Abstract
Current knowledge regarding acute regulation of adipocyte lipolysis is largely based on receptor-mediated activation or inhibition of pathways that influence intracellular levels of cAMP, thereby affecting protein kinase A (PKA) activity. We recently identified synthetic ligands of α-β-hydrolase domain containing 5 (ABHD5) that directly activate adipose triglyceride lipase (ATGL) by dissociating ABHD5 from its inhibitory regulator, perilipin-1 (PLIN1). In the current study, we used these novel ligands to determine the direct contribution of ABHD5 to various aspects of lipolysis control in white (3T3-L1) and brown adipocytes. ABHD5 ligands stimulated adipocyte lipolysis without affecting PKA-dependent phosphorylation on consensus sites of PLIN1 or hormone-sensitive lipase (HSL). Cotreatment of adipocytes with synthetic ABHD5 ligands did not alter the potency or maximal lipolysis efficacy of the β-adrenergic receptor (ADRB) agonist isoproterenol (ISO), indicating that both target a common pool of ABHD5. Reducing ADRB/PKA signaling with insulin or desensitizing ADRB suppressed lipolysis responses to a subsequent challenge with ISO, but not to ABHD5 ligands. Lastly, despite strong treatment differences in PKA-dependent phosphorylation of HSL, we found that ligand-mediated activation of ABHD5 led to complete triglyceride hydrolysis, which predominantly involved ATGL, but also HSL. These results indicate that the overall pattern of lipolysis controlled by ABHD5 ligands is similar to that of isoproterenol, and that ABHD5 plays a central role in the regulation of adipocyte lipolysis. As lipolysis is critical for adaptive thermogenesis and in catabolic tissue remodeling, ABHD5 ligands may provide a means of activating these processes under conditions where receptor signaling is compromised.
Introduction
The hydrolysis of stored triglycerides (TG) to free fatty acids in adipose tissue is an important mechanism for supplying energy to peripheral tissues in times of demand (Duncan et al., 2007). Lipolysis is under tight homeostatic control by numerous mechanisms that affect the expression and activity of core lipolytic proteins (Duncan et al., 2007; Jaworski et al., 2007; Nielsen et al., 2014). Dysfunctions in one or more of these factors can lead to elevated levels of circulating free fatty acids (FFA), which are associated with a number of metabolic disturbances, including insulin resistance, hepatic steatosis, and dyslipidemia (Roden et al., 1996; Boden, 1998; Kahn et al., 2006; Petta et al., 2016). Thus, a thorough understanding of both extrinsic and intrinsic regulation of lipolysis is of great interest for treating metabolic disease.
The current understanding of post-translational regulation of adipocyte lipolysis is largely based on acute regulation by activators and inhibitors of the protein kinase A (PKA) signaling pathway. In this model, extracellular signals including hormones and catecholamines bind surface G-protein receptors that couple to the generation, inhibition, or degradation of cAMP, a direct activator of PKA (Carmen and Victor, 2006; Granneman and Moore, 2008; Nielsen et al., 2014). Mechanistically, lipolysis is initiated when PKA phosphorylates a protein complex containing the lipid droplet scaffold protein perilipin-1 (PLIN1) and α-β-hydrolase domain containing 5 (ABHD5, also known as CGI-58), a critical activator of adipose triglyceride lipase (ATGL) (Lass et al., 2006; Schweiger et al., 2006). In the current model, PKA-dependent phosphorylation of PLIN1 releases ABHD5, thereby allowing activation of ATGL, the first and rate-limiting enzyme in TG hydrolysis (Zimmermann et al., 2004; Granneman et al., 2007, 2009a). PKA also phosphorylates hormone-sensitive lipase (HSL), which then translocates to the lipid droplet surface where it preferentially hydrolyzes diglyceride (DG) substrates (Egan et al., 1992; Clifford et al., 2000; Holm, 2003; Sztalryd et al., 2003). Thus, the current model postulates that PKA-dependent activation of ATGL and HSL comprises the central mechanism for regulating lipolysis by extracellular signals.
Our laboratories recently completed a high-throughput screen for compounds that activate lipolysis by directly dissociating complexes of ABHD5 with PLIN1 and PLIN5 (Sanders et al., 2015). We discovered two structurally distinct chemical scaffolds represented by the compounds SR-4995 [1-phenylpropyl-3-(10-methyl-11-oxo-10,11-dihydrodibenzo [b,f][1,4]thiazepin-8-yl)urea] and SR-4559 [1-(4-((2,3-dihydrobenzo[b][1,4]dioxin-6-yl)sulfonyl) piperazin-1-yl)-2-(6-methylbenzofuran-3-yl)ethanone] that directly bind ABHD5 and stimulate lipolysis by releasing it from PLIN, thereby mimicking the effects of extracellular signaling in the absence of PKA-dependent phosphorylation of HSL and PLIN. Subsequent work indicated that ABHD5 contains an allosteric binding site that regulates its interactions with PLIN proteins and provides a mechanism of lipolysis control that can be independent of extracellular signaling (Sanders et al., 2015).
In the present study, we used these novel synthetic ABHD5 ligands as well as a new structural analog of the thiaza-tricyclo-urea compound, SR-4995, to probe the integration of lipolysis controlled by canonical β-adrenergic receptors (ADRBs) and direct activators of ABHD5 in differentiated white (3T3-L1) and brown adipocytes (BA), as well as in adipose tissue explants. We further evaluated the relative involvement of HSL and ATGL in mediating ABHD5-dependent lipolysis using pharmacological inhibitors. Findings from this study provide evidence that direct activation of ABHD5 can lead to complete TG hydrolysis and lend further support for the central role of ABHD5-PLIN interactions in the control of basal and stimulated lipase activity.
Materials and Methods
Chemicals and Reagents.
Dulbecco's modified Eagle’s medium (DMEM) and phosphate-buffered saline (PBS) were purchased from HyClone (GE Lifesciences, Pittsburgh, PA); fetal bovine serum from Atlantis Biologicals (Flowery Branch, GA); indomethacin and cell culture supplements from Thermo Fisher Scientific (Grand Island, NY); and isoproterenol, Krebs-Ringer bicarbonate buffer (KRBB), 3-isobutyl-1-methylxanthine (IBMX), dexamethasone, free glycerol reagent, 3,3′,5-triiodo- l-thyronine (T3), and bovine insulin from Sigma-Aldrich (St. Louis, MO). Antibodies to detect phosphorylated HSL (Ser-563) and total HSL were purchased from Cell Signaling (Danvers, MA), a PLIN1 antibody was purchased from Everest Biotech (Ramona, CA), phospho-specific PLIN1 antibody (Ser-522) was from Vala Sciences (San Diego, CA), and secondary horseradish peroxidase antibodies were from Jackson Laboratories (Bar Harbor, ME).
The murine adipose triglyceride lipase inhibitor Atglistatin (N′-[4′-(dimethylamino)[1,1′-biphenyl]-3-yl]-N,N-dimethyl-urea) was purchased from Cayman Chemical (Ann Arbor, MI), and 4-isopropyl-3-methyl-2-(1-[3-(S)-methyl-piperidin-1-yl]-methanoyl)-2H-isoxalo-5-one (BAY 59-9435; BAY), a selective inhibitor of HSL (Lowe et al., 2004; Claus et al., 2005), was provided by Bayer (Whippany, NJ). The ABHD5 ligands SR-4995, SR-4559, and SR-3420 [1-(3-(3,5-bis(trifluoromethyl)phenyl)propyl)-3-(10-methyl-11-oxo-10,11-dihydrodibenzo[b,f][1,4] thiazepin-8-yl)urea] were synthesized at Scripps Research Institute (Jupiter, FL). SR-4995, a thiaza-tricyclo-urea, and SR-4559, a member of the sulfonyl piperazine class of compounds, have been described previously (Sanders et al., 2015). SR-3420 is a structural analog of SR-4995 and was designed to increase the lipolysis efficacy of ABHD5. The chemical structures for all three compounds are provided in Fig. 1, C–E. Additional sources of reagents are specified within the following text.
