Abstract
Black phosphorus nanosheets (BPs) show great potential for various applications including biomedicine, thus their potential side effects and corresponding improvement strategy deserve investigation. Here, in vitro and in vivo biological effects of BPs with and without titanium sulfonate ligand (TiL4) modification are investigated. Compared to bare BPs, BPs with TiL4 modification (TiL4@BPs) can efficiently escape from macrophages uptake, and reduce cytotoxicity and proinflammation. The corresponding mechanisms are also discussed. These findings may not only guide the applications of BPs, but also propose an efficient strategy to further improve the biocompatibility of BPs.
Keywords: biocompatibility, black phosphorus, cell uptake, inflammations, surface modification
Black phosphorus (BP), the most stable allotrope of the phosphorus element, is presently attracting intense research interests as a novel 2D material worldwide. In recent years, BP of a few atomic layers (BP nanosheets, BPs) has been produced by exfoliation techniques,1 and showed many unique properties such as high mobility, highly anisotropic charge transport, and distinct optical response properties.1, 2 As a result of these unique characteristics, BPs have been regarded as a promising nanomaterial not only in nanoscale electronic and optoelectronic devices, but also in various biomedical applications.3 Typically, BPs with excellent near‐infrared photothermal and photodynamic performances have shown great potential in cancer therapy.4 Combined with their phototherapy abilities, BPs have also been successfully applied as sensitive in vivo photoacoustic imaging agents.5 Furthermore, ascribed to their atomically thin 2D structure and large surface area, BPs have been proposed as efficient drug delivery platforms,6 and multifunctional theranostic agents in the treatment of cancer.5, 6, 7 However, the actual biological effects of few‐layered BPs as nanomaterials are difficult to predict in vivo, and the understanding towards the biological effects of BPs is extremely limited.8
As an important part of immune system, mononuclear phagocyte system (MPS) plays a crucial role in the clearance of nanomaterials from the circulation upon medical application.9 The engulfment of nanomaterials in MPS dramatically diminished the delivery of the circulated nanomaterials to target tissues (tumors or pathological organs, etc.). More importantly, when nanomaterials were accumulated in MPS, concentrated macrophages would inevitably initiate immune responses, and provoke various side effects (such as inflammation), which would also hamper therapeutic efficacy of nanomaterials.10 In recent years, various nanomaterials have been investigated and reported as active foreign particles that can stimulate inflammatory effects when administered to animals or humans.11 Severe or sustained inflammatory responses can lead to diseases due to damages to normal organs or tissues by the stimulated immune cells.12 Therefore, the reduction of macrophage uptake of the injected nanomaterials has been regarded as an important strategy to enhance corresponding therapeutic efficacy and reduce the potential toxicity caused by their nonspecific deposition in MPS.13 As a new biomaterial with great application potential, the inflammatory effects of BPs no doubt deserve detailed investigation. The corresponding strategy to improve their biocompatibility is also highly desired, not only in terms of environmental exposure but also as a mean to reduce the toxicity for biomedical uses.
Here, detailed in vitro and in vivo experiments were performed to investigate the inflammatory effects and biocompatibility of ultrasmall BPs (BPQDs) (named bare BPs) and ultrasmall BPs with titanium sulfonate ligand (TiL4) modification (named TiL4@BPs). The bare BPs and TiL4@BPs were synthesized using the strategies reported previously by our group.4b, 14 In short, BPs was synthesized using a liquid exfoliation technique in N‐methyl‐2‐pyrrolidone (NMP), then mixed with TiL4 in NMP at room temperature for 15 h to produce TiL4@BPs. The morphology of TiL4@BPs was examined using transmission electron microscopy (TEM), high‐resolution TEM (HR‐TEM), and atomic force microscopy (AFM). As shown in Figure S1 in the Supporting Information, TiL4@BPs had an average size and height of about 3.3 and 1.5 nm, respectively. The HR‐TEM image shows their lattice fringes of 0.21 nm, corresponding to the (014) plane of the crystal of BPs.15 Such chemical modification did not change the crystal morphology and structure of the BPs.14 High‐resolution X‐ray photoelectron spectroscopy (HR‐XPS) was performed to assess the chemical quality of bare BPs and TiL4@BPs. As shown in Figure S2a, bare BPs exhibited the P2p3/2 and P2p1/2 doublet at the 130.1 and 130.9 eV respectively, characteristic of crystalline BPs, and intense oxidized phosphorus sub‐bands were apparent at 134.0 eV as a result of partial oxidation. In contrast, the binding energy of P2p in TiL4@BPs was 132.4 eV, in accordance with the reported value of Ti‐P coordination, and oxidized phosphorus sub‐band was not found. The XPS thus confirmed successful coordination between P and Ti in TiL4@BPs. Furthermore, it was found that the zeta potential changed from −36.5±1.1 mV in bare BPs to +21.1±2.6 mV after TiL4 coordination in TiL4@BPs (Figure S3). In addition, TiL4@BPs exhibited higher stability against oxidation and degradation than bare BPs (Figure S4) and the corresponding mechanism has been reported previously.14
