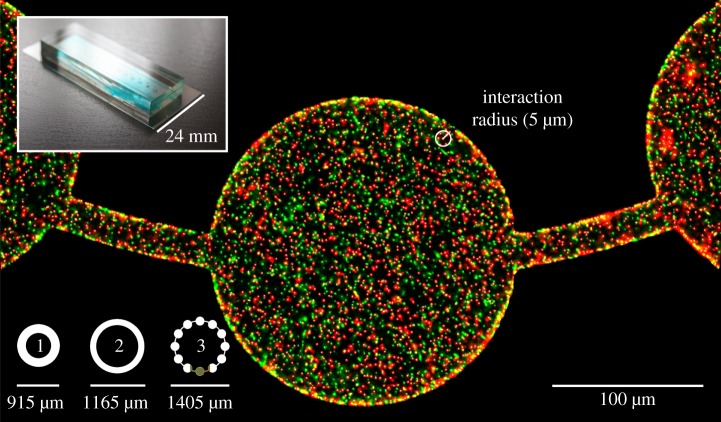
Figure 1.
Cooperator (green/light) and defector (red/dark), overlaid with grey spot detections, in a patchy habitat (T = 10 h). The three experimental microhabitats are illustrated in the lower left, and the device housing the microhabitats is pictured in the upper left.
2. Experimental set-up
(a). Device
Our experimental platform is a microhabitat device that uses microfluidics, an emerging technology that has led to important discoveries on microbial growth, interactions and spatial behaviour [21]. The microhabitat device is built from a silicon mould using photolithography, on which poly(dimethyl)siloxane (PDMS) was poured to 5 mm in thickness. Details on the design and fabrication of the microhabitat device can be found in [14]. The elastomer layer contains the three habitats shown in figure 1, which are 10 µm deep and 0.42 mm2 in area without corridors. For habitat 3, the addition of corridors brings the total area to 0.45 mm2. The diameters of habitat 1, 2 and 3 are 915, 1165 and 1405 µm, respectively. The microhabitat dimensions make it feasible to acquire the locations of all individual bacteria growing in two-dimensional space.
The microhabitat chip was filled with culture medium low in bioavailable iron [22] to mimic the iron-limiting conditions of a human host. The iron-limited environment rendered pyoverdine an effective common-pool resource [6]. The media consisted of casamino acids (CAA, 5 g l−1 with 0.005 M K2HPO4 and 0.001 M MgSO4), 50 mM NaHCO3 and 1 mg ml−1 human apo-transferrin (added 1 day prior to use). We tracked Pseudomonas aeruginosa cooperators, wild-type siderophore producers and defectors, mutants defective in producing the primary siderophore pyoverdine. The P. aeruginosa cooperators were the wild-type laboratory strain PAO1, and defectors were isogenic pvdA mutants [23] defective in pyoverdine production. Both strains were transformed with plasmids that constitutively expressed either green fluorescent protein GFP (pMRP9-1 [24]) or the red mCherry (pMKB1 [25]), which were alternated in each successive experiment to control for potential plasmid side effects (GFP cooperators versus mCherry defectors, GFP defectors versus mCherry cooperators). Starting from equal proportions, cooperators and defectors grew to equilibrium after 10–12 h.
We diluted 16-h overnight cultures of the cooperator and defector strains (LB medium, 37°C shaker incubator) to an optical density (600 nm) of 0.005 in the iron-limited CAA media; 0.7 µl of the mixed (1 : 1) or monoculture was pipetted directly onto each habitat, then the PDMS device was sealed onto a glass coverslip (24 × 60 mm #1.5H, Schott Nexterion). We focus on the 1 : 1 cooperator–defector ratio, which is theoretically an important population state from which to observe selection (see electronic supplementary material). The ratio also ensures that each strain does not stochastically go extinct given the small initial populations (approx. 500 individuals) required for about five generations to unfold in the device. The device was placed in a 30°C heated chamber (Chamlide TC, Live Cell Instrument) on the inverted robotic stage of a laser scanning confocal microscope (LSM 700, Zeiss) and was imaged every 57 min and 18 s, up to 20 h.
The position of each bacterium was analysed using the Imaris spot detection software. Corrections of biases for individual counts due to slight differences between GFP and mCherry fluorescences were implemented following Tekwa et al. [14], by comparing mono-fluorescent monocultures (either cooperators or defectors only, seven replicates) run simultaneously in pairs (GFP cooperators and mCherry cooperators; GFP defectors and mCherry defectors).
(b). Computing clustering
Consider a system with two morphs—cooperators and defectors with global densities (abundances divided by area) Xc and Xd. We introduce a set of clustering coefficients: Ccc, Ccd and Cdd with the subscripts describing the pairing (c for cooperator, d for defector). Given a spatial interaction scale that specifies the radius within which neighbours interact with the focal individual (and otherwise do not interact), the product CijXj gives the local densities Xij [26] of morph j around morph i—that is, the average number of j individuals that an i individual interacts with in its neighbourhood (figure 1). Local density is the demographically explicit and continuous-space version of pair-density (qj/i in [3,5,8,9,27,28]) or conditional probability of identity [29,30] and CccXc, the local density of cooperators, is approximately equal to relatedness if we assume rare invading cooperators [2,31]. However, relatedness is not the only important spatial factor due to other spatial effects through competition [7], so it is important to measure cooperator and defector clustering independently. See the electronic supplementary material for derivations of clustering coefficients and their relationships to relatedness in inclusive fitness theory. Cij > 1 indicates that the morphs cluster more than in a well-mixed, non-spatial, case such as an agitated liquid culture of bacteria. We use clustering coefficients Cij rather than local densities Xij as candidate predictors of cooperator frequency and population density because Xij are correlated with Xj even without spatial heterogeneity. In particular, Xcc would a priori correlate positively with Xc, and can thus falsely identify among-cooperator local density as benefiting cooperators. Clustering coefficients are normalized local densities and do not depend a priori on global densities (as local densities Xij or qj/i would be proportional to global densities Xj in the null case of no spatial patterns). Relatedness in inclusive fitness theory is also effectively a normalized metric that expresses clustering without a priori dependence on global densities, but focuses on the spatial relationship among cooperators (electronic supplementary material).
