Abstract
Background: Variations in intestinal antioxidant membrane transporters are implicated in the initiation and progression of inflammatory bowel disease (IBD). Facilitated glucose transporter member 14 (GLUT14), encoded by the solute carrier family 2 member 14 (SLC2A14) gene, is a putative transporter for dehydroascorbic acid and glucose. Although information on the gene is limited, shorter and longer GLUT14 isoforms have been identified. We hypothesized that GLUT14 mediates glucose and dehydroascorbic acid uptake. If this function could be validated, then genetic variations may associate with IBD.
Objective: This study aimed to determine the substrate(s) for the GLUT14 protein and interrogated genetic associations of SLC2A14 with IBD.
Design: The uptake of radiolabeled substrates into Xenopus laevis oocytes expressing the 2 GLUT14 isoforms was assessed. Examination of gene-targeted genetic association in the Manitoba Inflammatory Bowel Disease Cohort Study was conducted through the genotyping of single nucleotide polymorphisms (SNPs) representing linkage blocks of the SLC2A14 gene.
Results: Both GLUT14 isoforms mediated the uptake of dehydroascorbic acid and glucose into X. laevis oocytes. Three alleles in the SLC2A14 gene associated independently with IBD. The odds of having ulcerative colitis (UC) or Crohn disease (CD) were elevated in carriers of the SLC2A14 SNP rs2889504-T allele (OR: 3.60; 95% CI: 1.95, 6.64 and OR: 4.68; 95% CI: 2.78, 8.50, respectively). Similarly, the SNP rs10846086-G allele was associated with an increased risk of both UC and CD (OR: 2.91; 95% CI: 1.49, 5.68 and OR: 3.00; 95% CI: 1.55, 5.78, respectively). Moreover, the SNP rs12815313-T allele associated with increased susceptibility to CD and UC (OR: 2.12; 95% CI: 1.33, 3.36 and OR: 1.61; 95% CI: 1.01, 2.57, respectively).
Conclusion: These findings strengthen the hypothesis that genetically determined local dysregulation of dietary vitamin C or antioxidants transport contributes to IBD development. These transporter proteins are targetable by dietary interventions, opening the avenue to a precision intervention for patients of specific genotypes with IBD. This trial was registered at clinicaltrials.gov as NCT03262649.
Keywords: genetic association, SLC2A14, vitamin C, antioxidants, inflammatory bowel disease
See corresponding editorial on page 1335.
INTRODUCTION
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn disease (CD), are chronic, relapsing, immune-mediated intestinal inflammatory diseases of complex etiology (1–4), found predominantly in white people (1, 3) and incurring substantial socioeconomic costs; in excess of $1.7 billion/y in Canada (5). The exact contributions of environmental factors (6), immune dysregulations (7), and genetics (8) are unknown, which impairs therapeutic success. Current research efforts, therefore, aim to derive genetic markers to guide diagnostic and treatment decisions.
Variations in the solute carrier family 2 (SLC) genes SLC23A1 (9), SLC22A23 (10, 11), and SLC22A4/SLC22A5 (12–15), and ATP binding cassette subfamily B member 1 (ABCB1) (16–18) have been associated with IBD. They encode for intestinal membrane transporters of antioxidants, and it was hypothesized that genetically determined dysregulation of dietary antioxidants, prominently vitamin C, may contribute to IBD (9, 19, 20).
Vitamin C exists in 2 stable forms, reduced ascorbic acid and oxidized dehydroascorbic acid, each utilizing distinct pathways of transmembrane transport. The uptake of dehydroascorbic acid is mediated by selected facilitated hexose transporters (GLUT1–3, 8) (19); however, several genes in this family remain orphaned and need to be investigated in regard to function and disease association.
The primate-specific SLC2A14 gene encodes facilitated glucose transporter member 14 (GLUT14) (21), consists of 20 exons, and shows tissue-specific exon utilization in the 5′ portion (where the first 4 exons are neuron specific); and exons 5–9 are used exclusively in extraneural tissues, most prominently testis. Expression is limited to the testis, small intestine, colon, lung, ovaries, brain, skeletal muscle, heart, kidney, liver, blood, and placenta. Two major splice variants exist, and the resulting 2 protein isoforms have distinct N-termini; the longer form (L-GLUT14) has 29 amino acids and the shorter form (S-GLUT14) has 6 (22). Both isoforms locate to the cell membrane; however, no substrates are identified (21, 22). Because of its high similarity to GLUT3 it may be hypothesized that both mediate the transmembrane transport of glucose and dehydroascorbic acid (21, 22).