Macrophage, an important component of MPS, is the major cell type responsible for the clearance of nanomaterials from peripheral blood,9, 16 and the uptake of the high level of nanomaterials by macrophages has been regarded as the cause of serious side effects.17 Here, to evaluate their cellular uptake potential by macrophages, accumulation and localization of bare BPs and TiL4@BPs within raw 264.7 macrophages were examined by using Raman spectroscopy. As a label‐free intrinsic signature, Raman signal has been proven to be an effective tool for the detection of nanomaterials, and Raman spectroscopy has recently been used to determine the biological uptake of various nanomaterials.18 Because of the high background level of phosphorus in cells, it is difficult to determine intracellular BPs levels by measuring the amount of elemental phosphorus. However, BPs have three characteristic Raman peaks at about 359.5, 436.0, and 463.3 cm−1, respectively corresponding to the A1 g B2g, and A2 g vibrational modes of P atoms in BP.4 Therefore, Raman spectroscopy was performed to examine the cell uptake of BPs in vitro.
As shown in Figure 1 a and b, characteristic Raman spectra of the BPs can be apparently isolated from the background signals. Thus, two representative cells were mapped based on the intensity of the Raman peak at 463.3 cm−1 to reveal the intracellular localization of bare BPs or TiL4@BPs, and Raman spectra of four typical sites of the cells were given. It was observed that bare BPs predominantly accumulated in the intracellular compartment, while TiL4@BPs showed significantly lower potential to enter the cells. Furthermore, Raman spectra of the center site of 25 randomly selected cells treated with bare BPs or TiL4@BPs were collected, and their average Raman signal intensities at 463.3 cm−1 were calculated to determine their cell uptake levels (Figure 1 c). The relative Raman signal intensity of bare BPs group was about six times higher than that of TiL4@BPs group. These findings suggest that macrophages tend to engulf more bare BPs than TiL4@BPs.
Figure 1.

Uptake and accumulation levels of bare BPs and TiL4@BPs by raw 264.7 cells. a) Localization of bare BPs or TiL4@BPs (indicated with red arrow) associated with cells after 6 h exposure to 10 μg mL−1 of bare BPs or TiL4@BPs determined using bright‐field microscopy. Red arrow shows the aggregated BPs within or associated cells. b) Raman mapping based on the intensity at 463.3 cm−1 of the representative J774A.1 cell treated with bare BPs or TiL4@BPs at 10 μg mL−1 for 24 h. The Raman spectra acquired from 4 different sites are also shown. c) Raman spectra of the center site of 25 randomly selected cells treated with bare BPs or TiL4@BPs, and their average Raman signal intensities at 463.3 cm−1 are calculated to determine their cell uptake levels. *p<0.05.
The cytotoxicity of bare BPs and TiL4@BPs towards macrophage cell lines (J774A.1 and raw 264.7 cells) was examined by using the ATP assay. As shown in Figure 2 a, both bare BPs and TiL4@BPs showed no effect on the survival of raw 264.7 cells. While for J774A.1 cells, bare BPs resulted in a significant decrease of ATP content, while TiL4@BPs showed less effect. Lysosome is known to be an important intracellular compartment for nanomaterial accumulation, which implicated in cell energy metabolism, cell death or proinflammation.19 In the control group, concentrated red dots can be observed within the cytosol, suggesting the presence of cathepsin B within lysosomes (Figure 2 b). However, when treated with bare BPs, most cells displayed swelled lysosomes comparing to the control group. In contrast, TiL4@BPs treatment did not induce such morphological changes in lysosomes. To determine whether the cytotoxicity of bare BPs was due to cell uptake, ATP contents of J774A.1 cells treated with bare BPs were measured with or without cytochalasin D (Cyt D), an inhibitor of actin polymerization frequently applied to investigate cell uptake of nanomaterials as an inhibitor of classical phagocytosis.20 As shown in Figure 2 c, the presence of the inhibitor significantly inhibited the reduction of the ATP content upon bare BPs exposure, demonstrating that the reduction of ATP content of cells is mainly due to the uptake of bare BPs.