Because of Imaris’ spot detection limitations, bacteria of the same fluorescent colour cannot be reliably distinguished if they are closer than 4 µm. Thus, raw clustering estimates are biased. The correction procedure, involving comparisons of mono-fluorescent monocultures with mixed-fluorescent ‘monocultures’ (either cooperators or defectors only) is described in the electronic supplementary material.
Clustering coefficients are computed for 12 spatial scales, ranging from 4 µm (near individual scale) to 1280 µm (nearly covering the entire habitat, figure 3a).
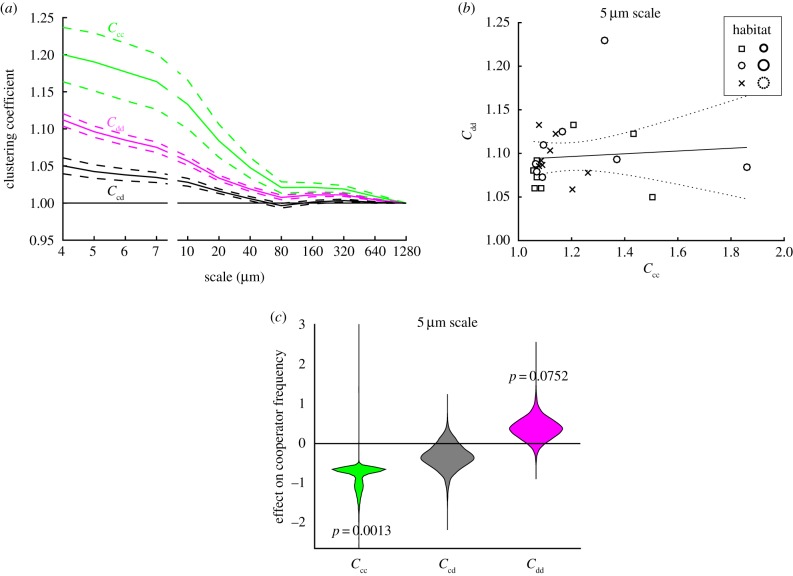
Figure 3.
(a) Clustering coefficients (Ccc, Ccd, Cdd) measured at different scales. Solid lines are means, and dotted lines are standard errors (n = 24). (b) Within-defector clustering Cdd versus within-cooperator clustering Ccc measured at the 5 µm scale. Data points are distinguished by their habitat types (see legend for corresponding habitat numbers with those shown in figure 1). The slope was not significant (slope = 0.016, s.e. = 0.041, F2,22 = 0.15, p = 0.70, R2 = 0.0068). The dotted lines represent the 95% confidence bounds. (c) Bootstrapped linear regression slopes (effects) of standardized cooperator frequency on standardized clustering coefficients (Cij/s.d.(Cij)). The clustering regression model at the 5 µm scale explains 0.79 of cooperator frequency and 0.81 of population density variations. (Online version in colour.)
(c). Experimental analyses
Control experiments were previously performed in [14] and are reviewed here. Monocultures and mixed cultures of cooperators and defectors were run in parallel eight times, and the strain count time series were each fitted (with least-squares maximum likelihood) to logistic growth curves dXi/Xidt = ri(1 − Xi/Ki), where Ki is the equilibrium density estimate of strain i. Monoculture experiments show no differences in initial growth (ri) but elevated densities (Ki) for cooperators when compared with defectors in all habitats (ANOVA F1,44 = 22, p = 2.9 × 10−5). This establishes pyoverdine production as a beneficial cooperative trait in the long run. Patchiness (F1,44 = 0.06, p = 0.81) and the interaction between patchiness and strain (F1,44 = 3.2, p = 0.081) did not have significant effects on densities. Mixed culture experiments (n = 24) showed no differences in initial growth (ri) but decreased densities (Ki) for cooperators when compared with defectors in all habitats (ANOVA F1,43 = 8.3, p = 0.0063). Thus, in later analyses of clustering effects, we focus on the equilibrium state, defined as values averaged over T = 10–12 h. Cooperation is costly and on average selected against in the microhabitats. Patchiness (F1,43 = 0.0024, p = 0.96) and the interaction between patchiness and strain (F1,43 = 0.047, p = 0.83) were found to have no direct effects (figure 2).
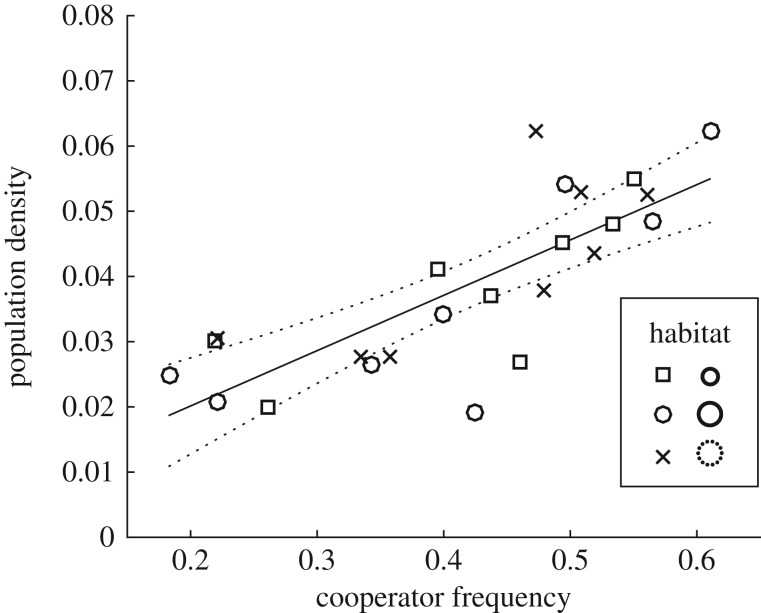
Figure 2.