Genetic variations of SLC2A14 are associated with diseases of the central nervous system (23, 24), lymphatic cancer (25), rheumatoid arthritis (26), and intraocular pressure in primary open-angle glaucoma (27). These diseases are consistent with tissue expression, and by virtue of expression in the intestinal tract, it can be hypothesized that variations in SLC2A14 could be associated with IBD.
Taken together, if dehydroascorbic acid could be confirmed as a GLUT14 substrate, SLC2A14 would be a candidate gene for an association with IBD. Here, we report on both hypotheses.
METHODS
Participants
The Manitoba Inflammatory Bowel Disease Cohort Study (clinicaltrials.gov; NCT03262649), initiated in 2002, included 388 individuals drawn from a population-based registry. Participants were required to be between 18 and 80 y old and diagnosed within the previous 7 y. The diagnosis and extent of IBD was determined based on surgical, endoscopic, radiologic, and histologic data, and participating individuals were phenotyped through use of the Montreal classification (28). Controls, drawn from the general population, included healthy individuals with no personal or first-degree relatives with chronic immune diseases. Of the 388 individuals in the cohort, 311 patients with IBD (162 with CD and 149 with UC) were white and 142 age- and sex-matched healthy white controls were included in the study. More details on the study design and creation of this study population are provided elsewhere (29). The Manitoba Inflammatory Bowel Disease Cohort Study was approved by the Research Ethics Board of the University of Manitoba.
Substrate transport
As previously described elsewhere, the 2 major GLUT14 isoforms were subcloned (21). [14C]Fructose was purchased from Amersham Biosciences. Radiolabeled [14C]dehydroascorbic acid was prepared from crystalline [14C]ascorbic acid (6.6 mCi/mmol; PerkinElmer Life Sciences) (30). Total conversion of [14C]ascorbic acid to DHA was confirmed through the use of HPLC with electrocoulometric detection (31). Radiolabeled deoxy-d-glucose,2-[1,2-3H(N)] (25–50 Ci/mmol; PerkinElmer Life Sciences) was adjusted to required concentrations in the transport buffer. Previously described Xenopus laevis oocyte isolation and injection techniques were used to express the GLUT14 isoforms (32).
The transport of radiolabeled substrates was determined with groups of 10–20 oocytes in OR2 buffer at 21°C. Individual oocytes were dissolved in 500 μL 10% SDS, and internalized radioactivity was determined with scintillation spectrometry. Transport was analyzed and plotted with Microsoft Excel, and Student’s t test was used to determine statistical differences. Data are expressed as the arithmetic means ± SDs of 10–20 oocytes analyzed at each data point.
Single nucleotide polymorphism selection, genotyping, and association analyses
A haplotype-based tag-single nucleotide polymorphism (SNP) (33) approach was implemented through the use of the 8 tagging SNPs rs10846086, rs12815313, rs2889504, rs10845990, rs11612319, rs7132415, rs2376904, and rs73007730. These 7 intronic and 1 noncoding exonic SNPs (5′ untranslated region) captured all of the common variations with a minor allelic frequency >5% in SLC2A14 (Supplemental Table 1).
Genomic DNA was isolated from peripheral white blood cells by absorption onto QIamp silica gel following QIAGEN protease digestion. After column elution, the purity and concentration of DNA was determined by UV spectroscopy with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific).
Genotyping was performed on all of the subjects with TaqMan Real-Time polymerase chain reaction assays (Applied Biosystems) (Supplemental Table 1). The genotype concordance rate was 100% in duplicate samples.
Statistical analysis
All of the data were initially processed with Microsoft Excel 2010. The functional data for uptake of the radiolabels in the Xenopus oocyte system were processed in Microsoft Excel 2010, and the T.TEST function was used to interrogate the probability associated with a Student’s t test.
Genetic data were analyzed through the use of SAS version 9.2 (SAS Institute Inc.); P < 0.05 was considered statistically significant. The association of genotypes with CD or UC risks was examined by logistic regression to estimate ORs and 95% CIs, by the use of a 3-level genotypic model (2 homozygotes and 1 heterozygote). Overdominance was tested when the heterozygote was not intermediate—in effect, between the 2 homozygotes. The genotype-phenotype association for individuals with CD and UC was determined with multinomial logistic regression. Haploview 4.2 software (Broad Institute) applying default parameters was used to determine linkage disequilibrium and haplotype blocks (34).