Figure 2.
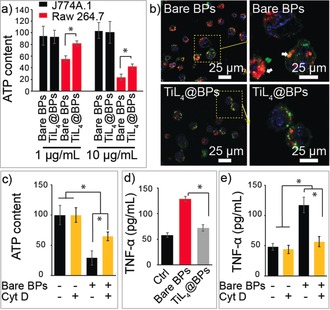
Cytotoxicity of bare BPs and TiL4@BPs. a) The cytotoxicity of bare BPs and TiL4@BPs towards raw 264.7 and J774A.1 cells evaluated with ATP assay after 24 h incubation. b) Confocal microscopy images of J774A.1 cells stained with Magic Red to characterize Cathepsin B location after incubation with bare BPs or TiL4@BPs for 6 h. Cell membrane, nuclear, and lysosomes was stained with PKH67 (green), H33342 (blue), and Magic red (red), respectively, and white arrows indicate the swelled lysosomes. c) ATP content of J774A.1 cells treated with 10 μg mL−1 of bare BPs with or without Cyt D at 24 h. d) TNF‐α concentration in the medium after the cells were treated with bare BPs or TiL4@BPs for 12 h. e) TNF‐α concentration released by raw 264.7 cells were evaluated after 24 h with the presence Cyt D treatment or not. *p<0.05.
The inflammatory effects induced by nanomaterials‐stimulated cytokines is a major concern for their biomedical applications.21 TNF‐α is among the typical cytokines secreted, and are frequently stimulated during the infection of pathogens or foreign particles.11 Because its ATP content was not affected according to previous experiments, raw 264.7 cells were selected to evaluate the proinflammatory activity of BPs by the assessment of TNF‐α release. As shown in Figure 2 d, the TNF‐α level of the cells treated with TiL4@BPs was much lower (71.9 pg mL−1) than that treated with bare BPs (128.4 pg mL−1), confirming better biocompatibility of TiL4@BPs. The presence of the inhibitor Cyt D significantly attenuated the promoted TNF‐α upon bare BPs incubation (Figure 2 e). These results together demonstrated that the TiL4 modification of BPs results in less cell uptake by macrophages, and thus renders lower cytotoxicity and proinflammation as compared to bare BPs.
Therefore, the possible mechanism of above biological effects of BPs with and without TiL4 modification might be due to both the intracellular level and the stability of BPs. On one hand, the negative zeta potential of bare BPs was reversed to a positive potential by the TiL4 modification. This alteration may tune the serum protein corona properties of BPs, and influence the cell uptake and corresponding biological effects.16 On the other hand, BPs undergo accelerated oxidation and degradation,22, 23 and when this process occurs within cells, the generation of O2 − could trigger the production of ROS, cause oxidative stresses, and finally induce cell death or inflammation.24 Comparing with bare BPs, TiL4@BPs had higher stability against oxidation and degradation (Figure S4). Correspondingly, TiL4@BPs may induce lower degree in ROS generation in cells than bare BPs, thus result in the reduced side effects.
To evaluate the potential inflammatory responses of BPs in vivo, mice were intravenously injected with bare BPs, TiL4@BPs, or PBS (solvent control) at 500 μg kg−1, then sacrificed at 1, 7, and 28 days for examinations (Figure 3 a). At day 1, compared to control group, bare BPs induced a distinct increase of neutrophils from 890 to 2470 cells μL−1. In contrast, TiL4@BPs exhibited no significant effect on the neutrophil population. It is known that an increase in the population of neutrophils is recognized as an important acute inflammatory response.25 Furthermore, a significant reduction in platelets (PLTs) in peripheral blood was observed in the bare BPs group, but not in the TiL4@BPs group. This phenomenon suggested that bare BPs are more toxic to mice than TiL4@BPs. After 7 or 28 days, all of these differences disappeared. These data implicated that bare BPs, rather than TiL4@BPs, acutely stimulated the increase of neutrophils population, which however recovered at a longer time point.