Cooperator frequencies versus total population densities in different habitat type (n = 8 each). Symbols represent different habitats (legend). The relationship was positive (slope = 0.085, s.e. = 0.014, F2,22 = 36, p = 5.1 × 10−6, R2 = 0.62). The dotted lines represent the 95% confidence bounds. MANOVA shows that habitat type did not significantly affect cooperator frequencies and population densities (χ2(4,n = 24) = 0.93, Wilk's Λ = 0.96, p = 0.92).
Clustering effects are quantified as the ordinary least-squares linear regression slopes (effects) of standardized cooperator frequency (Z/s.d.(Z) where s.d. is standard deviation) on standardized clustering coefficients (Cij/s.d.(Cij)) measured at different spatial scales. Using all data, the fit of the regression model at each scale is quantified as R2, or the portions of variations explained in cooperator frequency and population density, and as AIC score that accounts for both cooperator frequency and population density [32]. The lowest AIC indicates the interaction scale that best describes clustering effects on cooperator frequency and population density. For reference, a null model uses initial cooperator and defector densities as predictors of final cooperator frequency and population density. Clustering effect models with AIC greater than the reference AIC should be considered inadequate.
To obtain spreads of estimates for the clustering effects at each spatial scale, bootstrap least-squares linear regressions are performed over 10 000 resamples (with replacement) of the data, with each sample being 24 sets of clustering coefficients and corresponding cooperator frequency and population density drawn from the original 24 experimental replicates (see electronic supplementary material). Bootstrapping was used because effect sizes may not be normally distributed, and they were not in both the experiment and simulations (figures 3c and figure 4). As a non-parametric method, bootstrapping provides both a more accurate spread of effect size estimates and more precise confidence interval estimates [33]. The spreads are illustrated as violin plots, which are kernel densities for the effect estimates (figures 3c and figure 4). The 95% CIs and 25th/75th percentiles are reported in the electronic supplementary material. The one-sided p-value of each clustering effect is computed as the per cent of bootstrap regression coefficients in the opposite direction of the theoretical prediction. For example, the p-value of Ccc effect on cooperator frequency is the number of bootstrap regression estimates that are positive, divided by 10 000. The same bootstrap procedure is used to analyse clustering effects for the simulations.
Figure 4.
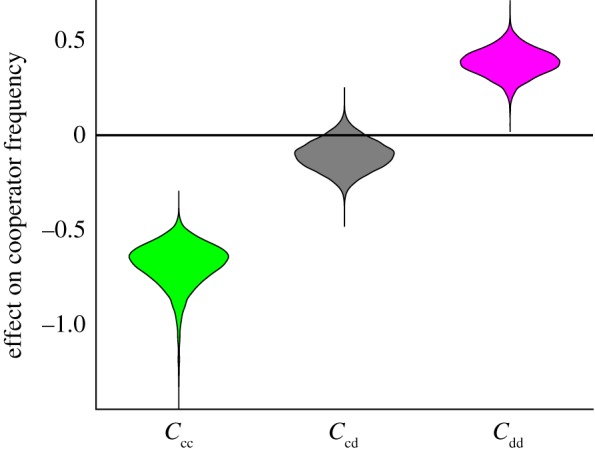
Spatial simulation outcomes on the competition of cooperators and defectors (n = 40, movement rate = 0.3). Spreads of effect estimates are from bootstrapped linear regression of cooperator frequency on standardized clustering coefficients (Cij/s.d.(Cij)). The model predicts (R2) 0.86 of cooperator frequency and 0.62 of population density variations. (Online version in colour.)
(d). Experimental results
In mixed cultures of cooperators and defectors, the mean initial density was 0.0013 (s.e. = 3.2 × 10−4) μm−2, and the mean initial cooperator frequency was 0.46 (s.e. = 0.036), which was not significantly different from 0.5 as desired (95% CI = (0.39, 0.53), t1,23 = −1.1, p = 0.26). The final cooperator frequency was 0.42 (s.e. = 0.026) and was significantly different from 0.5 (95% CI = (0.37,0.47), t1,23 = −3.2, p = 0.0043). Final cooperator frequency and population density exhibit a strong positive relationship (figure 2), showing that siderophore production is beneficial to the population in the mixed cultures. In the following, we investigate the effects of fine-scale spatial clustering on cooperator frequency and population density.
Clustering coefficients of cooperators and defectors declined nonlinearly with spatial scale (figure 3a), reaching values close to 1 at around 80 µm. This shows that clustering decays rapidly away from the individual, with very little clustering observed near the scale of habitat patchiness. We detected no correlation between Ccc and Cdd at the 5 µm scale (figure 3b), which supports the view that cooperator and defector clustering can develop separately, and enables their effects to be statistically distinguished. The 5 µm scale was displayed because according to AIC, clustering at this scale best predicted cooperator frequency (R2 = 0.79) and population density (R2 = 0.81; electronic supplementary material, figure S1). Strikingly, bootstrap regression analysis showed that cooperator clustering is negatively associated with cooperator frequency (p = 0.0013), while defector clustering is positively associated with cooperator frequency (p = 0.0752), at the 5 µm scale (figure 3c) and at all but the largest (and least predictive) scales tested (electronic supplementary material, figure S2). The fact that the best scale is not at the extremes tested (4 and 1280 µm), and that the same clustering effects were observed across scales, suggest 5 µm is close to the true interaction scale. These results support the hypothesis that cooperator clustering is detrimental to cooperation and that defector clustering is positively associated with cooperation.
