Significance
Viruses composed of a shell of coat proteins enclosing ssRNA genomes are among the simplest biological entities. Their lifecycles include a range of processes, such as specific genome encapsidation and efficient capsid self-assembly. Until recently, these were not linked, but we have shown that many viruses in this class encode multiple, degenerate RNA sequence/structure motifs that bind cognate coat proteins collectively. This simultaneously ensures specific genome packaging and efficient virion assembly via an RNA-encoded instruction manual. Here we extract essential features of this manual in a viral RNA genome, creating a synthetic sequence with an assembly substrate superior to the natural equivalent. Such RNAs have the potential for efficient production of stable virus-like particle vaccines and/or gene/drug delivery vehicles.
Keywords: satellite tobacco necrosis virus, packaging signals, viral assembly, synthetic virology
Abstract
Satellite tobacco necrosis virus (STNV) is one of the smallest viruses known. Its genome encodes only its coat protein (CP) subunit, relying on the polymerase of its helper virus TNV for replication. The genome has been shown to contain a cryptic set of dispersed assembly signals in the form of stem-loops that each present a minimal CP-binding motif AXXA in the loops. The genomic fragment encompassing nucleotides 1–127 is predicted to contain five such packaging signals (PSs). We have used mutagenesis to determine the critical assembly features in this region. These include the CP-binding motif, the relative placement of PS stem-loops, their number, and their folding propensity. CP binding has an electrostatic contribution, but assembly nucleation is dominated by the recognition of the folded PSs in the RNA fragment. Mutation to remove all AXXA motifs in PSs throughout the genome yields an RNA that is unable to assemble efficiently. In contrast, when a synthetic 127-nt fragment encompassing improved PSs is swapped onto the RNA otherwise lacking CP recognition motifs, assembly is partially restored, although the virus-like particles created are incomplete, implying that PSs outside this region are required for correct assembly. Swapping this improved region into the wild-type STNV1 sequence results in a better assembly substrate than the viral RNA, producing complete capsids and outcompeting the wild-type genome in head-to-head competition. These data confirm details of the PS-mediated assembly mechanism for STNV and identify an efficient approach for production of stable virus-like particles encapsidating nonnative RNAs or other cargoes.
Single-stranded (ss)RNA viruses comprise a large proportion of all viral pathogens. Our understanding of their capsid self-assembly has recently been transformed (1, 2). We have shown that, in contrast to previous ideas based on spontaneous RNA-coat protein (CP) coalescence via purely electrostatic contacts (3–9) or the presence of a single sequence-specific CP-genome interaction site leading to assembly initiation (10), assembly is regulated by multiple RNA-CP contacts (11–17). These contacts are sequence-specific and occur at many sequence/structure degenerate RNA sites scattered across all regions of the viral genome. We have termed the sites defining these CP interactions packaging signals (PSs), with the precise CP-binding sequence within each PS termed a recognition motif. The latter appear routinely as discontinuous nucleotides, e.g., in the loop or bulge of a potential stem-loop (SL) structure, making their identification by simple sequence analysis challenging. We have therefore developed a set of generic tools to facilitate identification of such PSs. Each individual PS site carries only limited sequence information, but commonly these sites have nanomolar affinities for cognate CPs. These affinities vary across the set of PSs in each genome creating a hierarchy, helping to define the assembly pathway and preventing kinetic trapping. PS-mediated assembly has been detected in viruses infecting bacteria (11), plants, (12) and humans (18–20), implying that it provides significant selective advantages, e.g., genetic robustness compared with a single assembly initiation site that would be vulnerable to the error-prone replication common to these pathogens. The precise relationship between the genomic RNA sequence and the CP shell created by defined CP-PS contacts leads to several predictions about the structure of the resultant virion. One is that the conformation of the RNA within every viral particle should be very similar in the vicinity of the protein shell (21, 22). This has been confirmed in multiple examples (23), most recently by direct asymmetric structure determination via high-resolution cryo-electron microscopy (EM) (24–27).
Viral CP shells (virus-like particles, VLPs) are currently attracting significant attention for their potential applications, including their use as noninfectious synthetic vaccines that display native antigenicity and their potential as gene/drug delivery vectors (28–30). Each of these applications faces technical problems that currently prevent their widespread exploitation. Neglecting to account for the PS-mediated assembly of natural virions may be one cause of such difficulties. We have therefore examined whether it is possible to identify the critical features of a natural viral RNA genome that contribute to its assembly via the PS-mediated mechanism. These features have been transferred to a nonviral RNA sequence and subsequently refined to improve assembly efficiency. The results suggest that it is possible to abstract most of the natural RNA features required for PS-mediated assembly, creating an RNA sequence that is encapsidated more efficiently by the cognate CP than the viral equivalent. Such synthetic viral RNA assembly substrates should lead to dramatic improvements in VLP technology.