Figure 3.
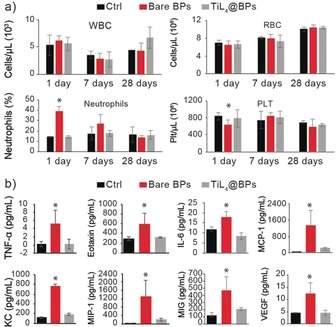
Peripheral blood cell count and cytokines concentration changes. a) WBCs, RBCs, neutrophils, lymphocytes, and PLTs cell numbers in peripheral blood of mice (n=5) sacrificed at 1, 7, and 28 days after administration of bare BPs, TiL4@BPs, or PBS (control). b) Concentrations of cytokines in mice (n=5) serum 1 day after i.v. injection of bare BPs, TiL4@BPs, or PBS (control). In serum, the concentrations of 32 cytokines were measured and the significantly increased cytokines were shown. *versus Ctrl, *p<0.05.
The variation of inflammatory cytokines in vivo was also evaluated in serum collected from the mice treated with above three samples at 1, 7 and 28 days. At day 1, among the 32 cytokines surveyed, TNF‐α, Eotaxin, IL‐6, MCP‐1, KC, MCP‐1β, MIG and VEGF showed a significant increase only in the bare BPs group, confirming a better biocompatibility of TiL4@BPs (Figure 3 b). Apoptosis of peripheral neutrophil populations is regulated by TNF‐α,26 which can increase the half‐life of neutrophils in circulation through inhibiting cell apoptosis. Therefore, the increased TNF‐α level could explain the increased neutrophil population. Correspondingly, the increased concentration of TNF‐α stimulates neutrophils to secrete growth factor (VEGF)27 as an endothelial cell mitogen,28 in line with the involvement of neutrophils in the bare BPs‐promoted inflammatory responses. The elevation of serum cytokines was not observed with longer exposures of 7 and 28 days (data not shown), suggesting an acute increase in peripheral cytokines upon a single administration.
It is known that the injection of foreign nanoparticles may cause the alterations in hematopoietic compartment (bone marrow (BM) and spleen, etc.).29 The injected nanoparticles tend to target BM because of the presence of high numbers of vessels and sinuses which contribute to a slower rate of blood flow and higher surface area, thus facilitate the accumulation of nanoparticles and their interactions with cells.9 Figure 4 a shows a significant decrease in the percentage of mature myeloid cells, as the Gr1+/CD11b+ percentage declined from 26.1 % in control group to 17.6 % in bare BPs group,30 while TiL4@ BPs showed no significant effect. Therefore, the increased population of peripheral neutrophils induced by bare BPs may also be attributed to the accelerated rate in myeloid cells driven out from BM.
Figure 4.
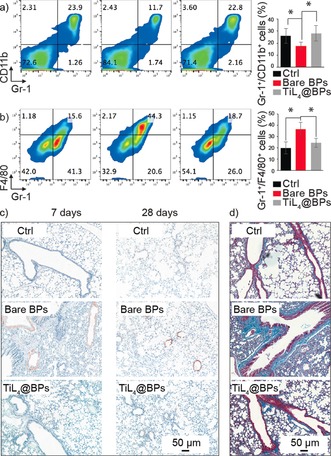
Alterations in the hematopoiesis compartment and histopathology. a) FACS analysis of the mature myeloid cells in BM after staining with CD11b and Gr1. b) FACS analysis of the F4/80+/CD11b+ monocytes/monocyte in spleen. c) Immunohistochemical staining of lung sections with a rat anti‐mouse CD68 followed by goat anti‐rat Masson's trichrome staining after 7 or 28‐day exposure. Original magnification, ×200 (n=6). d) Masson's trichrome staining of lungs after 28‐day exposure. *p<0.05.