3. Theory
(a). Model
Why has clustering often been found to favour the evolution of cooperation, and why did cooperator clustering act in the opposite direction in our experiment? We present a simple model that incorporates both cooperation and competition occurring at the same spatial scale, which is implied by the production and consumption of a single growth-limiting resource, the bioavailable iron.
Consider a dimorphic population of cooperators (c) and defectors (d). Each individual has an intrinsic growth rate ri according to its morph i. This rate is proportional to the concentration of bioavailable irons when there are no neighbours. All neighbours impose a competitive cost k to a focal individual, resulting from the neighbours withholding a number of bioavailable irons in the focal individual's neighbourhood. Cooperator neighbours indiscriminately bestow an additional benefit a to the focal individual, through the production of siderophores that add bioavailable iron to the neighbourhood. We assume k > a, such that the population is self-limiting. Let the cooperative character value of a cooperator be z = 1 and z = 0 for a defector. The density-dependent fitness (per capita growth rate) w of individuals with character z is given by
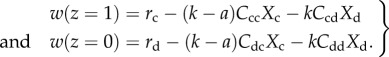 |
3.1 |
Xi is the global density of morph i, and Cij is the clustering coefficient between morphs i and j. Note that the population density is a function of a, because helping reduces competition. For instance, the monomorphic cooperator population density is rc/((k − a)Ccc), while the monomorphic defector population density is rd/(kCdd). The effect of cooperation on population density leads to a non-zero-sum game. See electronic supplementary material for the mathematical foundations of the dynamic system of equation (3.1).
We apply Price's covariance equation [34] to equation (3.1) and obtain an expression of how the population's mean cooperative character or cooperator frequency (Z) changes with genetic variance var(z) and selection factors (see electronic supplementary material, which also shows the equivalent inclusive fitness formulation):
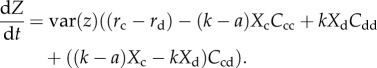 |
3.2 |
From the above equation, we recover three scenarios that parallel previous results. First, without clustering (all C's = 1), dZ/dt = var(z)(rc − rd), so cooperation does not evolve when cooperators pay a cost—a traditional expression of the cooperation dilemma [1,35]. Second, we consider any value of competition k but no demographic dynamics (habitat saturation). The latter forces cooperator and defector clusterings to be perfectly correlated (Ccc = Cdd = Cii) [3,36]. The rationale here is that any defector displaced from a cooperator's neighbourhood that increases Ccc must go to a defector's neighbourhood in the absence of an empty site, causing the same increase in defector clustering (and vice versa). Further, in the case of weak selection and sufficiently large populations (greater than 2000 as in our experiment), evolutionary dynamics are sufficiently described by what happens when Xc = Xd = Xi (see electronic supplementary material). Then, dZ/dt = var(z)((rc − rd) + aXi(Ccc − Ccd). Thus, within-morph clustering (or cooperator clustering) in this case is expected to favour cooperation.
Finally, with demographic dynamics, Ccc and Cdd are different, because a displaced neighbour does not have to enter another individual's neighbourhood. Equation (3.2) is the general form that shows Ccc is negatively associated with cooperation (dZ/dt), while Cdd is positively associated with cooperation. Owing to the generality of Price's equation [37], these clustering results are independent of cooperator frequency, population density and parameter values as long as k > a. For within-morph clustering to have a positive effect on cooperation under weak selection and sufficiently large populations (when Xc = Xd = Xi), it is required that (from equation (3.2)) −(k − a)XcCcc + kXdCdd > 0, or
| 3.3 |
In other words, the relative difference between cooperator and defector clustering must be sufficiently small (or negative) for within-morph clustering to favour cooperation. The uncorrelated clustering in our model leads to a novel finding: cooperator clustering lowers cooperation, while defector clustering favours cooperation. These predictions concur with our experimental results. The general result in equation (3.2) is actually identical to the case where cooperation affects survival in demographically explicit discrete-space models [5,8,9], but here it is generalized for strong selection and continuous-space, which is appropriate for our experiment. We have also taken a different identification approach by isolating clustering effects rather than life-history (r, a, k) effects (see the electronic supplementary material). In the electronic supplementary material, we also show that defector clustering Cdd already appeared in previous works in a competition term, but was not isolated as we have here.
We have chosen not to focus on the effect of between-morph clustering (Ccd) on cooperation, which is (k − a)Xc − kXd from equation (3.2), because it is frequency dependent (depends on the state of Xc relative to Xd) and thus cannot be easily tested using our current experimental set-up. When 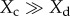 , the effect of Ccd is positive for cooperators; when
, the effect of Ccd is positive for cooperators; when 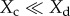 the effect is negative, thus overall between-morph clustering reinforces the prevailing cooperator frequency. With weak selection (Xc = Xd = Xi), the effect of Ccd should be negative (−aXc). In our experiment where strong selection occurs, we observe a negative effect of Ccd, but insignificantly (figure 3d). Between-morph clustering may destabilize the coexistence of cooperators and defectors, and can be investigated in the future using experiments with different initial frequencies.
the effect is negative, thus overall between-morph clustering reinforces the prevailing cooperator frequency. With weak selection (Xc = Xd = Xi), the effect of Ccd should be negative (−aXc). In our experiment where strong selection occurs, we observe a negative effect of Ccd, but insignificantly (figure 3d). Between-morph clustering may destabilize the coexistence of cooperators and defectors, and can be investigated in the future using experiments with different initial frequencies.