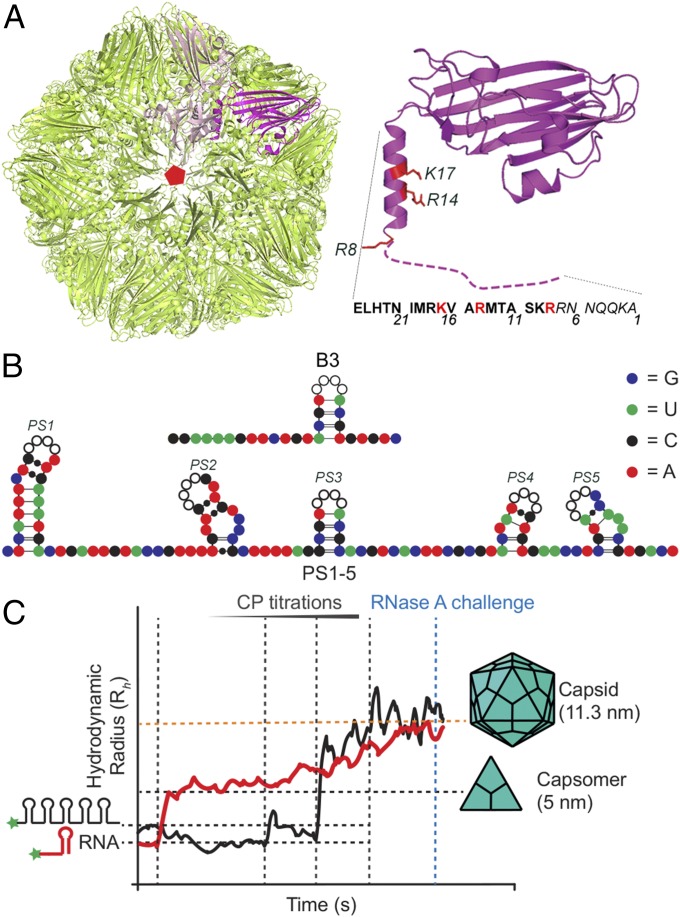
For these studies, we chose satellite tobacco necrosis virus (STNV), a small (∼23-nm diameter) plant virus (Fig. 1A), whose ∼1.2-kb positive-sense ssRNA genome only encodes its CP gene. Previously, we identified the highest-affinity STNV PS using RNA SELEX against the cognate CP (31). One aptamer, B3 (Fig. 1B), forms an SL with an ACAA sequence in the loop. This matches a similar potential SL in the 5′ region of the STNV CP gene. Analysis of the three known STNV strains suggests that up to 30 copies of such SLs could form across each genome, all with an AXXA loop motif (31). The 127-nt-long fragment (127-mer) located at the 5′ end of the STNV-1 genome spans both an untranslated region (UTR) and the start codon of the CP gene. It is predicted to encompass five PSs (PS1–PS5), including the match to the B3 aptamer (PS3, Fig. 1). Single-molecule fluorescence correlation spectroscopy (smFCS) reassembly assays can be carried out at low nanomolar concentrations. These conditions reveal the effects of PS-mediated assembly that are lost at higher concentration (13, 17). We showed previously by this method that the AXXA loop acts as a CP recognition motif and that all five of the PS sites act cooperatively to condense the RNA fragment and assemble a complete T = 1 CP shell around it (12) (Fig. 1C). The loop of the PS3 site is the most important for generating these effects, which require multiple PSs, consistent with the relative spacing to neighboring PSs being vital for accurate control of assembly. STNV is therefore an ideal test case for the design of a synthetic assembly substrate. Here, we analyze in depth the contribution of different molecular features to the cooperative assembly, arriving at an improved, synthetic RNA that surpasses the native sequence in assembly efficiency, paving the way for improved VLP production.
Fig. 1.