RESULTS
GLUT14 mediates cellular dehydroascorbic acid and glucose uptake
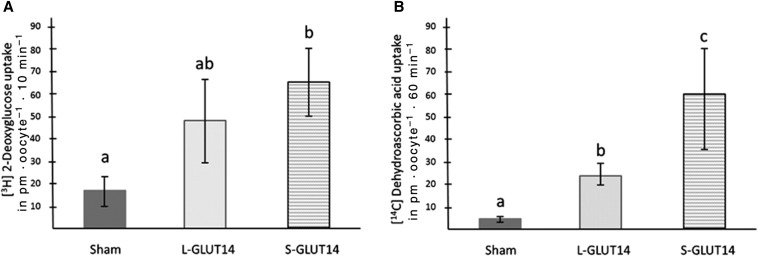
The shorter and longer GLUT14 isoforms (S-GLUT14 and L-GLUT14) locate to the plasma membrane in mammalian cells (21). On expression in X. laevis oocytes, these 2 major GLUT14 isoforms mediate the uptake of radiolabeled deoxyglucose and dehydroascorbic acid (Figure 1). Ascorbic acid and fructose did not get transported by GLUT14 (Supplemental Figure 1).
FIGURE 1.
GLUT14 isoforms mediate DHA and deoxyglucose uptake. Xenopus laevis oocytes expressing S-GLUT14 and L-GLUT14 isoforms and exhibit uptake of radiolabeled deoxyglucose (A) and dehydroascorbic acid (B). Incubations were performed with 300 μM [14C]DHA or 300 μM deoxy-d-glucose,2-[1,2-3H(N)] on 20 oocytes in each group in 3 independent experiments. Data are expressed as means ± SDs. Lowercase letters indicate statistically significant differences determined through 1-factor ANOVA, P < 0.05. GLUT14, facilitated glucose transporter 14; L-GLUT14, long GLUT14 isoform; S-GLUT, short GLUT14 isoform.
SLC2A14 SNPs independently associate with IBD
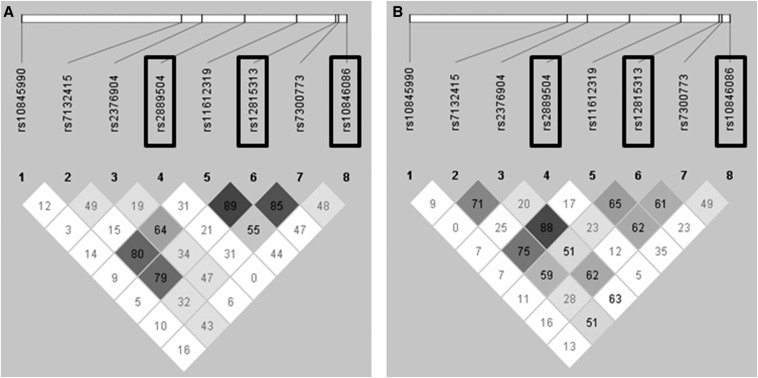
The baseline characteristics of the study participants are presented in Supplemental Table 2. Genetic variations in SNPs rs10846086, rs2889504, and rs12815313 associated with UC and CD (Table 1). No linkage was observed for the 8 tag-SNPs in the SLC2A14 gene (Figure 2), and the pattern of inheritance for the 3 SNPs associated with any disease phenotype differed substantially (Table 1).
TABLE 1.
Genetic associations of SNPs in the SLC2A14 gene to UC and CD1
| Controls, n = 142 | UC, n = 149 | UC vs. control, OR (95% CI) | CD, n = 162 | CD vs. control, OR (95% CI) | |
| rs12815313 | |||||
| CC | 73 (51.4) | 59 (39.6) | Ref | 54 (33.4) | Ref |
| CT | 55 (38.7) | 71 (47.6) | 1.59 (0.98, 2.61) | 83 (51.2) | 2.04 (1.25, 3.33)* |
| TT | 14 (9.9) | 19 (12.7) | 1.68 (0.78, 3.63) | 25 (15.4) | 2.41 (1.15, 5.07)* |
| T carrier | 69 (48.59) | 90 (60.4) | 1.61 (1.01, 2.57)* | 108 (66.7) | 2.12 (1.33, 3.36)* |
| rs10845990 | |||||
| TT | 34 (23.9) | 29 (19.5) | Ref | 32 (19.7) | Ref |
| GT | 68 (47.9) | 64 (42.9) | 1.10 (0.60, 2.01) | 76 (46.9) | 1.19 (0.66, 2.13) |
| GG | 40 (28.2) | 56 (37.6) | 1.64 (0.86, 3.11) | 54 (33.4) | 1.43 (0.76, 2.70) |
| G carrier | 108 (76.1) | 120 (80.5) | 1.30 (0.74, 2.28) | 130 (80.25) | 1.28 (0.74, 2.21) |
| rs11612319 | |||||
| GG | 66 (46.5) | 65 (43.6) | Ref | 79 (48.8) | Ref |
| GA | 66 (46.5) | 69 (46.3) | 1.06 (0.66, 1.72) | 65 (40.1) | 0.82 (0.51, 1.32) |
| AA | 10 (7.0) | 15 (10.1) | 1.52 (0.64, 3.64) | 18 (11.1) | 1.50 (0.65, 3.48) |
| A carrier | 79 (53.5) | 84 (56.4) | 1.12 (0.71, 1.78) | 83 (51.2) | 0.91 (0.58, 1.43) |