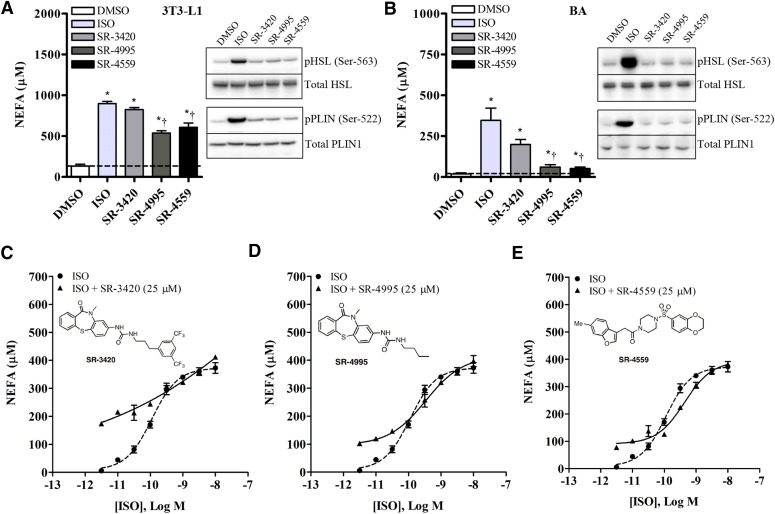
Fig. 1.
Isoproterenol and ABHD5 ligands (SR-3420, SR-4995, and SR-4559) differentially activate PKA-dependent phosphorylation of HSL and PLIN1, but both target a common pool of ABHD5. Differentiated 3T3-L1 (A) and BA cells (B) were incubated in KRBB medium containing 1% FF-BSA and one of the following treatments: DMSO (0.1%), ISO [10 nM (BA) or 10 µM (3T3-L1)], or an ABHD5 ligand (20 µM) for 1 hour. Relative levels of phospho-HSL (pHSL; Ser-563) and phoso-PLIN1 (pPLIN; Ser-522) were evaluated in whole-cell lysates by Western blotting and NEFA quantified in the culture medium. Shown are representative images from at least three independent experiments. Data are presented as the mean ± S.E.M. *Significantly different from vehicle (DMSO)-treated controls (P < 0.05). †Significantly different from isoproterenol-treated cells (P < 0.05). (C–E) Differentiated BA cells were incubated in KRBB medium containing 1% FF-BSA and 25 μM of an ABHD5 ligand in the presence or absence of increasing concentrations of ISO (0.003–10 nM). NEFA were quantified in the culture medium after 1 hour. As shown, the presence of submaximal levels of the ABHD5 ligands SR-3420 (C), SR-4995 (D), or SR-4559 (E) had no discernible effect on the potency or maximal lipolysis efficacy induced by ISO.
3T3-L1 Cell Culture and Differentiation.
3T3-L1 preadipocytes were purchased from the American Type Culture Collection (Manassas, VA) and maintained in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (growth medium). For each experiment, cells were subcultured using 0.25% trypsin-EDTA and seeded onto tissue culture plates. Forty-eight hours after reaching confluency, differentiation was initiated by treating cells with growth medium containing 1 µg/ml insulin, 0.5 mM IBMX, and 1 µM dexamethasone. At 2 and 4 days postinduction, medium was replaced with growth medium containing 1 µg/ml insulin, and on day 6 with growth medium alone. Unless otherwise specified, treatments were initiated 8 days postinduction.
BA Culture and Differentiation.
A brown preadipocyte cell line was generated as previously described (Klein et al., 1999; Mottillo et al., 2016) and provided by the laboratory of Dr. Gregory Steinberg (McMaster University, Hamilton, ON, Canada). In brief, preadipocytes were isolated from the interscapular brown adipose tissue of 3–4-day-old FVB/NJ female pups using collagenase digestion. Isolated cells were then cultured and immortalized by infection with the SV40 large T-antigen, followed by selection with puromycin. Preadipocytes were cultured and maintained in DMEM growth medium as described earlier. To induce differentiation, cells were trypsinized and then subcultured onto plates in growth medium supplemented with 1 nM T3 and 20 nM insulin (differentiation medium). Upon reaching confluency, preadipocytes were treated in differentiation medium containing 5 µM dexamethasone, 0.5 mM IBMX, and 0.125 mM indomethacin (induction medium) for 48 hours. Thereafter, cultures were maintained in differentiation medium, and treatments were initiated 7 days postinduction.
Cell Treatments and Lipolysis Assays.
To evaluate the effect of ABHD5 compounds on lipolysis, 3T3-L1 and BA cells were cultured and differentiated as described earlier. At 7 or 8 days postinduction, cells were washed once in PBS, then incubated in KRBB medium containing 1% fatty acid–free bovine serum albumin (FF-BSA; Gemini Bio-Products, West Sacramento, CA) and one of the following treatments: dimethylsulfoxide (DMSO; 0.2%), isoproterenol [ISO; 10 nM (BA) or 10µM (3T3-L1)], or 20 µM of an ABHD5 ligand (SR-4995, SR-4559, or SR-3420) either alone or in the presence of Atglistatin (10 μM), BAY (5 μM), or Atglistatin (10 µM) and BAY (5 µM). The concentrations of Atglistatin and BAY were selected based on previous dose-response studies and demonstrated to inhibit both basal and stimulated fatty acid release (Claus et al., 2005; Mayer et al., 2013). At 1 and 3 hours following the initial treatment, an aliquot of culture medium was collected for determination of nonesterified free fatty acids (NEFAs) and glycerol.
To evaluate the effect of insulin pretreatment on compound-mediated lipolysis, differentiated 3T3-L1 and BA cells were washed once in PBS and incubated in serum-free DMEM for 2 hours. Cells were washed again in PBS and then preincubated in KRBB medium containing 1% FF-BSA either alone or with 100 nM insulin for 30 minutes, after which DMSO, isoproterenol, or ABHD5 ligands were added directly to the medium at concentrations indicated in the individual figure legends. The effect of β-adrenergic receptor prestimulation on compound-mediated lipolysis was also assessed. In brief, 100 nM isoproterenol or vehicle control was added directly to wells containing DMEM. Following a preincubation period of 2.5 (3T3-L1) or 3 hours (BA), cells were washed three times in PBS, and then treated with DMSO (0.1%), ISO [10 (3T3-L1) or 3 nM (BA)], or ABHD5 ligands (20 µM) diluted in KRBB medium containing 1% FF-BSA. For both procedures, an aliquot of the culture medium was collected for NEFA determination 1 hour following the initial treatment.
Nonesterified fatty acids were quantified in the medium using a NEFA assay kit (Wako Diagnostics, Mountain View, CA) and the fluorogenic substrate Amplex Red (Cayman Chemical). Fluorescence intensities were quantified using a Clariostar plate reader (BMG LabTech, Ortenberg, Germany) with an excitation/emission maximum of 545/600 nm. Glycerol release was estimated in the culture medium using free glycerol reagent (Sigma-Aldrich) according to the manufacturers’ instructions.
Organ Culture of Mouse Adipose Tissues.
Male (n = 7) and female (n = 4) C57BL/6J mice (8–12 weeks of age) were obtained from a breeding colony maintained in an Association for Assessment and Accreditation of Laboratory Animal Care–approved animal facility at Wayne State University (Detroit, MI). Animals were weaned onto a regular chow diet (Envigo, Indianapolis, IN) and allowed free access to food and water. Procedures were performed according to the regulatory guidelines of the Division of Laboratory Animal Resources at Wayne State University. In brief, to evaluate the effects of ABHD5 ligands on lipolysis in white and brown adipose tissue ex vivo, nonfasted animals were euthanized by CO2 asphyxiation followed by cervical dislocation. The gonadal white adipose tissue (gWAT) and interscapular brown adipose tissue (iBAT) were then removed surgically, carefully trimmed, and rinsed several times in PBS.
Tissues were further sectioned (∼25 mg of gWAT or ∼10 mg of iBAT), placed into individual wells of tissue culture plates, and incubated in KRBB medium containing 1% FF-BSA and 0.1% DMSO, 10 µM ISO, or 20 µM of an ABHD5 ligand (SR-4995, SR-4559, or SR-3420) for 20 minutes at 37°C. Following this preincubation period, medium was removed and replaced with fresh medium containing identical treatments. Tissues were further incubated for a period of 3 hours at 37°C with gentle agitation every 20 minutes. Aliquots of the medium were collected at 1 and 3 hours for NEFA and glycerol determination, and tissue weights were measured after 3 hours for normalization of values. Results are expressed as nmol NEFA or glycerol per milligram of tissue. For each animal, individual tissue sections obtained from gWAT and iBAT were distributed equally across all experimental treatments.