(b). Simulations
We further conducted individual-based simulations to corroborate our findings (see electronic supplementary material for detail and Dryad repository [38] for Matlab code and data). Cooperators and defectors occupy a 36 × 36 patch space and follow the eco-evolutionary dynamics specified in equation (3.1) within each patch. Cooperators and defectors move to an adjacent patch probabilistically with no cost or intrinsic difference between morphs. We again observed that cooperator clustering lowers cooperation, while defector clustering favours cooperation using the same bootstrap regression procedure outlined for the experimental analyses (figure 4). Variation in movement rate, which effectively changes the scale of interaction without changing the measurement scale (the patch), yielded the same conclusions with respect to clustering effects (see electronic supplementary material, figure S5–S7).
Interestingly, in all simulation cases, there is a very slight but significant negative relationship between cooperator and defector clustering (electronic supplementary material, figure S5–S7), in contrast to the experimental result that showed no relationships (figure 3b). With greater movement rate, the relationship between Ccc and Cdd appears to weaken: the slopes of Cdd/Ccc go from −0.086 (p = 0.0030) to −0.0095 (p = 0.013) for the lowest to highest movement rates tested. The negative relationship between the two among-morph clustering may be due to the fact that stochastically higher clustering for one morph limits its growth, leaving more habitat for the other morph to spread into (thus reducing its clustering). It may well be that we could see a slight negative relationship in the experiment with more replicates; but so long as Ccc and Cdd are not strongly correlated, they remain identifiable and are expected to exhibit the predicted effects. The observed clustering effects in both the simulations and the experiment agree with this line of reasoning.
4. Discussion
Together our experiment and models show that cooperator clustering is negatively associated with cooperation, contrary to previous theoretical [1,3,36] and experimental results [6,11], but defector clustering is positively associated with cooperation.
Two uncommon but desirable features of the microhabitat chip contribute to our novel empirical findings. First, the chip and image analysis make it possible to track every individual bacterium in space through morph-specific fluorescent tags, allowing clustering patterns to be measured at various scales. At an aggregate level, fluorescent bacteria have been frequently tracked to study biofilm development and community interactions [39–41], as well as the evolution of cooperation [12,42]. However, the scales of interest in these studies were relatively large. Our individual-resolution analysis (figure 3c) suggests that the interaction scale is 5 µm—close to the individual size—consistent with recent spatial genetic studies of another cooperative bacterium Myxococcus xanthus [43] and the social amoeba Dictyostelium discoideum [44]. Even though siderophores can diffuse in liquid medium at the millimetre scale over periods similar to the duration of our experiment [11], the effective interaction scale in the presence of bacteria is much smaller. This small interaction scale also explains why we did not see significant 100-μm scale habitat patchiness effects on cooperator frequency. The proper identification of spatial interaction scale can be essential to infer clustering effects.
Second, the chip provides an enclosed environment where spatial patterns emerge. Other microbial experiments on cooperation serially transfers bacteria through cycles of liquid cultures [6,45], thereby preventing cooperators and defectors from clustering differently, and imposing artificial interaction scales. Our experiment shows that cooperator and defector clustering are uncorrelated (figure 3b). This makes it statistically possible to distinguish what aspect of clustering favours or hampers cooperation. It is important to recognize that our experiment only provides a first description of the statistical association between multiple clustering aspects and the evolution of cooperation; control of cooperator and defector clustering (for example through motility mutants [39]) in longer experiments with other model organisms would be necessary to generalize these findings. Nevertheless, the theoretical and simulation models we developed merit further investigation.
Our models predict that cooperator clustering is detrimental to cooperation, but that defector clustering favours cooperation. This agrees with our experiment and generalizes previously known theoretical results by deriving their equivalences in continuous-space without assuming weak selection (cf. equation (3.2) to [5,8,9]). Importantly, both our model and experiment improve our understanding of kin competition. Kin competition is the potential for among-cooperator competition to impede cooperation in space [3,4,7,28,46,47], but it was previously only inferred from the overall decrease in cooperation level due to spatial competition, not through direct observation or theoretical isolation of the cooperator clustering effect. This is a result of previous focus on isolating life-history effects, in particular cost and benefit, leaving relatedness to be a compound metric that is mainly but not solely driven by cooperator clustering (see [2,31]; electronic supplementary material). Our model and clustering coefficients highlight the causality of cooperator versus defector clustering effects on cooperation in a demographically dynamic setting. As in previous work, kin competition does not necessarily preclude the evolution of cooperation, but here we identified a novel factor, defector clustering, as a spatial aspect that counters the detrimental effect of cooperator clustering on cooperation (equation (3.3)).
The clustering effects on cooperator frequency are strikingly similar in the experiment (figure 3c) and in the model simulations (figure 4), even though the latter only qualitatively corresponds to the experiment in that both exhibit demographic dynamics driven by cooperation and competition. The simulations also showed that no motility differences between morphs are required for cooperators and defectors to cluster differently. This absence of motility differences matches what is previously known in our cooperator and defector strains [48], although this is difficult to establish within the microhabitats where movement and clustering are confounded with growth and interaction dynamics. Motility differences, in particular the increased motility of defectors, is expected to differentiate cooperator and defector clustering [49]; but even without motility differences, we showed that cooperator and defector clustering effects can be distinct and consequential to the evolution of cooperation.