In spleen, F4/80+/CD11b+ monocytes/monocyte in bare BPs group was 31.4 %, which was much higher than that of control group (15.1 %) and TiL4@ BPs group (21.6 %) (Figure 4 b). Spleen is the most important part of MPS, and displays high accumulation of foreign nanoparticles including BPs.9 After exposure to bare BPs, the accumulation of BPs in spleen can stimulate inflammation and recruit immune cells into this organ. These increased splenic F4/80+/CD11b+myloid cells upon BPs treatment is consistent with the notion that immune cells such as myeloid cells could be recruited to regions of infection or inflammation to engulf and kill infected pathogens.29c
No difference can be observed in individual erythroid precursors or progenitors, T cells and B cells for the three groups, indicating there was no effect on the other lineages (Figure S5), and suggesting the treatment of bare BPs predominantly resulted in myeloid linage alteration. These results further revealed the underlying mechanisms of different inflammatory responses observed in bare BPs and TiL4@BPs groups, and bare BPs displayed a higher inflammatory potential than TiL4@ BPs. To further evaluate potential tissue alterations, the histology of major organs including heart, liver, lungs, spleen, and kidneys was examined. There were no observable changes in organ index (data not shown). Furthermore, no histopathological alteration can be observed in the bare BPs or TiL4@BPs groups (Figure S6).
CD68 is a surface marker identified predominantly on macrophages and white blood cells, and is frequently applied as a marker for evaluation of inflammation in organs including the proinflammatory effects triggered by foreign nanoparticles. Upon examination of CD68 level in organs, bare BPs resulted in greater accumulation of CD68+ cells in lungs after 7‐day and 28‐day treatment, while TiL4@BPs produced almost no obvious increase in CD68+ (Figure 4 c). Furthermore, compared with TiL4@BPs group, the increased collagen in the bronchioles can be observed for bare BPs group, confirming that TiL4 modification improved the biocompatibility of BPs. Our results were also in line with the notion that the lungs are among the most sensitive target organs upon various exposure routes.21a
In conclusion, in vitro and in vivo biological effects of BPs with and without TiL4 modification have been investigated in detail. In vitro experiments demonstrated that BPs with TiL4 modification can escape the cell uptake by macrophages, and further reduce the cytotoxicity and proinflammation. When intravenously injected into mice, bare BPs trigger significant inflammatory responses, including an increase in peripheral neutrophils accompanied by elevation of a group of inflammatory cytokines. In contrast, TiL4@BPs exhibit no such adverse immune responses. The mechanisms of the BPs’ inflammatory effects and the improved biocompatibility caused by the TiL4 modification have been discussed. The detailed investigation of biological effects of BPs may guide their applications as foreign particles administered to animals or humans. In addition, our findings proposed an efficient strategy to improve the biocompatibility of BPs, and the synthesized TiL4@BPs may have a great potential in biomedical applications.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Nos. 21527901 and 51672305), the Youth Foundation of Chinese Academy of Inspection and Quarantine (2016JK025), Frontier Research Key Project of the Chinese Academy of Sciences (QYZDB‐SSW‐SLH034), and Leading Talents of Guangdong Province Program No. 00201520.
G. Qu, W. Liu, Y. Zhao, J. Gao, T. Xia, J. Shi, L. Hu, W. Zhou, J. Gao, H. Wang, Q. Luo, Q. Zhou, S. Liu, X.-F. Yu, G. Jiang, Angew. Chem. Int. Ed. 2017, 56, 14488.
Contributor Information
Xue‐Feng Yu, Email: xf.yu@siat.ac.cn.
Guibin Jiang, Email: gbjiang@rcees.ac.cn.
References
- 1. Churchill H. O., Jarillo-Herrero P., Nat. Nanotechnol. 2014, 9, 330–331. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. Rodin A. S., Carvalho A., Castro Neto A. H., Phys. Rev. Lett. 2014, 112, 176801; [DOI] [PubMed] [Google Scholar]
- 2b. Doganov R. A., O'Farrell E. C. T., Koenig S. P., Yeo Y. T., Ziletti A., Carvalho A., Campbell D. K., Coker D. F., Watanabe K., Taniguchi T., Neto A. H. C., Ozyilmaz B., Nat. Commun. 2015, 6, 6647; [DOI] [PubMed] [Google Scholar]
- 2c. Wang X. M., Jones A. M., Seyler K. L., Tran V., Jia Y. C., Zhao H., Wang H., Yang L., Xu X. D., Xia F. N., Nat. Nanotechnol. 2015, 10, 517–521; [DOI] [PubMed] [Google Scholar]
- 2d. Xiang D., Han C., Wu J., Zhong S., Liu Y. Y., Lin J. D., Zhang X. A., Hu W. P., Ozyilmaz B., Neto A. H. C., Wee A. T. S., Chen W., Nat. Commun. 2015, 6, 6485. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Engel M., Steiner M., Avouris P., Nano Lett. 2014, 14, 6414–6417; [DOI] [PubMed] [Google Scholar]
- 3b. Low T., Rodin A. S., Carvalho A., Jiang Y. J., Wang H., Xia F. N., Neto A. H. C., Phys. Rev. B 2014, 90, 075437; [Google Scholar]
- 3c. Mu H. R., Lin S. H., Wang Z. C., Xiao S., Li P. F., Chen Y., Zhang H., Bao H. F., Lau S. P., Pan C. X., Fan D. Y., Bao Q. L., Adv. Opt. Mater. 2015, 3, 1447–1453; [Google Scholar]
- 3d. Guo Z. N., Zhang H., Lu S. B., Wang Z. T., Tang S. Y., Shao J. D., Sun Z. B., Xie H. H., Wang H. Y., Yu X. F., Chu P. K., Adv. Funct. Mater. 2015, 25, 6996–7002. [Google Scholar]
- 4.