Western Blotting.
3T3-L1 and BA cells were subcultured and differentiated onto six-well plates as described earlier. Seven (BA) or 8 days (3T3-L1) following induction of preadipocyte differentiation, cells were washed with PBS, and medium was replaced with KRBB containing 1% FF-BSA and one of the following treatments: DMSO (0.1%), ISO [10 nM (BA) or 10 µM (3T3-L1)], or an ABHD5 ligand (20 µM). One hour following the initial treatment, cells were washed once with PBS, then scraped and collected in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris, 150 mM NaCl, 0.2% sodium dodecyl sulfate, 0.25% sodium deoxycholate, 1% Triton X-100, 1 mM EDTA) containing Halt protease and phosphatase inhibitors (Thermo Fisher Scientific). Cells were sonicated briefly, and lysates were clarified by centrifugation at 13,000g for 15 minutes at 4°C. Protein concentrations were determined in the lysates using the bicinchoninic acid assay (Thermo Fisher), and immunoblotting was performed using Nusep gradient gels (VWR, Radnor, PA), as described previously (Rondini et al., 2014). Blots were then probed with antibodies to detect phospho-HSL (Ser-563), total HSL, phospho-specific PLIN1 (Ser-522), or total PLIN1 diluted in Tris-buffered saline containing 5% FF-BSA. Following an overnight incubation, immunoreactive bands were detected using peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Thermo Fisher Scientific), and images were captured with a digital imaging system (Azure Biosystems, Dublin, CA). For each experiment, an aliquot of the medium was saved for NEFA determination.
Statistical Analysis.
Statistical analyses were conducted using SigmaPlot software (version 10.0; Systat Software, Inc., Point Richmond, CA). Lipolysis data were analyzed using a one- or two-way analysis of variance, and when significant differences were detected (P < 0.05), individual comparisons were made using the Student-Newman-Keuls test. Where appropriate, data were log-transformed prior to analysis to fit the assumptions of analysis of variance. Percentage inhibition was analyzed using the Kruskal-Wallis test, and individual comparisons were made using the Student-Newman-Keuls test. All results are presented as the mean ± S.E.M.
Results
ADRB and ABHD5 Converge on a Common Mechanism for TG Hydrolysis.
To determine whether ABHD5 ligands stimulate lipolysis independently of cAMP/PKA signaling, differentiated 3T3-L1 adipocytes (a model of white adipocytes) and differentiated immortalized BA were treated with vehicle, isoproterenol [10 nM (BA) or 10 µM (3T3-L1)], or 20 µM of an ABHD5 ligand for 1 hour, after which medium was collected for NEFA determination, and cell lysates were probed for PKA-dependent phosphorylation of PLIN1 (Ser-522) and HSL (Ser-563). As shown in Fig. 1, A and B, ISO and ABHD5 ligands stimulated NEFA release from 3T3-L1 and BA cells at 1 hour, with ISO and SR-3420 being more potent inducers of lipolysis than SR-4995 or SR-4559. However, unlike the strong PKA-dependent phosphorylation observed following ISO stimulation in both cell types, the ABHD5 ligands SR-4995, SR-4559, and SR-3420 did not promote phosphorylation of PLIN1 or HSL (Fig. 1B). These observations corroborate previous findings with SR-4995 and SR-4559 and further indicate that the more effective ABHD5 ligand SR-3420 also stimulates lipolysis through a mechanism different from that of ISO, as further supported by the following experiments.
We next performed concentration-response analyses for ISO and the three ABHD5 ligands, representing two distinct structures (thiaza-tricyclo-ureas and sulfonyl piperazines) and levels of intrinsic activity. Dose-response curves for the ABHD5 ligands are presented in Supplemental Fig. 1. SR-3420 was found to be a more potent and effective agonist, eliciting maximal lipolysis responses that were 2- to 5-fold higher than those observed with either SR-4995 or SR-4559. We next performed experiments with the respective extracellular (ISO) activator in the absence and presence of intracellular activators (ABHD5). As shown in Fig. 1, C–E, the presence of submaximal concentrations of ABHD5 ligands had no discernible effect on the potency or maximal lipolysis efficacy of isoproterenol. Importantly, nearly saturating concentrations of the “partial agonists” SR-4995 and SR-4559 did not reduce maximal stimulation induced by isoproterenol (Fig. 1, C–E). These data support a mechanism whereby ABHD5 ligands act to dissociate ABHD5/PLIN1 complexes but do not themselves directly affect the ability of dissociated ABHD5 to activate ATGL (Sanders et al., 2015). Further, our data indicate that isoproterenol and synthetic ABHD5 ligands activate lipolysis by targeting a common pool of ABHD5.
Ligand-Mediated Activation of ABHD5 Leads to Complete TG Hydrolysis and Involves Both ATGL and HSL.
Triglyceride lipolysis is thought to be mediated by the combined action of ATGL and HSL, which comprise >90% of TG activity in adipose tissue (Schweiger et al., 2006). ATGL exhibits high specificity for TG and, therefore, is thought to be the first and rate-limiting enzyme in TG hydrolysis (Zimmermann et al., 2004). Although HSL can hydrolyze both TG and DG, the activity toward the former is approximately 11-fold lower than that for DG substrates (Osterlund et al., 1996; Holm, 2003). Importantly, each of these steps is thought to require PKA-dependent phosphorylation of PLIN1 and HSL, respectively (Granneman and Moore, 2008). Based on this model, it would be predicted that ABHD5 ligands would only activate ATGL, and thus generate a fraction of the NEFA released by isoproterenol and no glycerol, which requires PKA-dependent HSL activation.
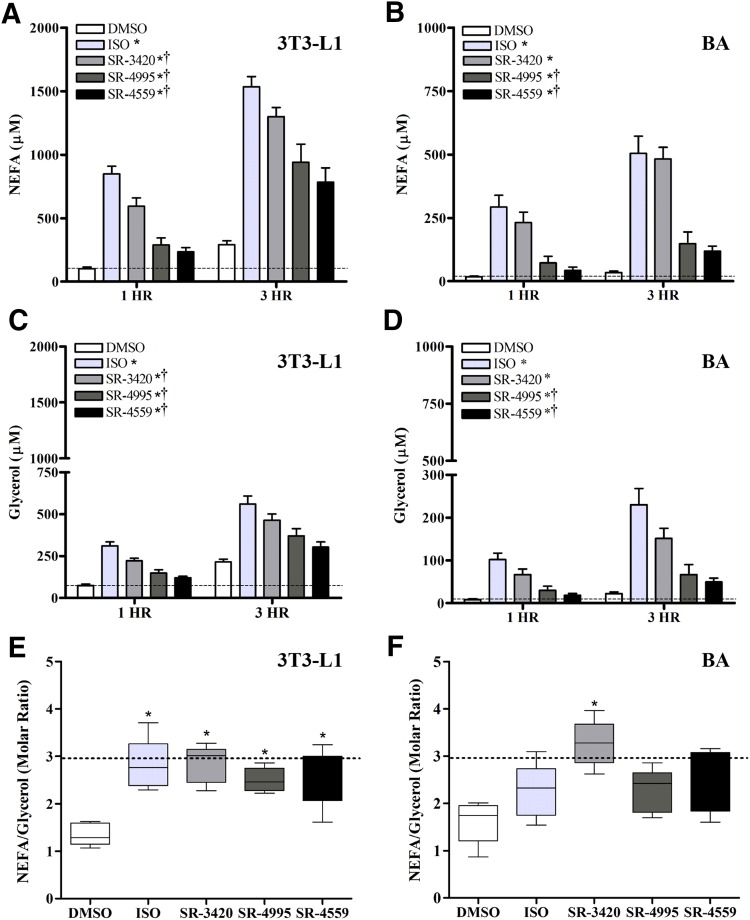
We therefore evaluated the effect of ISO and ABHD5 ligands on NEFA and glycerol release in differentiated 3T3-L1 and BA cells stimulated in culture for 3 hours, and the results are presented in Fig. 2. We found that ABHD5 ligands generated significant amounts of NEFA and glycerol in 3T3-L1 adipocytes (Fig. 2, A and C) as well as BA cells (Fig. 2, B and D) in a manner that was highly similar to that produced by isoproterenol. In general, both basal and stimulated lipolysis were higher in 3T3-L1 cells, and further increased by ∼2.5- and ∼2-fold in 3T3-L1 and BA cells, respectively, after 3 hours of stimulation (Fig. 2, A and B). Complete hydrolysis of triglyceride would theoretically generate 3 mol of FFA per mole of glycerol in the absence of significant re-esterification. In the case of 3T3-L1 cells, activation of lipolysis by isoproterenol or ABHD5 ligands resulted in the expected molar ratio of 3:1 (Fig. 2E). In BA, the ABHD5 ligands SR-3420 and SR-4559 also elevated the FFA/glycerol ratio compared with vehicle control, whereas isoproterenol and SR-4995 were somewhat less effective (Fig. 2F).