These experimental and theoretical results suggest a new research focus: defector clustering is important for the evolution of indiscriminate cooperation. Defector clustering must be sufficiently large relative to cooperator clustering for within-morph clustering to favour cooperation (equation (3.3)). As a corollary, whenever defector clustering is not perfectly correlated to cooperator clustering (l.h.s. of equation (3.3) ≠ 0), the benefit from cooperation may not be great enough to compensate for the elevated competition from clustering (quantified by a/k in equation (3.3)). Indeed, the experiment shows that at low cooperator clustering, defector clustering is similarly low (figure 3b); but as cooperator clustering increases, defector clustering does not increase, leading to a larger disparity that selects against cooperation. In other words, our experiment exhibits a lack of regulation on defector clustering, such that within-morph clustering does not favour cooperation. This contrasts with the fact that P. aeruginosa produces siderophores in nature and hints at the importance of defector clustering regulation for cooperation. We next discuss cases where cooperation is impaired by a lack of regulation on defector clustering, as well as examples where the regulation on defector clustering appears to maintain cooperation.
The human lung is highly heterogeneous at a wide range of scales, leading bacterial populations in chronic infections to colonize and adapt to micrometre-scale features of the lower respiratory tract [50], which potentially restrict movement. In CF patients with chronic infection, P. aeruginosa tends to exhibit impaired siderophore production and other loss-of-function mutations, including lowered motility [19,51]. In addition, CF patients exhibit abnormally thick airway secretions that drastically impair bacterial motility and clearance [51]. These features suggest a relatively static environment and high clustering, which according to our experiment can increase the differentiation between cooperator and defector clustering (figure 3b). Some clustering is required for siderophore cooperation to evolve, but very high clustering, if associated with the deregulation of defector clustering, is detrimental to cooperation (equation (3.3)). In another experiment that manipulated P. aeruginosa clustering without disturbing pattern formation (by varying agar concentration), very high clustering appears less conducive to siderophore cooperation than intermediate clustering [11], although clustering could not be directly measured. While our experiment suggests that in stagnant, constrained space, defector clustering is deregulated, it remains unknown whether this is the case in vivo.
In another example of cooperator clustering acting against the evolution of cooperation, the production of biofilm EPS by Vibrio cholerae benefits neighbours but limits cooperator dispersal [52]. Thus, cooperators experience higher clustering, which was observed to prevent their evolution when success depends on colonizing areas outside of biofilms [52].
Some examples exhibit relatively clear defector clustering regulation resulting in a high degree of cooperation. In multicellular organisms, cancerous defector cells are kept clustered through the developmental regulations of cell adhesion and repulsion, which usually prevents tumours from metastasis and further spread [53]. For example, the naked mole rat possesses especially strong cell contact inhibition mechanisms, which would regulate defector cell clustering [54] and has resulted in the absence of cancer (in contrast with most other mammals). In the social amoeba Dictyostelium discoideum, defectors are less able to produce dispersal-facilitating stalks [55], which increases defector clustering, in addition to the overall clustering required to produce the cooperative stalks. The hypothesis that defector clustering can be critical to cooperation provides new interpretations of known phenomena and merits further investigations.
In conclusion, although previous results on the generally positive effect of clustering on cooperation might apply to some systems, our work suggests that more intricate spatial mechanisms may critically contribute to the evolution of cooperation. With increased clustering, cooperators help each other more, but compete even more. Thus, evolution of indiscriminate cooperation may require mechanisms that specifically increase defector clustering.
Supplementary Material
Supplementary Material
Acknowledgements
We thank D. Juncker, G.A. McKay and M. Nannini for help with the experimental device M.R. Parsek and S. Moskowitz for providing the pMRP9-1 and pMKB1 plasmids, and the reviewers for clarifying some theoretical connections with the existing literature.
Data accessibility
The data for the experimental analyses are summarized in the electronic supplementary material, Dataset spreadsheet. All experimental and simulation data and Matlab code can be found in the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.fq1sj [38]. Additional information and videos of the experimental device can be found in [56].
Authors' contributions
E.W.T., A.G. and M.L. conceived the study. A.G. and E.W.T. conceived the device. E.W.T performed the experiment, derived the equations and implemented the simulations. D.N. provided guidance, strains and equipment for the experiment. E.W.T. wrote the first draft, and all authors contributed substantially to revisions.
Competing interests
We declare we have no competing interests.
Funding
E.W.T was supported by the Fonds Québécois de la Recherche sur la Nature et les Technologies and the Québec Centre for Biodiversity Science. D.N. was supported by a CFI Leaders Opportunity Fund (25636), a Burroughs Wellcome Fund CAMS award (1006827.01) and Canadian Institutes of Health research and Fonds de Recherche Quebec Santé salary awards. A.G. was supported by the Canada Research Chair programme and an NSERC Discovery grant. M.L. was supported by the TULIP Laboratory of Excellence (ANR-10-LABX-41).