- 4a. Wang H., Yang X. Z., Shao W., Chen S. C., Xie J. F., Zhang X. D., Wang J., Xie Y., J. Am. Chem. Soc. 2015, 137, 11376–11382; [DOI] [PubMed] [Google Scholar]
- 4b. Sun Z. B., Xie H. H., Tang S. Y., Yu X. F., Guo Z. N., Shao J. D., Zhang H., Huang H., Wang H. Y., Chu P. K., Angew. Chem. Int. Ed. 2015, 54, 11526–11530; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 11688–11692; [Google Scholar]
- 4c. Shao J. D., Xie H. H., Huang H., Li Z. B., Sun Z. B., Xu Y. H., Xiao Q. L., Yu X. F., Zhao Y. T., Zhang H., Wang H. Y., Chu P. K., Nat. Commun. 2016, 7, 12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.W. Tao, X. B. Zhu, X. H. Yu, X. W. Zeng, Q. L. Xiao, X. D. Zhang, X. Y. Ji, X. S. Wang, J. J. Shi, H. Zhang, L. Mei, Adv. Mater 2017, DOI: https://doi.org/10.1002/adma.201603276. [DOI] [PMC free article] [PubMed]
- 6.W. Chen, J. Ouyang, H. Liu, M. Chen, K. Zeng, J. Sheng, Z. Liu, Y. Han, L. Wang, J. Li, L. Deng, Y. N. Liu, S. Guo, Adv. Mater 2017, DOI: https://doi.org/10.1002/adma.201603864. [DOI] [PubMed]
- 7.D. Yang, G. X. Yang, P. P. Yang, R. C. Lv, S. L. Gai, C. X. Li, F. He, J. Lin, Adv. Funct. Mater 2017, DOI: https://doi.org/10.1002/adfm.201700371.
- 8. Ling X., Wang H., Huang S. X., Xia F. N., Dresselhaus M. S., Proc. Natl. Acad. Sci. USA 2015, 112, 4523–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weissleder R., Nahrendorf M., Pittet M. J., Nat. Mater. 2014, 13, 125–138. [DOI] [PubMed] [Google Scholar]
- 10. Brubaker S. W., Bonham K. S., Zanoni I., Kagan J. C., Annu. Rev. Immunol. 2015, 33, 257–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng Z. J., Liang M. T., Monteiro M., Toth I., Minchin R. F., Nat. Nanotechnol. 2011, 6, 39–44. [DOI] [PubMed] [Google Scholar]
- 12.
- 12a. Karin M., Greten F. R., Nat. Rev. Immunol. 2005, 5, 749–759; [DOI] [PubMed] [Google Scholar]
- 12b. Chawla A., Nguyen K. D., Goh Y. P. S., Nat. Rev. Immunol. 2011, 11, 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.