| rs7132415 | |||||
| GG | 37 (26.1) | 36 (24.2) | Ref | 31 (19.1) | Ref |
| GT | 70 (49.3) | 76 (51.0) | 1.12 (0.64, 1.96) | 72 (44.5) | 1.23 (0.69, 2.19) |
| TT | 35 (24.6) | 37 (24.8) | 1.09 (0.57, 2.08) | 59 (36.4) | 2.01 (1.07, 3.79) |
| T carrier | 105 (73.9) | 113 (75.8) | 1.11 (0.65, 1.88) | 131 (80.9) | 1.49 (0.87, 2.56) |
| rs10846086 | |||||
| AA | 128 (90.1) | 113 (75.8) | Ref | 122 (75.4) | Ref |
| AG | 9 (6.3) | 17 (11.4) | 2.14 (0.92, 4.99) | 20 (12.3) | 2.33 (1.02, 5.32)* |
| GG | 5 (3.5) | 19 (12.8) | 4.30 (1.56, 11.9)* | 20 (12.3) | 4.20 (1.53, 11.5)* |
| G carrier | 14 (9.9) | 36 (24.2) | 2.91 (1.49, 5.68)* | 40 (24.7) | 3.00 (1.55, 5.78)* |
| rs2376904 | |||||
| GG | 93 (65.5) | 83 (55.7) | Ref | 113 (69.8) | Ref |
| GA | 43 (30.3) | 58 (38.9) | 1.51 (0.92, 2.47) | 37 (22.8) | 0.70 (0.42, 1.19) |
| AA | 6 (4.2) | 8 (5.4) | 1.49 (0.50, 4.48) | 12 (7.4) | 1.65 (0.59, 4.55) |
| A carrier | 49 (34.5) | 66 (44.3) | 1.51 (0.94, 2.42) | 49 (30.2) | 0.82 (0.51, 1.33) |
| rs7300773 | |||||
| TT | 50 (35.2) | 47 (31.5) | Ref | 57 (35.2) | Ref |
| CT | 74 (52.1) | 74 (49.7) | 1.06 (0.64, 1.78) | 76 (46.9) | 0.90 (0.54, 1.48) |
| CC | 18 (12.7) | 28 (18.8) | 1.65 (0.81, 3.37) | 29 (17.9) | 1.41 (0.70, 2.85) |
| CT + CC | 92 (64.8) | 102 (68.5) | 1.18 (0.72, 1.92) | 105 (64.8) | 1.00 (0.62, 1.60) |
| rs2889504 | |||||
| GG | 125 (88.0) | 100 (67.1) | Ref | 99 (61.1) | Ref |
| GT | 7 (4.9) | 30 (20.1) | 5.36 (2.26, 12.71)* | 36 (22.2) | 6.49 (2.77, 15.2)* |
| TT | 10 (7.1) | 19 (12.8) | 2.37 (1.06, 5.34)* | 27 (16.7) | 3.41 (1.57, 7.38)* |
| T carrier | 17 (12.0) | 49 (32.9) | 3.60 (1.95, 6.64)* | 63 (38.9) | 4.68 (2.78, 8.50)* |
Values are n (%) unless otherwise indicated. Per-allele effects were derived from binary logistic regression performed with SAS version 9.2 (SAS Institute Inc.) through the use of a 3-level genotypic model (2 homozygotes and 1 heterozygote). Overdominance was tested when the heterozygote was not intermediate in effect between the 2 homozygotes. *Significant differences in the ORs to the reference SNPs. CD, Crohn disease; Ref, reference; SLC2, solute carrier family 2; SNP, single nucleotide polymorphism; UC, ulcerative colitis.
FIGURE 2.
Genetic linkage across the SLC2A14 locus in individuals with Crohn disease (A) and ulcerative colitis (B). The degree of linkage (diamonds) is given as a percentage, and the darker color indicates a higher degree of linkage. SNPs with relevant associations to any form of inflammatory bowel disease are indicated by rectangles. The single nucleotide polymorphisms associated with inflammatory bowel disease are not in genetic linkage. SLC2, solute carrier family 2; SNP, single nucleotide polymorphism.