References
- 1.Hamilton WD. 1964. The genetical evolution of social behaviour. I. J. Theor. Biol. 7, 1–16. ( 10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 2.Lion S, Baalen MV. 2008. Self-structuring in spatial evolutionary ecology. Ecol. Lett. 11, 277–295. ( 10.1111/j.1461-0248.2007.01132.x) [DOI] [PubMed] [Google Scholar]
- 3.Débarre F, Hauert C, Doebeli M. 2014. Social evolution in structured populations. Nat. Commun. 5, 3409 ( 10.1038/ncomms4409) [DOI] [PubMed] [Google Scholar]
- 4.Taylor PD. 1992. Altruism in viscous populations — an inclusive fitness model. Evol. Ecol. 6, 352–356. ( 10.1007/BF02270971) [DOI] [Google Scholar]
- 5.Lion S, Gandon S. 2009. Habitat saturation and the spatial evolutionary ecology of altruism. J. Evol. Biol. 22, 1487–1502. ( 10.1111/j.1420-9101.2009.01769.x) [DOI] [PubMed] [Google Scholar]
- 6.Griffin AS, West SA, Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027. ( 10.1038/nature02744) [DOI] [PubMed] [Google Scholar]
- 7.Lion S, Gandon S. 2010. Life history, habitat saturation and the evolution of fecundity and survival altruism. Evolution 64, 1594–1606. ( 10.1111/j.1558-5646.2009.00933.x) [DOI] [PubMed] [Google Scholar]
- 8.Matsuda H, Ogita N, Sasaki A, Sato K. 1992. Statistical mechanics of population. Prog. Theor. Phys. 88, 1035–1049. ( 10.1143/PTP.88.1035) [DOI] [Google Scholar]
- 9.van Baalen M, Rand D. 1998. The unit of selection in viscous populations and the evolution of altruism. J. Theor. Biol. 193, 631–648. ( 10.1006/jtbi.1998.0730) [DOI] [PubMed] [Google Scholar]
- 10.Kümmerli R, Gardner A, West SA, Griffin AS. 2009. Limited dispersal, budding dispersal, and cooperation: an experimental study. Evolution 63, 939–949. ( 10.1111/j.1558-5646.2008.00548.x) [DOI] [PubMed] [Google Scholar]
- 11.Kümmerli R, Griffin AS, West SA, Buckling A, Harrison F. 2009. Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc. R. Soc. B 276, 3531–3538. ( 10.1098/rspb.2009.0861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hol FJH, Galajda P, Nagy K, Woolthuis RG, Dekker C, Keymer JE. 2013. Spatial structure facilitates cooperation in a social dilemma: empirical evidence from a bacterial community. PLoS ONE 8, e77402 ( 10.1371/journal.pone.0077042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media—a review. Adv. Water Resour. 30, 1505–1527. ( 10.1016/j.advwatres.2006.05.025) [DOI] [Google Scholar]
- 14.Tekwa EW, Nguyen D, Juncker D, Loreau M, Gonzalez A. 2015. Patchiness in a microhabitat chip affects evolutionary dynamics of bacterial cooperation. Lab. Chip 15, 3723–3729. ( 10.1039/C5LC00576K) [DOI] [PubMed] [Google Scholar]
- 15.Poole K, McKay GA. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8, 661–686. ( 10.2741/1051) [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Rainey PB. 2013. Exploring the sociobiology of pyoverdin-producing Pseudomonas. Evolution 67, 3161–3174. ( 10.1111/evo.12183) [DOI] [PubMed] [Google Scholar]
- 17.Dionisio F, Gordo I. 2006. The tragedy of the commons, the public goods dilemma, and the meaning of rivalry and excludability in evolutionary biology. Evol. Ecol. Res. 8, 321–332. [Google Scholar]
- 18.Kümmerli R, Jiricny N, Clarke LS, West SA, Griffin AS. 2009. Phenotypic plasticity of a cooperative behaviour in bacteria. J. Evol. Biol. 22, 589–598. ( 10.1111/j.1420-9101.2008.01666.x) [DOI] [PubMed] [Google Scholar]
- 19.Smith EE, et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl Acad. Sci. USA 103, 8487–8492. ( 10.1073/pnas.0602138103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crespi B, Foster K, Úbeda F. 2014. First principles of Hamiltonian medicine. Phil. Trans. R. Soc. B 369, 20130366 ( 10.1098/rstb.2013.0366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hol FJH, Dekker C. 2014. Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346, 1251821 ( 10.1126/science.1251821) [DOI] [PubMed] [Google Scholar]
- 22.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64, 518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 194, 6387–6389. ( 10.1128/JB.01479-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. ( 10.1126/science.280.5361.295) [DOI] [PubMed] [Google Scholar]
- 25.Brannon MK, Davis JM, Mathias JR, Hall CJ, Emerson JC, Crosier PS, Huttenlocher A, Ramakrishnan L, Moskowitz SM. 2009. Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell. Microbiol. 11, 755–768. ( 10.1111/j.1462-5822.2009.01288.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin SA, Pacala SW. 1997. Theories of simplification and scaling of spatially distributed processes. In Spatial ecology: the role of space in population dynamics and interspecific interactions (eds Tilman D, Kareiva P), pp. 271–296. Princeton, NJ: Princeton University Press. [Google Scholar]
- 27.Van Baalen M. 2000. Pair approximations for different spatial geometries. In The geometry of ecological interactions: simplifying spatial complexity (eds Dieckmann U, Metz JAJ, Law R), pp. 359–387. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 28.Taylor PD, Day T, Wild G. 2007. Evolution of cooperation in a finite homogeneous graph. Nature 447, 469–472. ( 10.1038/nature05784) [DOI] [PubMed] [Google Scholar]
- 29.Tekwa EW, Gonzalez A, Loreau M. 2015. Local densities connect spatial ecology to game, multilevel selection and inclusive fitness theories of cooperation. J. Theor. Biol. 380, 414–425. ( 10.1016/j.jtbi.2015.06.016) [DOI] [PubMed] [Google Scholar]
- 30.Rousset F. 2004. Genetic structure and selection in subdivided populations. Princeton, NJ: Princeton University Press. [Google Scholar]