The STNV system. (A) Ribbon diagram of the STNV T = 1 capsid (green) (Left, PDB 3S4G) viewed along a fivefold axis with a trimeric capsomer highlighted (magenta/pink) and (Right) a CP monomer (magenta, PDB 4BCU). Sidechains mutated here are shown and labeled. The disordered N-terminal amino acid sequence is shown as a dashed line, next to the sequence of the first 25 amino acids. (B) Sequence and putative secondary structure of the 127-nt 5′ STNV-1 genomic fragment showing the locations of the PS SLs, named 5′ to 3′ as PS1 to PS5, respectively. Each contains the CP recognition motif, AXXA, in their loops (white circles, black outline). The B3 aptamer is shown similarly above. Nucleotides are color-coded as indicated, here and throughout. (C) Example smFCS assays. Hydrodynamic radius (Rh) values for CP-free, fluorescently labeled RNAs (black line for PS1–PS5 and red line for B3) are determined before and during STNV CP titration at fixed time points (vertical dashed lines). The Rh values were allowed to equilibrate after each step. The PS1–PS5 Rh initially collapses by up to 30% until the CP concentration reaches a threshold, triggering cooperative assembly to T = 1 VLPs (Rh ∼ 11 nm). At the end of each titration, the complexes formed are challenged by addition of RNase A. Largely unchanged Rh values were assumed to indicate that the RNA is in a closed VLP.
Results
Sequence-Specific Recognition of Individual PS Sites.
There are multiple consequences of sequence-specific RNA-CP recognition in the STNV system (Fig. 1). Titration of CP into oligonucleotides encompassing only PS3 (or B3) initially results in formation of a trimeric capsomer (Rh ∼ 5 nm), followed by formation of T = 1 VLPs (Rh ∼11.3 nm) as the CP concentration is raised gradually. Rh distribution plots of the smFCS data at the end of the titration suggest that the VLPs formed are homogeneous, while EM images and RNase challenge assays suggest that they are composed of complete protein shells. A similar titration with a PS3/B3 variant having a loop sequence of UUUU showed that CP binds such SLs, but the complex formed is unable to assemble to VLPs (12). The natural 127-mer, encompassing PS1–PS5, shows more complex behavior. Addition of low CP concentrations triggers a collapse in its Rh by about 20–30%, mimicking the behavior seen for the full-length genome (11). Subsequent CP additions result in cooperative conversion to T = 1 VLPs with the same properties as those formed around PS3 alone. PS variants within this fragment confirm that AXXA is a CP recognition motif. Its presence is only absolutely required in PS3. However, the variants no longer assemble with wild-type cooperativity (12). STNV-1 CP alone does not aggregate below 15 µM under these conditions, and therefore everything in the titrations shown here is a consequence of RNA-CP interaction.
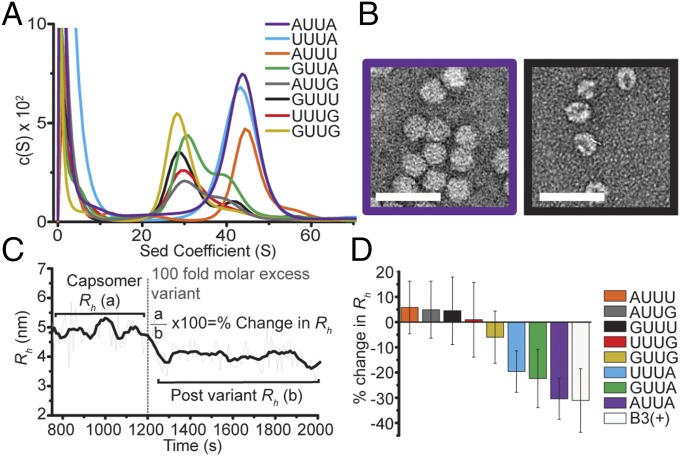
To identify the critical features of PS3 recognition, we produced a series of SLs encompassing variant loop sequences but retaining the PS3 stem sequence (SI Appendix, Fig. S1 and Table S1). The variants have altered nucleotides in the “inner” two positions (CC, AA, GG, GU, and UG) compared with the wild-type CA of PS3. “Outer” variants (AUUA, AUUG, GUUA, GUUG, GUUU, UUUG, UUUA, and AUUU), in which both inner nucleotides were altered to uridines, were also tested. Our expectation was that there would be no base specificity at the middle positions, while the adenines would be preferred at the first and last positions of the tetraloop. We examined their abilities to support assembly of both the T = 1 shell and the trimeric capsomer. Capsid reassembly assays (SI Appendix) were carried out at a molar ratio of 1:3 RNA:CP with a final CP concentration of 6 µM. Note, reassembly remains sensitive to the loop sequence at these concentrations (17). The results were assayed by velocity sedimentation analysis and in EM images, and yields were quantitated by quasi-elastic light scattering (QELS) chromatography. The inner nucleotide variants form T = 1 capsids with roughly similar efficiency as PS3, confirming that their identities are not part of the CP recognition motif (Fig. 2 A and B and SI Appendix, Fig. S1). The outer nucleotide variants showed differing behavior, with only the AUUU, UUUA, and AUUA variants having a peak in a similar position to PS3, confirming that the outer adenines are part of the CP recognition motif.