First, the susceptibility for CD and UC was elevated in individuals carrying the SNP rs12815313-T allele (ORs: 2.12 and 1.62 respectively; Table 1), with equal impact sizes for T homozygotes and CT heterozygotes. Elevated susceptibility for CD was established for rs12815313 T homozygotes and CT heterozygotes (OR: 2.04 for CT and OR: 2.41 for TT; Table 1).
Second, the rs10846086-G allele elevated risks for UC and CD (ORs: 2.91 and 3.00, respectively; Table 1). An additive allele dosage effect was demonstrated, wherein highest susceptibility for UC and CD was observed for rs10846086-GG homozygotes (ORs: 4.30 and 4.20, respectively; Table 1), whereas the impact size was ∼50% for rs10846086-AG heterozygotes (ORs: 2.14 and 2.33, respectively; Table 1).
Third, the presence of the SNP rs2889504-T allele increased susceptibility to UC and CD (ORs: 3.60 and 4.68, respectively; Table 1). rs2889504-GT heterozygotes exhibited the highest risk for UC and CD (ORs: 5.36 and 6.49, respectively; Table 1) compared with T-allele homozygotes (ORs: 2.37, 3.41, respectively; Table 1). The rs2889504-GT heterozygosity was not overly dominant, however, indicating similar effect sizes of the T allele in both genotypes. No significant correlations were found to specific CD and UC subphenotypes as defined in the Montreal Classification (data not shown).
DISCUSSION
The presented data establish GLUT14 as a membrane transporter for glucose and dehydroascorbic acid. Moreover, 3 SLC2A14 SNPs associate independently with IBD in a well-phenotyped white cohort of moderate size, in which major genetic associations with IBD identified by a genome-wide association study had been replicated (35). All SNPs interrogated in this targeted genetic association study are noncoding and were chosen based on their ability to tag haplotype blocks in the SLC2A14 gene. This method of selection did not consider the possible impact of the SNPs on the functions of the genes or proteins, such as transcription, translation, or protein activity, but on their ability to serve as markers for disease association. In this regard, it was somewhat remarkable that 3 variations in distinct linkage blocks strongly associated with IBD (ORs 2.1–4.3), strengthening the validity of each association. This association, however, does not indicate the causality of these SNPs, which is unlikely for the intronic SNPs rs10846086 and rs2889504, which do not fall into regions of high genetic conservation or known genetic enhancers of transcription (36). Neither does SNP rs12815313 in the 5′ untranslated region (NM_001286233.1:c.-171G > A) affect any known transcription factors or enhancer binding sites (36, 37). As such, future studies to identify causal SNPs are warranted.
These findings strengthen the evidence that a local mucosal vitamin C imbalance could contribute to IBD, because previously SLC23A1, the gene encoding an intestinal trans-epithelial ascorbic acid transporter, was also associated with CD (9). Low mucosal tissue levels and lower plasma concentrations of vitamin C have been reported in individuals with IBD, even when their dietary intake was adequate (38–41), suggesting increased consumption of antioxidants during the inflammation process. Consequently, because genetic variations in the 2 intestinal vitamin C transporters genes SLC23A1 (9) and SLC2A14 are associated with IBD, we speculate that a localized vitamin C deficiency caused by decreased transmembrane transport could be a causative or contributing factor in the etiology of intestinal inflammation.
Although this study was not intended to define the underlying mechanisms in how mutations in these transporter genes lead to IBD, 2 scenarios could contribute to the weakening of the intestinal barrier function. First, variations in SLC2A14 could decrease the capacity to provide dehydroascorbic acid to the enterocytes during the inflammatory oxidative burst, where dehydroascorbic acid is produced in the extracellular environment, immediately transported into the cell and reduced to ascorbic acid (30, 42, 43). This mechanism, called ascorbate recycling or the bystander effect, greatly elevates the antioxidant capacity of the cells (44, 45), and if compromised, it could decrease the intestinal barrier function, leading to increased bacterial invasion and inflammation. Second, a decrease in vitamin C transport could affect the functioning of immune cells themselves through the redox changes described above or changes in gene expression, enhancing the severity of an existing inflammation (46–48).
In either case, the local vitamin C imbalance caused by impaired transport of ascorbic acid through SLC23A1 or dehydroascorbic acid through GLUT14 could be remedied by dietary supplementation of the complementary forms. In the future, individuals who are suitable for such interventions could be identified by their genotypes. This may be relevant for only a small number of individuals affected by IBD; however, genotype-specific dietary prevention and intervention strategies would likely be easy to implement and less costly than the current treatments.
We emphasize the fact that the underlying mechanism for the presented genetic associations ought to be validated through biological and clinical intervention studies to devise precise genotype-specific IBD intervention strategies.