Fig. 2.
Time course of NEFA and glycerol release by ISO and ABHD ligands in 3T3-L1 and BA cells. Differentiated 3T3-L1 (A, C, and E) and BA cells (B, D, and F) were incubated in KRBB medium containing 1% FF-BSA and one of the following treatments: DMSO (0.2%), isoproterenol [10 nM (BA) or 10 µM (3T3-L1)], or an ABHD5 ligand (20 µM) for 3 hours, as described in Materials and Methods. Free fatty acids and glycerol were then measured in an aliquot of the culture medium after 1 and 3 hours. Each bar represents the mean ± S.E.M. Shown are representative results from five independent experiments per cell line. For both cell lines, there were significant differences detected among treatments and across time on both NEFA and glycerol levels (P < 0.05), but no significant interaction. In general, NEFA and glycerol levels were significantly higher at 3 hours compared with 1 hour (P < 0.05). Symbols denote statistically significant differences that were detected among the treatment groups (independent of time): *significantly different compared with DMSO controls (P < 0.05); †significantly different compared with isoproterenol (P < 0.05).
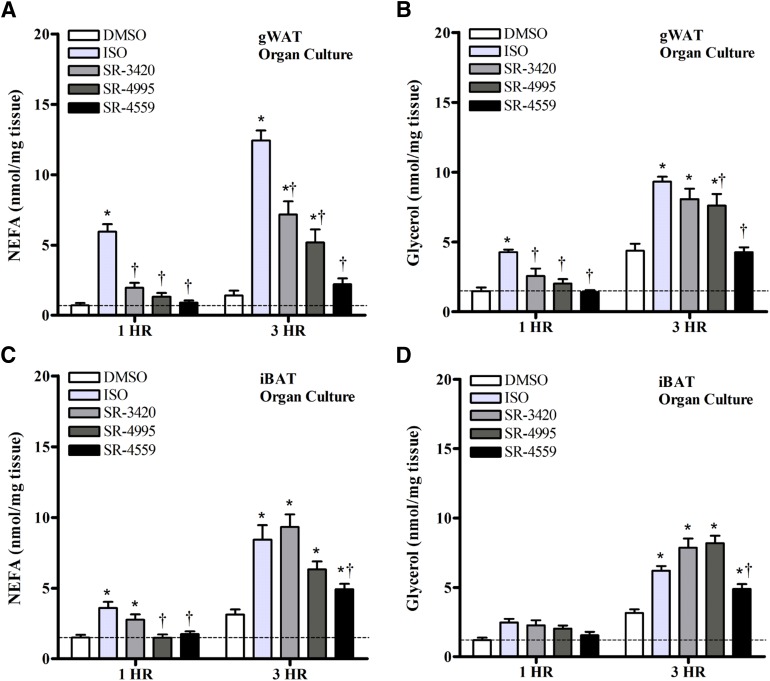
To further corroborate findings from our in vitro cellular models, we tested the effects of ISO and ABHD5 ligands on lipolysis in explants of gWAT and iBAT isolated from male (n = 7) and female (n = 4) mice (Fig. 3). In general, gWAT cultures from females tended to have lower basal NEFA release than males; however, overall responses to treatments were similar between sexes, and therefore, results were combined for analysis. There was no effect of sex on NEFA levels in iBAT cultures or on glycerol release from either tissue (data not shown). As shown in Fig. 3, both isoproterenol and ABHD5 ligands stimulated NEFA release in a manner similar to that observed in cultured cells (Fig. 3, A and C), with ISO responses generally stronger than ABHD5 ligands in gWAT. Glycerol measurements closely paralleled the changes observed with NEFA, with ISO, SR-3420, and SR-4995 being more potent in stimulating glycerol release than SR-4559 (Fig. 3, B and D). The molar ratio of FFA to glycerol observed in organ culture was closer to 1:1 for all treatments, suggesting a more efficient re-esterification and/or metabolism of FFA in tissues compared with that observed in cultured cells. Nonetheless, findings of complete TG hydrolysis as evidenced by an increase in glycerol further support the results from cultured adipocytes.
Fig. 3.
ABHD5 ligands stimulate NEFA and glycerol release in organ cultures of gWAT and iBAT. Adipose tissue sections were isolated from the gWAT (A and B) and iBAT (C and D) of nonfasted C57BL/6J mice [n = 11 (seven males, four females); 8–12 weeks of age]. Tissues were then incubated in KRBB medium containing 1% FF-BSA and one of the following treatments: DMSO (0.1%), isoproterenol (10 µM), or an ABHD5 ligand (20 µM) at 37°C for 3 hours, as described in Materials and Methods. Free fatty acids and glycerol were measured in an aliquot of the culture medium after 1 and 3 hours, and tissue weights were recorded upon termination of the experiment. Results are expressed as nanomoles per milligram of tissue. Each bar represents the mean ± S.E.M. For both tissue types, there was a significant treatment × time interaction on NEFA and glycerol levels (P < 0.05), with levels generally higher at 3 hours compared with 1 hour. Symbols denote significant differences detected among treatments within each time point: *significantly different compared with time-matched DMSO controls (P < 0.05); †significantly different compared with time-matched ISO-treated cells (P < 0.05).
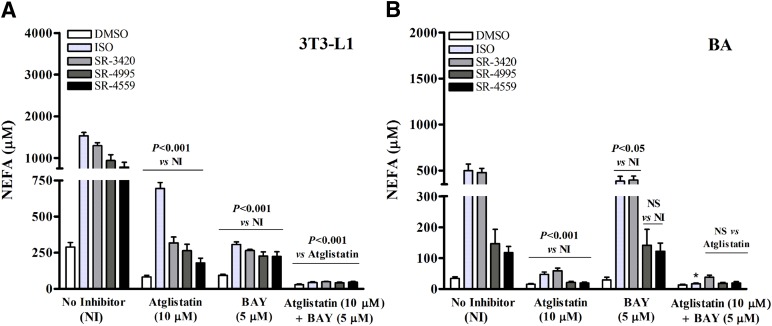
Because ATGL is thought to be a specific TG lipase (i.e., lacking DG or monoglyceride lipase activity), observations from our in vitro and ex vivo studies indicate that additional lipases may contribute to ABHD5-mediated release of NEFA and glycerol. To investigate this possibility further, we used selective pharmacological inhibitors of ATGL (Atglistatin) and HSL (BAY) and further evaluated the effect of ABHD5 ligands on NEFA release in 3T3-L1 and BA cells (Fig. 4; Table 1). As expected, the combined inhibition of ATGL and HSL virtually eliminated NEFA efflux induced by ISO and ABHD5 ligands in both 3T3-L1 and BA cells (control values were redrawn from Fig. 2, which was performed as part of the same experiment). BAY alone was sufficient to strongly inhibit NEFA efflux from 3T3-L1 cells, and this effect was indistinguishable between ISO and ABHD5 ligands (Fig. 4A). Interestingly, whereas Atglistatin strongly suppressed ABHD5 ligand–induced lipolysis (70%–77%) after 3 hours, it was significantly less effective (54%) in suppressing that induced by ISO (Fig. 4A; Table 1). The Atglistatin-resistant component of NEFA release was fully suppressed by BAY, indicating that HSL contributes significantly to TG hydrolysis following PKA activation, and similarly, that basal HSL activity is sufficient to hydrolyze DG generated by selective ABHD5/ATGL activation. Further, under conditions where basal or stimulated HSL activity was suppressed (i.e., in the presence of BAY), synthetic ABHD5 ligands were as effective as ISO in inducing ATGL-dependent lipolysis. In BA cells, lipolysis was generally more sensitive to ATGL inhibition compared with that observed in 3T3-L1 cells and significantly less sensitive to HSL inhibition (Fig. 4B). We note that 3T3-L1 adipocytes (but not BA cells) exhibited a significant component (17%–25%) of compound-induced lipolysis that was resistant to Atglistatin, yet sensitive to BAY, indicating HSL-dependent, ATGL-independent stimulation of lipolysis by ABHD5. Nonetheless, the overall pattern of NEFA release during selective lipase inhibition was highly similar between ISO and ABHD5 ligands, especially for SR-3420. Together, these results indicate that ATGL and HSL are critical lipases for ISO- and ABHD5 ligand–induced activation, and that the relative contribution of these lipases is remarkably similar for a given cell type.