- 31.Day T, Taylor PD. 1998. Unifying genetic and game theoretic models of kin selection for continuous traits. J. Theor. Biol. 194, 391–407. ( 10.1006/jtbi.1998.0762) [DOI] [PubMed] [Google Scholar]
- 32.Akaike H. 1974. A new look at the statistical model identification. Autom. Control. IEEE Trans. 19, 716–723. ( 10.1109/TAC.1974.1100705) [DOI] [Google Scholar]
- 33.Efron B, Tibshirani R. 1986. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1, 54–77. ( 10.1214/ss/1177013815) [DOI] [Google Scholar]
- 34.Price GR. 1970. Selection and covariance. Nature 227, 520–521. ( 10.1038/227520a0) [DOI] [PubMed] [Google Scholar]
- 35.Nowak MA. 2006. Evolutionary dynamics. Cambridge, MA: Harvard University Press. [Google Scholar]
- 36.Nathanson CG, Tarnita CE, Nowak MA. 2009. Calculating evolutionary dynamics in structured populations. PLoS Comput. Biol. 5, e1000615 ( 10.1371/journal.pcbi.1000615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Queller DC. 2017. Fundamental theorems of evolution. Am. Nat. 189, 345–353. ( 10.1086/690937) [DOI] [PubMed] [Google Scholar]
- 38.Tekwa E, Nguyen D, Loreau M, Gonzalez A. 2017. Data from: Defector clustering is linked to cooperation in a pathogenic bacterium Dryad Digital Repository. ( 10.5061/dryad.fq1sj) [DOI] [PMC free article] [PubMed]
- 39.Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2009. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 74, 1380–1392. ( 10.1111/j.1365-2958.2009.06934.x) [DOI] [PubMed] [Google Scholar]
- 40.Nguyen D, et al. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. ( 10.1126/science.1211037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 2013. 3D printing of microscopic bacterial communities. Proc. Natl Acad. Sci. USA 110, 18 380–18 385. ( 10.1073/pnas.1309729110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keymer JE, Galajda P, Lambert G, Liao D, Austin RH. 2008. Computation of mutual fitness by competing bacteria. Proc. Natl Acad. Sci. USA 105, 20 269–20 273. ( 10.1073/pnas.0810792105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kraemer SA, Wielgoss S, Fiegna F, Velicer GJ. 2016. The biogeography of kin discrimination across microbial neighborhoods. Mol. Ecol. 25, 4875–4888. ( 10.1111/mec.13803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith J, Strassmann JE, Queller DC. 2016. Fine-scale spatial ecology drives kin selection relatedness among cooperating amoebae. Evolution 70, 848–859. ( 10.1111/evo.12895) [DOI] [PubMed] [Google Scholar]
- 45.Chuang JS, Rivoire O, Leibler S. 2009. Simpson's paradox in a synthetic microbial system. Science 323, 272–275. ( 10.1126/science.1166739) [DOI] [PubMed] [Google Scholar]
- 46.Wilson DS, Pollock GB, Dugatkin LA. 1992. Can altruism evolve in purely viscous populations? Evol. Ecol. 6, 331–341. ( 10.1007/BF02270969) [DOI] [Google Scholar]
- 47.Grafen A, Archetti M. 2008. Natural selection of altruism in inelastic viscous homogeneous populations. J. Theor. Biol. 252, 694–710. ( 10.1016/j.jtbi.2008.01.021) [DOI] [PubMed] [Google Scholar]
- 48.Banin E, Vasil ML, Greenberg EP. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl Acad. Sci. USA 102, 11 076–11 081. ( 10.1073/pnas.0504266102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakano JY, Nowak MA, Hauert C. 2009. Spatial dynamics of ecological public goods. Proc. Natl Acad. Sci. USA 106, 7910–7914. ( 10.1073/pnas.0812644106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10, 841–851. ( 10.1038/nrmicro2907) [DOI] [PubMed] [Google Scholar]
- 51.Gellatly SL, Hancock RE. W. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173. ( 10.1111/2049-632X.12033) [DOI] [PubMed] [Google Scholar]
- 52.Nadell CD, Bassler BL. 2011. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc. Natl Acad. Sci. USA 108, 14 181–14 185. ( 10.1073/pnas.1111147108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batlle E, Wilkinson DG. 2012. Molecular mechanisms of cell segregation and boundary formation in development and tumorigenesis. Cold Spring Harb. Perspect. Biol. 4, 1–14. ( 10.1101/cshperspect.a008227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, Mao Z, Catania KC, Gorbunova V. 2009. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl Acad. Sci. USA 106, 19 352–19 357. ( 10.1073/pnas.0905252106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Queller DC, Ponte E, Bozzaro S, Strassmann JE. 2003. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science 299, 105–106. ( 10.1126/science.1077742) [DOI] [PubMed] [Google Scholar]
- 56.Greiner A, Tekwa EW, Gonzalez A, Nguyen D. 2017. Rapid inoculation and recovery of microbes in a microfluidic device Chips Tips. See http://blogs.rsc.org/chipsandtips/2017/10/04/rapid-inoculation-and-recovery-of-microbes-in-a-microfluidic-device/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Tekwa E, Nguyen D, Loreau M, Gonzalez A. 2017. Data from: Defector clustering is linked to cooperation in a pathogenic bacterium Dryad Digital Repository. ( 10.5061/dryad.fq1sj) [DOI] [PMC free article] [PubMed]
- Greiner A, Tekwa EW, Gonzalez A, Nguyen D. 2017. Rapid inoculation and recovery of microbes in a microfluidic device Chips Tips. See http://blogs.rsc.org/chipsandtips/2017/10/04/rapid-inoculation-and-recovery-of-microbes-in-a-microfluidic-device/.
Supplementary Materials
Data Availability Statement
The data for the experimental analyses are summarized in the electronic supplementary material, Dataset spreadsheet. All experimental and simulation data and Matlab code can be found in the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.fq1sj [38]. Additional information and videos of the experimental device can be found in [56].