Acknowledgments
The authors’ responsibilities were as follows—PE: was the principal investigator, responsible for the conception and design of the project, the acquisition of financial support, and the writing and final editing of the manuscript; MAS: conducted the genetic association studies and created the first draft of the manuscript; HZ: conducted the glucose uptake studies and added the results to the manuscript; CNB: was responsible for conducting the Manitoba Inflammatory Bowel Disease Cohort Study and collecting the samples, and contributed to revisions of the final manuscript; HE-G: was responsible for sample collection from healthy controls; HT: conducted the dehydroascorbic acid uptake experiments; GHC: consulted on the static analyses; ML: contributed to the experimental design data interpretation and manuscript writing; and all authors: reviewed, read, and approved the final manuscript. CNB has served on the advisory boards of or consulted to Abbott Canada, Abbvie Canada, Bristol-Myers Squibb, Forest Canada, Takeda Canada, Janssen Canada, Hospira, and Vertex Pharmaceuticals, and has received research grants from Abbott Canada and Abbvie Canada, and an educational grant from Aptalis. He is supported in part by the Bingham Chair in Gastroenterology. None of the authors reported a conflict of interest related to the study.
Footnotes
Abbreviations used: ABCB1, ATP binding cassette subfamily B member 1; CD, Crohn disease; GLUT14, facilitated glucose transporter member 14; IBD, inflammatory bowel disease; L-GLUT, long form of the facilitated glucose transporter; S-GLUT14, short form of the facilitated glucose transporter; SLC2A14, Solute Carrier Family 2 member 14; SNP, single nucleotide polymorphism; UC, ulcerative colitis.
REFERENCES
- 1.Green C, Elliott L, Beaudoin C, Bernstein CN. A population-based ecologic study of inflammatory bowel disease: searching for etiologic clues. Am J Epidemiol 2006;164:615–23. [DOI] [PubMed] [Google Scholar]
- 2.Hong SN, Park C, Park SJ, Lee CK, Ye BD, Kim YS, Lee S, Chae J, Kim JI, Kim YH. Deep resequencing of 131 Crohn’s disease associated genes in pooled DNA confirmed three reported variants and identified eight novel variants. Gut 2016;65:788–96. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Bernstein CN, Vatn MH, Lakatos PL, Loftus EV Jr., Tysk C, O’Morain C, Moum B, Colombel JF. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut 2013;62:630–49. [DOI] [PubMed] [Google Scholar]
- 4.Nørgård BM, Larsen PV, Fedder J, De Silva PS, Larsen MD, Friedman S. Live birth and adverse birth outcomes in women with ulcerative colitis and Crohn’s disease receiving assisted reproduction: a 20-year nationwide cohort study. Gut 2016;65:767–76. [DOI] [PubMed] [Google Scholar]
- 5.Canadian Digestive Health Foundation. Statistics [Internet]. [cited 2017 Aug 8]. Available from: http://www.cdhf.ca/en/statistics.
- 6.Torres J, Colombel J-F. Genetics and phenotypes in inflammatory bowel disease. Lancet 2016;387:98–100. [DOI] [PubMed] [Google Scholar]
- 7.Blumberg RS. Environment and genes: what is the interaction? Dig Dis 2016;34:20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleynen I, Boucher G, Jostins L, Schumm LP, Zeissig S, Ahmad T, Andersen V, Andrews JM, Annese V, Brand S, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amir Shaghaghi M, Bernstein CN, Serrano Leon A, El-Gabalawy H, Eck P. Polymorphisms in the sodium-dependent ascorbate transporter gene SLC23A1 are associated with susceptibility to Crohn disease. Am J Clin Nutr 2014;99:378–83. [DOI] [PubMed] [Google Scholar]
- 10.León AS, Shaghaghi MA, Yurkova N, Bernstein CN, El-Gabalawy H, Eck P. Single-nucleotide polymorphisms in SLC22A23 are associated with ulcerative colitis in a Canadian white cohort. Am J Clin Nutr 2014;100:289–94. [DOI] [PubMed] [Google Scholar]
- 11.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 2008;40:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakahara S, Arimura Y, Saito K, Goto A, Motoya S, Shinomura Y, Miyamoto A, Imai K. Association of SLC22A4/5 polymorphisms with steroid responsiveness of inflammatory bowel disease in Japan. Dis Colon Rectum 2008;51:598–603. [DOI] [PubMed] [Google Scholar]
- 13.Ferraris A, Torres B, Knafelz D, Barabino A, Lionetti P, de Angelis GL, Iacono G, Papadatou B, D’Amato G, Di Cionmo V, et al. Relationship between CARD15, SLC22A4/5, and DLG5 polymorphisms and early-onset inflammatory bowel diseases: an Italian multicentric study. Inflamm Bowel Dis 2006;12:355–61. [DOI] [PubMed] [Google Scholar]