Fig. 4.
Effect of ATGL and HSL inhibitors on lipolysis mediated by isoproterenol and ABHD5 ligands. Differentiated 3T3-L1 (A) and BA cells (B) were incubated in KRBB medium containing 1% FF-BSA and one of the following treatments: DMSO (0.2%), isoproterenol [10 nM (BA) or 10 µM (3T3-L1)], or an ABHD5 ligand (20 µM) either alone or in the presence of Atglistatin (10 μM), BAY (5 μM), or Atglistatin (10 µM) and BAY (5 µM). Free fatty acids were then estimated in an aliquot of the culture medium at 3 hours following the initial treatment. Shown are representative results from five independent experiments per cell line. Control values are redrawn from Fig. 2, which was performed as part of the same experiment. (A) In 3T3-L1 cells, the presence of Atglistatin or BAY significantly reduced NEFA levels from treatment-matched control values, and the combination of Atglistatin and BAY significantly reduced levels compared with controls and that of Atglistatin alone (P < 0.001). (B) In BA cells, Atglistatin significantly reduced NEFA levels compared with treatment-matched controls not containing inhibitors (P < 0.001), and BAY reduced NEFA levels in ISO- and SR-3420–treated cells compared with treatment-matched controls. *The combination of inhibitors significantly reduced NEFA levels in the ISO-treated group only when compared with Atglistatin-treated cells.
TABLE 1.
Effect of ATGL and HSL inhibitors on NEFA release in 3T3-L1 and BA cells 3 hours following incubation with isoproterenol or ABHD5 ligands.
All values are expressed as a percentage of time-matched treatments without inhibitors.
| Compound | Atglistatin (10 μM) | BAY (5 μM) | Atglistatin (10 μM) + BAY (5 μM) | |||
|---|---|---|---|---|---|---|
| % NEFA Inhibition (3T3-L1) | % NEFA Inhibition (BA) | % NEFA Inhibition (3T3-L1) | % NEFA Inhibition (BA) | % NEFA Inhibition (3T3-L1) | % NEFA Inhibition (BA) | |
| DMSO | 72 ± 1.7b | 51 ± 8.3a | 67 ± 2.9 | 19 ± 11 | 90 ± 2.1a | 58 ± 6.2a |
| ISO | 54 ± 4.9a | 90 ± 2.1b | 80 ± 1.6 | 23 ± 3.9 | 97 ± 0.37c | 96 ± 0.92c |
| SR-3420 | 75 ± 3.7b | 87 ± 2.1b | 79 ± 1.7 | 18 ± 2.2 | 96 ± 0.22b | 92 ± 1.4b |
| SR-4995 | 70 ± 7.1b | 80 ± 5.5b | 73 ± 6.6 | 9.2 ± 5.2 | 95 ± 1.1b | 80 ± 8.0b |
| SR-4559 | 77 ± 2.0b | 83 ± 2.4b | 71 ± 0.59 | 0.92 ± 5.6 | 94 ± 1.2b | 83 ± 4.2b |
Letters denote significant differences in NEFA levels among treatments within each cell line and inhibitor class (P < 0.05).
Insulin Differentially Inhibits Lipolysis Mediated by cAMP and ABHD5 Ligands in 3T3-L1 and BA Cells.
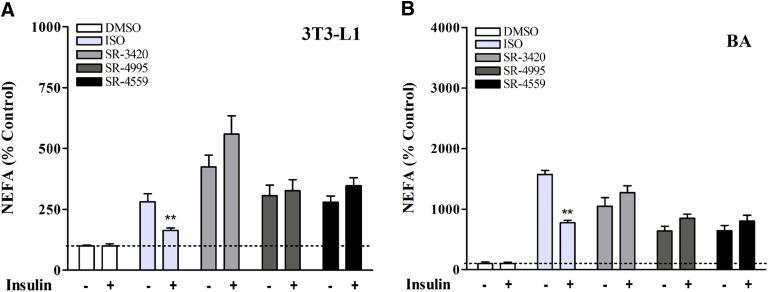
Adipocyte lipolysis is under inhibitory control by insulin receptor signaling, and it is thought that this suppression is mediated largely, if not exclusively, by phosphodiesterase 3B, which degrades cAMP, thereby suppressing PKA signaling (Lonnroth and Smith, 1986; Degerman et al., 1998). The availability of ABHD5 ligands that activate lipolysis downstream of PKA allowed us to probe the insulin signaling pathway for possible cAMP-independent regulation. Insulin pretreatment suppressed basal fatty acid efflux in 3T3-L1 cells by 42% (P < 0.05, data not shown) but did not affect basal lipolysis in BA cells. To account for differential effects of insulin on basal lipolysis between cell lines and to allow comparison among activators, results from both cell lines were normalized to the mean of pretreatment control levels, and values are expressed as a percentage of these controls. As shown in Fig. 5, insulin sharply suppressed activation of lipolysis by isoproterenol in both 3T3-L1 (Fig. 5A) and BA cells (Fig. 5B) (P < 0.01). In contrast, insulin failed to suppress lipolysis stimulated by synthetic ABHD5 ligands. These data demonstrate that insulin signaling does not acutely impact lipolysis distal to PLIN1 phosphorylation, and further imply that ABHD5 ligands regulate lipolysis through a mechanism independent of cAMP-mediated signaling.
Fig. 5.
Insulin pretreatment differentially affects lipolysis mediated by ISO and ABHD5 ligands. (A and B) Differentiated adipocytes were washed with PBS and incubated in serum-free DMEM. After 2 hours, cells were washed again in PBS and further preincubated in KRBB medium containing 1% FF-BSA with or without 100 nM insulin for 30 minutes, after which treatments [isoproterenol, 1 (3T3-L1 cells), 10 nM (BA cells); or ABHD5 ligands, 20 µM] were added directly to the culture medium. NEFAs were then measured in the medium after 1 hour. Shown are results from four to five independent experiments. Insulin pretreatment reduced basal lipolysis in 3T3-L1 cells (P < 0.05), but not in BA cells (data not shown). Lipolysis results from both cell lines were therefore normalized to the mean of pretreatment-matched controls to further differentiate individual treatment responses to insulin. Values are expressed as a percentage of controls (controls are 100%). As shown, insulin suppressed lipolysis stimulated by submaximal concentrations of ISO in both 3T3-L1 (A) and BA (B). In contrast, insulin failed to suppress lipolysis stimulated by synthetic ABHD5 ligands. **Significant effect of insulin on NEFA levels within the ISO-treated group (P < 0.01).
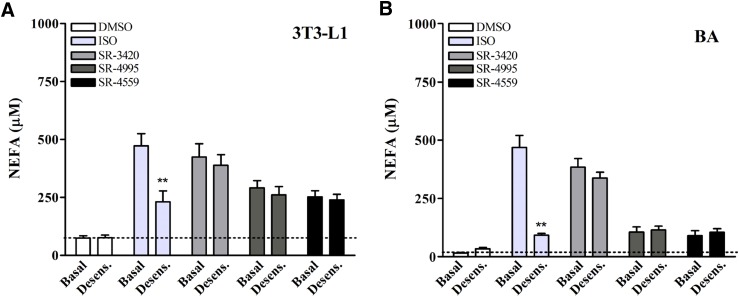
Lipolysis Induced by ABHD5 Ligands Is Unaffected by Receptor-Mediated Desensitization in 3T3-L1 and BA Cells.