- 13a. Walkey C. D., Chan W. C. W., Chem. Soc. Rev. 2012, 41, 2780–2799; [DOI] [PubMed] [Google Scholar]
- 13b. Yamamoto Y., Nagasaki Y., Kato Y., Sugiyama Y., Kataoka K., J. Controlled Release 2001, 77, 27–38. [DOI] [PubMed] [Google Scholar]
- 14. Zhao Y. T., Wang H. Y., Huang H., Xiao Q. L., Xu Y. H., Guo Z. N., Xie H. H., Shao J. D., Sun Z. B., Han W. J., Yu X. F., Li P. H., Chu P. K., Angew. Chem. Int. Ed. 2016, 55, 5003–5007; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 5087–5091. [Google Scholar]
- 15. Hultgren R., Gingrich N. S., Warren B. E., J. Chem. Phys. 1935, 3, 351–355. [Google Scholar]
- 16. Nel A. E., Madler L., Velegol D., Xia T., Hoek E. M. V., Somasundaran P., Klaessig F., Castranova V., Thompson M., Nat. Mater. 2009, 8, 543–557. [DOI] [PubMed] [Google Scholar]
- 17. Nel A., Xia T., Madler L., Li N., Science 2006, 311, 622–627. [DOI] [PubMed] [Google Scholar]
- 18.
- 18a. Qu G. B., Liu S. J., Zhang S. P., Wang L., Wang X. Y., Sun B. B., Yin N. Y., Gao X., Xia T., Chen J. J., Jiang G. B., ACS Nano 2013, 7, 5732–5745; [DOI] [PubMed] [Google Scholar]
- 18b. Liu Z., Davis C., Cai W. B., He L., Chen X. Y., Dai H. J., Proc. Natl. Acad. Sci. USA 2008, 105, 1410–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J., Nature 2008, 451, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bartneck M., Keul H. A., Zwadlo-Klarwasser G., Groll J., Nano Lett. 2010, 10, 59–63. [DOI] [PubMed] [Google Scholar]
- 21.
- 21a. Nel A., Xia T., Meng H., Wang X., Lin S. J., Ji Z. X., Zhang H. Y., Acc. Chem. Res. 2013, 46, 607–621; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21b. Zhu M. T., Nie G. J., Meng H., Xia T., Nel A., Zhao Y. L., Acc. Chem. Res. 2013, 46, 622–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Q. H., Chen Q., Tong Y. L., Wang J. L., Angew. Chem. Int. Ed. 2016, 55, 11437–11441; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 11609–11613. [Google Scholar]
- 23. Favron A., Gaufres E., Fossard F., Phaneuf-L'Heureux A. L., Tang N. Y. W., Levesque P. L., Loiseau A., Leonelli R., Francoeur S., Martel R., Nat. Mater. 2015, 14, 826–832. [DOI] [PubMed] [Google Scholar]
- 24.
- 24a. Scarfi S., Magnone M., Ferraris C., Pozzolini M., Benvenuto F., Benatti U., Giovine M., Respir. Res. 2009, 10, 25; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24b.X. Mu, J. Y. Wang, X. Bai, F. Xu, H. Liu, J. Yang, Y. Jing, L. Liu, X. Xue, H. Dai, Q. Liu, Y. M. Sun, C. Liu, X. D. Zhang, ACS Appl Mater Interfaces 2017, https://doi.org/10.1021/acsami.7b02900. [DOI] [PubMed]
- 25.
- 25a. Ehlken B., Berger K., Leithauser M., Koester U., Schmid C., Thalheimer M., Plesnila-Frank C., Freund M., Junghanss C., Value Health 2008, 11, A638–A638; [Google Scholar]
- 25b. Stock W., Hoffman R., Lancet 2000, 355, 1351–1357. [DOI] [PubMed] [Google Scholar]
- 26. Fox S., Leitch A. E., Duffin R., Haslett C., Rossi A. G., J. Innate Immun. 2010, 2, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Webb N. J. A., Myers C. R., Watson C. J., Bottomley M. J., Brenchley P. E. C., Cytokine 1998, 10, 254–257. [DOI] [PubMed] [Google Scholar]
- 28.
- 28a. Gospodarowicz D., Abraham J. A., Schilling J., Proc. Natl. Acad. Sci. USA 1989, 86, 7311–7315; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28b. Senger D. R., Galli S. J., Dvorak A. M., Perruzzi C. A., Harvey V. S., Dvorak H. F., Science 1983, 219, 983–985. [DOI] [PubMed] [Google Scholar]
- 29.
- 29a. King K. Y., Goodell M. A., Nat. Rev. Immunol. 2011, 11, 685–692; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29b. Furze R. C., Rankin S. M., Immunology 2008, 125, 281–288; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29c. von Vietinghoff S., Ley K., J. Immunol. 2008, 181, 5183–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lagasse E., Weissman I. L., J. Immunol. Methods 1996, 197, 139–150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