- 14.Newman B, Gu X, Wintle R, Cescon D, Yazdanpanah M, Liu X, Peltekova V, Van Oene M, Amos CI, Siminovitch KA. A risk haplotype in the solute carrier family 22A4/22A5 gene cluster influences phenotypic expression of Crohn’s disease. Gastroenterology 2005;128:260–9. [DOI] [PubMed] [Google Scholar]
- 15.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet 2004;36:471–5. [DOI] [PubMed] [Google Scholar]
- 16.Andersen V, Svenningsen K, Knudsen LA, Hansen AK, Holmskov U, Stensballe A, Vogel U. Novel understanding of ABC transporters ABCB1/MDR/ P-glycoprotein, ABCC2/MRP2, and ABCG2/BCRP in colorectal pathophysiology. World J Gastroenterol 2015;21:11862–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urcelay E, Mendoza JL, Martín MC, Mas A, Martínez A, Taxonera C, Fernandez-Arquero M, Díaz-Rubio M, De La Concha EG. MDR1 gene: susceptibility in Spanish Crohn’s disease and ulcerative colitis patients. Inflamm Bowel Dis 2006;12:33–7. [DOI] [PubMed] [Google Scholar]
- 18.Brant SR, Panhuysen CIM, Nicolae D, Reddy DM, Bonen DK, Karaliukas R, Zhang L, Swanson E, Datta LW, Moran T, et al. MDR1 Ala893 polymorphism is associated with inflammatory bowel disease. Am J Hum Genet 2003;73:1282–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corpe CP, Eck P, Wang J, Al-Hasani H, Levine M. Intestinal dehydroascorbic acid (DHA) transport mediated by the facilitative sugar transporters, GLUT2 and GLUT8. J Biol Chem 2013;288:9092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padayatty SJ, Katz A, Wang Y, Eck P, Kwon O, Lee JH, Chen S, Corpe C, Dutta A, Dutta SK, et al. Vitamin C as an antioxidant: evaluation of its role in disease prevention. J Am Coll Nutr 2003;22:18–35. [DOI] [PubMed] [Google Scholar]
- 21.Amir Shaghaghi M, Murphy B, Eck P. The SLC2A14 gene: genomic locus, tissue expression, splice variants, and subcellular localization of the protein. Biochem Cell Biol 2016;94:331–5. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Freeze HH. GLUT14, a duplicon of GLUT3, is specifically expressed in testis as alternative splice forms. Genomics 2002;80:553–7. [DOI] [PubMed] [Google Scholar]
- 23.Shulman JM, Chipendo P, Chibnik LB, Aubin C, Tran D, Keenan BT, Kramer PL, Schneider JA, Bennett DA, Feany MB, et al. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am J Hum Genet 2011;88:232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Yu JT, Zhang W, Cui WZ, Wu ZC, Zhang Q, Tan L. Genetic association of SLC2A14 polymorphism with Alzheimer’s disease in a Han Chinese population. J Mol Neurosci 2012;47:481–4. [DOI] [PubMed] [Google Scholar]
- 25.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, Rahmatpanah FB, Sjahputera O, Caldwell CW. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res 2007;67:2617–25. [DOI] [PubMed] [Google Scholar]
- 26.Veal CD, Reekie KE, Lorentzen JC, Gregersen PK, Padyukov L, Brookes AJ. A 129-kb deletion on chromosome 12 confers substantial protection against rheumatoid arthritis, implicating the gene SLC2A3. Hum Mutat 2014;35:248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nag A, Venturini C, Hysi PG, Arno M, Aldecoa-Otalora Astarloa E, Macgregor S, Hewitt AW, Young TL, Mitchell P, Viswanathan AC, et al. Copy number variation at chromosome 5q21.2 is associated with intraocular pressure. Invest Ophthalmol Vis Sci 2013;54:3607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverberg MS, Satsangi J, Ahmad T, Arnott IDR, Bernstein CN, Brant SR, Caprilli R, Colombel JF, Gasche C, Geboes K, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol 2005;19:5A–36A. [DOI] [PubMed] [Google Scholar]
- 29.Ediger JP, Walker JR, Graff L, Lix L, Clara I, Rawsthorne P, Rogala L, Miller N, McPhail C, Deering K, et al. Predictors of medication adherence in inflammatory bowel disease. Am J Gastroenterol 2007;102:1417–26. [DOI] [PubMed] [Google Scholar]
- 30.Rumsey SC, Kwon O, Xu GW, Burant CF, Simpson I, Levine M. Glucose transporter isoforms GLUT1 and GLUT3 transport dehydroascorbic acid. J Biol Chem 1997;272:18982–9. [DOI] [PubMed] [Google Scholar]
- 31.Washko PW, Hartzell WO, Levine M. Ascorbic acid analysis using high-performance liquid chromatography with coulometric electrochemical detection. Anal Biochem 1989;181:276–82. [DOI] [PubMed] [Google Scholar]
- 32.Soreq H, Seidman S. Xenopus oocyte microinjection - from gene to protein. Methods Enzymol 1992;207:225–65. [DOI] [PubMed] [Google Scholar]
- 33.National Institute of Environmental Health Sciences, LD TAG SNP Selection. [Internet]. [cited 2013 Jan 5]. Available from: https://snpinfo.niehs.nih.gov/snpinfo/snptag.html.