Sustained stimulation of β-adrenergic signaling causes tachyphylaxis of functional responses due to the desensitization of various components in the PKA signaling pathway (Benovic et al., 1988; Lohse et al., 1990). It was therefore of interest to determine whether ABHD5 ligands could stimulate lipolysis when cells have undergone desensitization to β-adrenergic activation. As shown in Fig. 6, pretreatment of 3T3-L1 (Fig. 6A) and BA cells (Fig. 6B) with 100 nM ISO significantly suppressed the lipolysis response to a subsequent challenge by ISO in both cell lines (P < 0.05). In sharp contrast, previous exposure to ISO did not affect stimulation of lipolysis by the synthetic ABHD5 ligands.
Fig. 6.
β-Adrenergic receptor desensitization differentially affects lipolysis mediated by ISO and ABHD5 ligands. (A and B) Differentiated 3T3-L1 preadipocytes and BA were pretreated with 100 nM ISO or DMSO in DMEM for 2.5–3 hours as described in Materials and Methods. Cells were then washed with PBS and incubated in KRBB medium containing 1% FF-BSA in the presence of vehicle (DMSO), ISO (10 nM 3T3-L1 or 3 nM BA), or ABHD5 ligands (20 μM). NEFAs were then measured in the medium 1 hour following treatment. Shown are results from six independent experiments. Preincubation with 100 nM ISO desensitized cells to further lipolysis induced by ISO but had no inhibitory effect on lipolysis in the presence of ABHD5 ligands. For both graphs, there was a significant interaction between prestimulated and basal conditions (P < 0.05) on NEFA levels. **Significant effect of prestimulation on NEFA levels within the ISO-treated group (P < 0.001).
Discussion
ABHD5 (also called CGI-58) is a member of the α/β-hydrolase family of proteins (Lefevre et al., 2001). A central role for ABHD5 in lipolysis was revealed by the finding that inactivating mutations in the CGI-58 gene are causative for Chanarin-Dorfman syndrome, a rare form of neutral lipid storage disease in humans (Lefevre et al., 2001). Subsequent studies demonstrated that targeted knockdown of ABHD5 in mice increased TG accumulation in various tissues, including liver (Brown et al., 2010; Radner et al., 2010), macrophages (Goeritzer et al., 2014), and muscle (Zierler et al., 2013; Xie et al., 2015), whereas overexpression reduced cellular TG content in some (Lass et al., 2006; Brown et al., 2007; Badin et al., 2012) but not all studies (Caviglia et al., 2011). ABHD5 does not possess intrinsic hydrolytic activity (Lass et al., 2006) and is thought to function predominantly as a protein coactivator for ATGL (Lass et al., 2006; Schweiger et al., 2006), although ATGL-independent functions have also been suggested (Lord and Brown, 2012; Lord et al., 2016). In the basal state, ABHD5 is localized on the surface of the lipid droplet through direct interaction with PLIN proteins (Subramanian et al., 2004; Yamaguchi et al., 2004; Granneman et al., 2009a). Activation of PKA by extracellular signals leads to phosphorylation of PLIN1 and ABHD5 (Granneman et al., 2009a; Sahu-Osen et al., 2015), thereby dissociating ABHD5, which can then bind to and stimulate the activity of ATGL (Subramanian et al., 2004; Yamaguchi et al., 2004).
Most of what is known regarding ABHD5 is through loss- and gain-of-function studies as well as PKA-dependent regulatory mechanisms. Our laboratory recently reported that ABHD5 is also subject to direct allosteric regulation by endogenous and synthetic ligands that modulate lipolysis independently of extracellular signaling by mediating the interaction of ABHD5 with PLIN proteins (Sanders et al., 2015). Although endogenous activating ligands have not been identified, long-chain acyl-CoAs are potent antagonist regulators that promote PLIN-ABHD5 interactions, thereby reducing lipolysis (Granneman et al., 2009b; Sanders et al., 2015). The discovery of synthetic ABHD5 ligands therefore allows further dissection of PKA-dependent and allosteric-mediated effects of ABHD5 on TG catabolism, including how activation by extracellular and intracellular signals is integrated, the lipases that are engaged, and the products that are generated.
ABHD5 is abundantly expressed in differentiated adipocytes (Subramanian et al., 2004; Yamaguchi et al., 2004; Lass et al., 2006), and in the current study, we used cellular models of both white (3T3-L1) and brown adipocytes (BA) to determine whether extracellular and intracellular signals interact in mediating ABHD5-dependent lipolysis. We found that synthetic ABHD5 ligands had no effect on the potency or maximal lipolysis efficacy of the ADRB agonist ISO, suggesting both converge on a common pathway for lipolysis regulation. Stated differently, these results indicate that ABHD5 plays a central role in integrating extracellular and intracellular signals that control adipocyte lipolysis. Because ABHD5 clearly lies downstream of PKA activation, its allosteric regulation by synthetic and endogenous ligands provides a means of rapidly adjusting fatty acid supply to internal demand that can occur independently of extracellular signals.
Results from the current study also indicate that direct activation of ABHD5 results in the complete hydrolysis of TG to free fatty acids and glycerol. Indeed, the relative efflux of fatty acids and glycerol induced by the more potent ABHD5 ligand, SR-3420, was equal to that produced by isoproterenol, despite differences in PKA-dependent phosphorylation of HSL. The findings that ABHD5 ligands stimulate complete TG hydrolysis were also further corroborated ex vivo in organ cultures of gWAT and iBAT. Although ABHD5 is integral for ATGL lipase activity (Lass et al., 2006), pharmacological analysis demonstrated that HSL can account for a significant portion of mobilized fatty acids released under basal conditions and during direct ABHD5 activation in 3T3-L1 adipocytes. By comparison, pharmacological inhibition experiments indicated that basal NEFA release was more reliant on ATGL than HSL in BA cells. Nonetheless, the combined inhibition of ATGL and HSL reduced NEFA efflux induced by ISO or ABHD5 ligands by >94% in 3T3-L1 and 80%–96% in BA cells, confirming the importance of these lipases in TG metabolism (Schweiger et al., 2006). Importantly, that synthetic ABHD5 ligands elicited lipolysis responses to an extent similar to that observed with isoproterenol further reinforces the importance of ABHD5-PLIN interactions in controlling lipase activity.
In our study, the finding that HSL can contribute significantly to both basal and ABHD5-dependent lipolysis was somewhat surprising given previous work indicating the importance of PKA-dependent phosphorylation on HSL translocation and cellular activity (Egan et al., 1992; Clifford et al., 2000; Sztalryd et al., 2003). Nonetheless, significant TG hydrolase activity has been reported in cytosolic preparations and WAT organ cultures from ATGL-deficient mice (Haemmerle et al., 2006; Schweiger et al., 2006), and HSL activity accounted for ∼40% of basal lipolysis in human differentiated preadipocytes (Ryden et al., 2007). Based on our findings, it seems likely that basal activity of HSL is sufficient to allow substantial NEFA mobilization from DG generated by ABHD5-dependent activation of ATGL, especially in the cellular model of white adipocytes. Additionally, our experiments revealed a significant (17%–25%) component of ABHD5 ligand–induced lipolysis in 3T3-L1 cells that was dependent on HSL (BAY-sensitive), but independent of ATGL (Atglistatin-resistant). The mechanisms involved are presently uncertain; however, given that neither compounds nor ABHD5 directly interacts with HSL (Sanders et al., 2015), the mechanism is likely indirect and possibly cell type–specific. Lipolysis, which is dependent on ABHD5 yet independent of ATGL, is thought to exist in the skin (Radner et al., 2010), and we speculate that ABHD5 ligands might be useful in uncovering the mechanisms involved.
Collectively, our work demonstrates that ABHD5 is a dominant locus of acute lipolysis regulation in adipocytes, and that ABHD5 is integral for both PKA- and ligand-mediated hydrolysis of TG. Further, ABHD5-induced lipolysis is fully intact under conditions in which receptor-mediated signaling is suppressed by insulin or desensitization. As lipolysis is necessary and sufficient for brown adipocyte thermogenesis (Guerra et al., 1998; Ahmadian et al., 2011), and plays a critical role in catabolic adipose tissue remodeling (Granneman et al., 2005; Li et al., 2005; Mottillo et al., 2007), it seems reasonable to suggest that ABHD5 ligands might be developed as sustained activators that promote fatty acid catabolism in the presence of hyperinsulinemia, or where receptor-dependent signaling is compromised.