- 34.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- 35.Ryan JD, Silverberg MS, Xu W, Graff LA, Targownik LE, Walker JR, Carr R, Clara I, Miller N, Rogala L, et al. Predicting complicated Crohn’s disease and surgery: phenotypes, genetics, serology and psychological characteristics of a population-based cohort. Aliment Pharmacol Ther 2013;38:274–83. [DOI] [PubMed] [Google Scholar]
- 36.Gao T, He B, Liu S, Zhu H, Tan K, Qian J. EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics 2016;32:3543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 2006;34:D108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendland BE, Aghdassi E, Tam C, Carrrier J, Steinhart AH, Wolman SL, Baron D, Allard JP. Lipid peroxidation and plasma antioxidant micronutrients in Crohn disease. Am J Clin Nutr 2001;74:259–64. [DOI] [PubMed] [Google Scholar]
- 39.Hoffenberg EJ, Deutsch J, Smith S, Sokol RJ. Circulating antioxidant concentrations in children with inflammatory bowel disease. Am J Clin Nutr 1997;65:1482–8. [DOI] [PubMed] [Google Scholar]
- 40.Fernandez-Banares F, Abad-lacruz A, Xiol X, Gine JJ, Dolz C, Cabre E, Esteve M, Gonzalez-Huix F, Gassull MA. Vitamin status in patients with inflammatory bowel-disease. Am J Gastroenterol 1989;84:744–8. [PubMed] [Google Scholar]
- 41.Buffinton GD, Doe WF. Altered ascorbic-acid status in the mucosa from inflammatory bowel-disease patients. Free Radic Res 1995;22:131–43. [DOI] [PubMed] [Google Scholar]
- 42.Rumsey SC, Daruwala R, Al-Hasani H, Zarnowski MJ, Simpson IA, Levine M. Dehydroascorbic acid transport by GLUT4 xenopus oocytes and isolated rat adipocytes. J Biol Chem 2000;275:28246–53. [DOI] [PubMed] [Google Scholar]
- 43.Rumsey SC, Levine M. Absorption, transport, and disposition of ascorbic acid in humans. J Nutr Biochem 1998;9:116–30. [Google Scholar]
- 44.Washko PW, Wang Y, Levine M. Ascorbic acid recycling in human neutrophils. J Biol Chem 1993;268:15531–5. [PubMed] [Google Scholar]
- 45.Nualart FJ, Rivas CI, Montecinos VP, Godoy AS, Guaiquil VH, Golde DW, Vera JC. Recycling of vitamin C by a bystander effect. J Biol Chem 2003;278:10128–33. [DOI] [PubMed] [Google Scholar]
- 46.Ki MR, Lee HR, Park JK, Hong IH, Han SY, You SY, Lee EM, Kim AY, Lee SS, Jeong KS. Ascorbate promotes carbon tetrachloride-induced hepatic injury in senescence marker protein 30-deficient mice by enhancing inflammation. J Nutr Biochem 2011;22:535–42. [DOI] [PubMed] [Google Scholar]
- 47.Chatterjee M, Saluja R, Kumar V, Jyoti A, Jain GK, Barthwal MK, Dikshit M. Ascorbate sustains neutrophil NOS expression, catalysis, and oxidative burst. Free Radic Biol Med 2008;45:1084–93. [DOI] [PubMed] [Google Scholar]
- 48.Chapple ILC, Matthews JB, Wright HJ, Scott AE, Griffiths HR, Grant MM. Ascorbate and alpha-tocopherol differentially modulate reactive oxygen species generation by neutrophils in response to Fc gamma R and TLR agonists. Innate Immun-London 2013;19:152–9. [DOI] [PubMed] [Google Scholar]