Several ATGL-independent roles for ABHD5 have also been described (Lord and Brown, 2012; Brown and Brown, 2017). For example, it has been suggested that ABHD5 has tumor suppressor functions in colorectal carcinogenesis, potentially by regulating autophagic flux and AMP-activated protein kinase signaling (Ou et al., 2014; Peng et al., 2016). ABHD5 is also involved in skin barrier function by participating in ω-O-acylceramide biosynthesis (Radner et al., 2010; Grond et al., 2017), regulating hepatic triacylglyceride metabolism and steatosis (Radner et al., 2010; Lord et al., 2016), as well as macrophage phenotype/function (Goeritzer et al., 2014; Miao et al., 2014). Thus, synthetic ligands may also help delineate additional ABHD5-mediated signaling events in a variety of conditions with potential therapeutic implications.
Limitations were that our study was conducted using murine models of adipose tissue lipolysis. Although preclinical and cellular models are integral in understanding protein function in a more controlled fashion, it is possible that additional, and currently unknown, species-dependent regulatory mechanisms may exist for eliciting full stimulation of ABHD5. Additionally, although synthetic ABHD5 ligands clearly act downstream of cAMP and PKA-dependent phosphorylation, it is conceivable that ABHD5 ligands may indirectly activate other phosphorylation-dependent signaling pathways that were not evaluated in the current study. Notwithstanding these limitations, however, our findings help contribute to a better understanding of allosteric regulation of ABHD5 in adipocyte lipolysis.
Acknowledgments
The authors thank Dr. Gregory Steinberg and Dr. Emilio Mottillo (McMaster University) for helping to establish and generously providing the brown preadipocyte cell line used in the current study. The authors also thank Dr. Emilio Mottillo, Dr. Todd Leff, and Dr. Vanesa Ramseyer-Payant (Wayne State University) for useful manuscript comments.
Abbreviations
- ABHD5
α-β-hydrolase domain containing 5
- ADRB
β-adrenergic receptor
- ATGL
adipose triglyceride lipase
- BA
brown adipocytes
- BAY 59-9435 (BAY)
4-isopropyl-3-methyl-2-(1-[3-(S)-methyl-piperidin-1-yl]-methanoyl)-2H-isoxalo-5-one
- DG
diglyceride
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethylsulfoxide
- FFA
free fatty acids
- FF-BSA
fatty acid–free bovine serum albumin
- gWAT
gonadal white adipose tissue
- HSL
hormone-sensitive lipase
- iBAT
interscapular brown adipose tissue
- IBMX
3-isobutyl-1-methylxanthine
- ISO
isoproterenol
- KRBB
Krebs-Ringer bicarbonate buffer
- NEFA
nonesterified fatty acid
- PBS
phosphate-buffered saline
- PKA
protein kinase A
- PLIN1
perilipin-1
- SR-3420
1-(3-(3,5-bis(trifluoromethyl)phenyl)propyl)-3-(10-methyl-11-oxo-10,11-dihydrodibenzo[b,f][1,4] thiazepin-8-yl)urea
- SR-4559
1-(4-((2,3-dihydrobenzo[b][1,4]dioxin-6-yl)sulfonyl) piperazin-1-yl)-2-(6-methylbenzofuran-3-yl)ethanone
- SR-4995
1-phenylpropyl-3-(10-methyl-11-oxo-10,11-dihydrodibenzo [b,f][1,4]thiazepin-8-yl)urea
- T3
3,3′,5-triiodo-l-thyronine
- TG
triglyceride (triacylglycerol)
Authorship Contributions
Participated in research design: Rondini, Granneman.
Conducted experiments: Rondini, Mladenovic-Lucas.
Contributed new reagents or analytic tools: Green, Halvorsen, Roush.
Performed data analysis: Rondini, Granneman.
Wrote or contributed to the writing of the manuscript: Rondini, Granneman.
Footnotes
This research was supported by the National Institutes of Health [Grants RO1 DK76629 and RO1 DK105963 to J.G.G.].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, et al. (2011) Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab 13:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badin PM, Loubière C, Coonen M, Louche K, Tavernier G, Bourlier V, Mairal A, Rustan AC, Smith SR, Langin D, et al. (2012) Regulation of skeletal muscle lipolysis and oxidative metabolism by the co-lipase CGI-58. J Lipid Res 53:839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovic JL, Bouvier M, Caron MG, Lefkowitz RJ. (1988) Regulation of adenylyl cyclase-coupled beta-adrenergic receptors. Annu Rev Cell Biol 4:405–428. [DOI] [PubMed] [Google Scholar]
- Boden G. (1998) Free fatty acids (FFA), a link between obesity and insulin resistance. Front Biosci 3:d169–175. [DOI] [PubMed] [Google Scholar]
- Brown AL, Brown MJ. (2017) Critical roles for α/β hydrolase domain 5 (ABHD5)/comparative gene identification-58 (CGI-58) at the lipid droplet interface and beyond. Biochim Biophys Acta 1862:1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Betters JL, Lord C, Ma Y, Han X, Yang K, Alger HM, Melchior J, Sawyer J, Shah R, et al. (2010) CGI-58 knockdown in mice causes hepatic steatosis but prevents diet-induced obesity and glucose intolerance. J Lipid Res 51:3306–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Chung S, Das A, Shelness GS, Rudel LL, Yu L. (2007) CGI-58 facilitates the mobilization of cytoplasmic triglyceride for lipoprotein secretion in hepatoma cells. J Lipid Res 48:2295–2305. [DOI] [PubMed] [Google Scholar]
- Carmen GY, Víctor SM. (2006) Signalling mechanisms regulating lipolysis. Cell Signal 18:401–408. [DOI] [PubMed] [Google Scholar]
- Caviglia JM, Betters JL, Dapito DH, Lord CC, Sullivan S, Chua S, Yin T, Sekowski A, Mu H, Shapiro L, et al. (2011) Adipose-selective overexpression of ABHD5/CGI-58 does not increase lipolysis or protect against diet-induced obesity. J Lipid Res 52:2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus TH, Lowe DB, Liang Y, Salhanick AI, Lubeski CK, Yang L, Lemoine L, Zhu J, Clairmont KB. (2005) Specific inhibition of hormone-sensitive lipase improves lipid profile while reducing plasma glucose. J Pharmacol Exp Ther 315:1396–1402. [DOI] [PubMed] [Google Scholar]
- Clifford GM, Londos C, Kraemer FB, Vernon RG, Yeaman SJ. (2000) Translocation of hormone-sensitive lipase and perilipin upon lipolytic stimulation of rat adipocytes. J Biol Chem 275:5011–5015. [DOI] [PubMed] [Google Scholar]
- Degerman E, Landström TR, Wijkander J, Holst LS, Ahmad F, Belfrage P, Manganiello V. (1998) Phosphorylation and activation of hormone-sensitive adipocyte phosphodiesterase type 3B. Methods 14:43–53. [DOI] [PubMed] [Google Scholar]
- Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS. (2007) Regulation of lipolysis in adipocytes. Annu Rev Nutr 27:79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan JJ, Greenberg AS, Chang MK, Wek SA, Moos MC, Jr, Londos C. (1992) Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc Natl Acad Sci USA 89:8537–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeritzer M, Schlager S, Radovic B, Madreiter CT, Rainer S, Thomas G, Lord CC, Sacks J, Brown AL, Vujic N, et al. (2014) Deletion of CGI-58 or adipose triglyceride lipase differently affects macrophage function and atherosclerosis. J Lipid Res 55:2562–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Li P, Zhu Z, Lu Y. (2005) Metabolic and cellular plasticity in white adipose tissue I: effects of beta3-adrenergic receptor activation. Am J Physiol Endocrinol Metab 289:E608–E616. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Moore HP. (2008) Location, location: protein trafficking and lipolysis in adipocytes. Trends Endocrinol Metab 19:3–9. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Granneman RL, Greenberg AS, Obin MS, Zhu Z. (2007) Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem 282:5726–5735. [DOI] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Krishnamoorthy R, Rathod M. (2009a) Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J Biol Chem 284:34538–34544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Mottillo EP, Zhu Z. (2009b) Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J Biol Chem 284:3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grond S, Radner FP, Eichmann TO, Kolb D, Grabner GF, Wolinski H, Gruber R, Hofer P, Heier C, Schauer S, et al. (2017) Skin barrier development depends on CGI-58 protein expression during late-stage keratinocyte differentiation. J Invest Dermatol 137:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Koza RA, Walsh K, Kurtz DM, Wood PA, Kozak LP. (1998) Abnormal nonshivering thermogenesis in mice with inherited defects of fatty acid oxidation. J Clin Invest 102:1724–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, et al. (2006) Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312:734–737. [DOI] [PubMed] [Google Scholar]
- Holm C. (2003) Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans 31:1120–1124. [DOI] [PubMed] [Google Scholar]
- Jaworski K, Sarkadi-Nagy E, Duncan RE, Ahmadian M, Sul HS. (2007) Regulation of triglyceride metabolism. IV. Hormonal regulation of lipolysis in adipose tissue. Am J Physiol Gastrointest Liver Physiol 293:G1–G4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn SE, Hull RL, Utzschneider KM. (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444:840–846. [DOI] [PubMed] [Google Scholar]
- Klein J, Fasshauer M, Ito M, Lowell BB, Benito M, Kahn CR. (1999) beta(3)-adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in brown adipocytes. J Biol Chem 274:34795–34802. [DOI] [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R. (2006) Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab 3:309–319. [DOI] [PubMed] [Google Scholar]
- Lefèvre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, Lakhdar H, Wollenberg A, Verret JL, Weissenbach J, et al. (2001) Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet 69:1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zhu Z, Lu Y, Granneman JG. (2005) Metabolic and cellular plasticity in white adipose tissue II: role of peroxisome proliferator-activated receptor-alpha. Am J Physiol Endocrinol Metab 289:E617–E626. [DOI] [PubMed] [Google Scholar]
- Lohse MJ, Benovic JL, Caron MG, Lefkowitz RJ. (1990) Multiple pathways of rapid beta 2-adrenergic receptor desensitization. Delineation with specific inhibitors. J Biol Chem 265:3202–3211. [PubMed] [Google Scholar]
- Lönnroth P, Smith U. (1986) The antilipolytic effect of insulin in human adipocytes requires activation of the phosphodiesterase. Biochem Biophys Res Commun 141:1157–1161. [DOI] [PubMed] [Google Scholar]
- Lord CC, Brown JM. (2012) Distinct roles for alpha-beta hydrolase domain 5 (ABHD5/CGI-58) and adipose triglyceride lipase (ATGL/PNPLA2) in lipid metabolism and signaling. Adipocyte 1:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CC, Ferguson D, Thomas G, Brown AL, Schugar RC, Burrows A, Gromovsky AD, Betters J, Neumann C, Sacks J, et al. (2016) Regulation of hepatic triacylglycerol metabolism by CGI-58 does not require ATGL co-activation. Cell Reports 16:939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DB, Magnuson S, Qi N, Campbell AM, Cook J, Hong Z, Wang M, Rodriguez M, Achebe F, Kluender H, et al. (2004) In vitro SAR of (5-(2H)-isoxazolonyl) ureas, potent inhibitors of hormone-sensitive lipase. Bioorg Med Chem Lett 14:3155–3159. [DOI] [PubMed] [Google Scholar]
- Mayer N, Schweiger M, Romauch M, Grabner GF, Eichmann TO, Fuchs E, Ivkovic J, Heier C, Mrak I, Lass A, et al. (2013) Development of small-molecule inhibitors targeting adipose triglyceride lipase. Nat Chem Biol 9:785–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Ou J, Ma Y, Guo F, Yang Z, Wiggins M, Liu C, Song W, Han X, Wang M, et al. (2014) Macrophage CGI-58 deficiency activates ROS-inflammasome pathway to promote insulin resistance in mice. Cell Reports 7:223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottillo EP, Desjardins EM, Crane JD, Smith BK, Green AE, Ducommun S, Henriksen TI, Rebalka IA, Razi A, Sakamoto K, et al. (2016) Lack of adipocyte AMPK exacerbates insulin resistance and hepatic steatosis through brown and beige adipose tissue function. Cell Metab 24:118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottillo EP, Shen XJ, Granneman JG. (2007) Role of hormone-sensitive lipase in beta-adrenergic remodeling of white adipose tissue. Am J Physiol Endocrinol Metab 293:E1188–E1197. [DOI] [PubMed] [Google Scholar]
- Nielsen TS, Jessen N, Jørgensen JO, Møller N, Lund S. (2014) Dissecting adipose tissue lipolysis: molecular regulation and implications for metabolic disease. J Mol Endocrinol 52:R199–R222. [DOI] [PubMed] [Google Scholar]
- Osterlund T, Danielsson B, Degerman E, Contreras JA, Edgren G, Davis RC, Schotz MC, Holm C. (1996) Domain-structure analysis of recombinant rat hormone-sensitive lipase. Biochem J 319:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Miao H, Ma Y, Guo F, Deng J, Wei X, Zhou J, Xie G, Shi H, Xue B, et al. (2014) Loss of abhd5 promotes colorectal tumor development and progression by inducing aerobic glycolysis and epithelial-mesenchymal transition. Cell Reports 9:1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Miao H, Wu S, Yang W, Zhang Y, Xie G, Xie X, Li J, Shi C, Ye L, et al. (2016) ABHD5 interacts with BECN1 to regulate autophagy and tumorigenesis of colon cancer independent of PNPLA2. Autophagy 12:2167–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petta S, Gastaldelli A, Rebelos E, Bugianesi E, Messa P, Miele L, Svegliati-Baroni G, Valenti L, Bonino F. (2016) Pathophysiology of non alcoholic fatty liver disease. Int J Mol Sci 1b7:2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radner FP, Streith IE, Schoiswohl G, Schweiger M, Kumari M, Eichmann TO, Rechberger G, Koefeler HC, Eder S, Schauer S, et al. (2010) Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58). J Biol Chem 285:7300–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden M, Price TB, Perseghin G, Petersen KF, Rothman DL, Cline GW, Shulman GI. (1996) Mechanism of free fatty acid-induced insulin resistance in humans. J Clin Invest 97:2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondini EA, Fang H, Runge-Morris M, Kocarek TA. (2014) Regulation of human cytosolic sulfotransferases 1C2 and 1C3 by nuclear signaling pathways in LS180 colorectal adenocarcinoma cells. Drug Metab Dispos 42:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydén M, Jocken J, van Harmelen V, Dicker A, Hoffstedt J, Wirén M, Blomqvist L, Mairal A, Langin D, Blaak E, et al. (2007) Comparative studies of the role of hormone-sensitive lipase and adipose triglyceride lipase in human fat cell lipolysis. Am J Physiol Endocrinol Metab 292:E1847–E1855. [DOI] [PubMed] [Google Scholar]
- Sahu-Osen A, Montero-Moran G, Schittmayer M, Fritz K, Dinh A, Chang YF, McMahon D, Boeszoermenyi A, Cornaciu I, Russell D, et al. (2015) CGI-58/ABHD5 is phosphorylated on Ser239 by protein kinase A: control of subcellular localization. J Lipid Res 56:109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MA, Madoux F, Mladenovic L, Zhang H, Ye X, Angrish M, Mottillo EP, Caruso JA, Halvorsen G, Roush WR, et al. (2015) Endogenous and synthetic ABHD5 ligands regulate ABHD5-perilipin interactions and lipolysis in fat and muscle. Cell Metab 22:851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M, Schreiber R, Haemmerle G, Lass A, Fledelius C, Jacobsen P, Tornqvist H, Zechner R, Zimmermann R. (2006) Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem 281:40236–40241. [DOI] [PubMed] [Google Scholar]
- Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, et al. (2004) Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem 279:42062–42071. [DOI] [PubMed] [Google Scholar]
- Sztalryd C, Xu G, Dorward H, Tansey JT, Contreras JA, Kimmel AR, Londos C. (2003) Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol 161:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Kadegowda AK, Ma Y, Guo F, Han X, Wang M, Groban L, Xue B, Shi H, Li H, et al. (2015) Muscle-specific deletion of comparative gene identification-58 (CGI-58) causes muscle steatosis but improves insulin sensitivity in male mice. Endocrinology 156:1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Omatsu N, Matsushita S, Osumi T. (2004) CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem 279:30490–30497. [DOI] [PubMed] [Google Scholar]
- Zierler KA, Jaeger D, Pollak NM, Eder S, Rechberger GN, Radner FP, Woelkart G, Kolb D, Schmidt A, Kumari M, et al. (2013) Functional cardiac lipolysis in mice critically depends on comparative gene identification-58. J Biol Chem 288:9892–9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, et al. (2004) Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306:1383–1386. [DOI] [PubMed] [Google Scholar]