Fig. 2.
Defining the CP recognition motif. (A) Ensemble reassembly of variant B3 RNAs, analyzed by sedimentation velocity (variant RNAs are color-coded as given in Inset). The expected T = 1 VLP sediments at ∼42 S, where S is the sedimentation (Sed) coefficient. c(S) is a continuous distribution from the Lamm equation model (SI Appendix). (B) EM images of representative assembly products. (Scale bar, 50 nm, here and throughout.) See also SI Appendix, Fig. S1B. (C) Example variant RNA smFCS displacement assay for results plotted in D. (D) Percent change in Rh following addition of 100-fold molar excess of variant RNAs (color-coded) to a capsomer (Rh ∼ 5 nm) formed with 1-nM AF488-labeled B3. Error bars indicate SEM.
To examine the relative importance of the loop sequence for CP affinity, we adapted the smFCS assay (Fig. 1C). Labeled B3 was titrated with CP to form the trimer, as judged by the Rh value, and then a 100-fold molar excess of each outer sequence variant was added to see if this would displace the bound dye-label. Variants that do not bind with a similar affinity to B3 fail to displace the labeled RNA, whereas B3 and other variants displace the labeled species restoring the Rh to that of CP-free RNA (Fig. 2C). The results (Fig. 2D) show the percentage Rh change following this challenge, revealing a wide variation between loop sequence variants. All those with guanine substitutions and the AUUU variant fail to displace B3. The superior performance of the UUUA variant suggests that the 3′ A is the most important for CP recognition. Alternatively, the A-U base pair at the top of the adjacent stem may break and present an AUUUAU variant of the B3 motif that is still recognized by the CP. Either way, AXXA outperforms all variants, suggesting that SLs carrying tetraloop motifs of AXXA encompass the best CP recognition motif for assembly into VLPs.
Roles of Electrostatics and PS Cooperativity in VLP Assembly.
The results above demonstrate that sequence specificity of RNA-CP interaction is the major determinant of assembly. Previous work with other plant viruses having similar positively charged N-terminal tails has, in contrast, suggested that the major assembly driving force is electrostatic neutralization. STNV-1 CP is typical of many viruses having many basic amino acids in its N-terminal tail giving it a net +8 charge, including the N terminus; cf. cowpea chlorotic mosaic virus (3), which has a charge of +10. Three of these sidechains, R8, R14, and K17 (Fig. 1), are close to the RNA duplex seen in the crystal structure of the B3 VLP (17). To examine the effects of these positive charges on assembly we produced mutations with A or D in place of K or R. Since R14 and K17 are adjacent in three dimensions, their variants were made as the double mutants, i.e., R14A/K17A and R14D/K17D. All the mutant CPs express normally (SI Appendix, Fig. S2 and Table S2), but only the ones at R8 form VLPs equivalent to those seen with wild type (SI Appendix), consistent with the basic sidechains contributing positively to assembly. All the variant proteins were examined for their abilities to bind RNA oligos encompassing either a single PS (B3) or the 127-mer fragment (Fig. 3 and SI Appendix, Fig. S3). Neither double mutant bound either RNA under these conditions. R8A assembles around B3 but requires a much higher (>10-fold) CP concentration to do so, consistent with it having a lowered affinity for the RNA. By 1 µM CP it forms T = 1 shells that are resistant to RNase challenge. The R8D variant fails to form any stable higher-order species with either RNA (SI Appendix, Fig. S3B). In contrast, the R8A variant binds to the 127-mer very similarly to wild-type CP, including undergoing an initial collapse in Rh , implying that reduced intrinsic RNA affinity can be compensated by cooperative PS binding. Unfavorable electrostatic interaction presumably explains the lack of assembly when R8D is titrated against the 127-mer. If we assume that the mutations do not significantly alter the unliganded CP conformation, these effects probe the role(s) of electrostatic interactions during assembly. They imply that charge neutralization is not an absolute requirement for assembly on longer natural RNA fragments. This is consistent with the PS-mediated, but not purely electrostatic assembly mechanism for this virus.
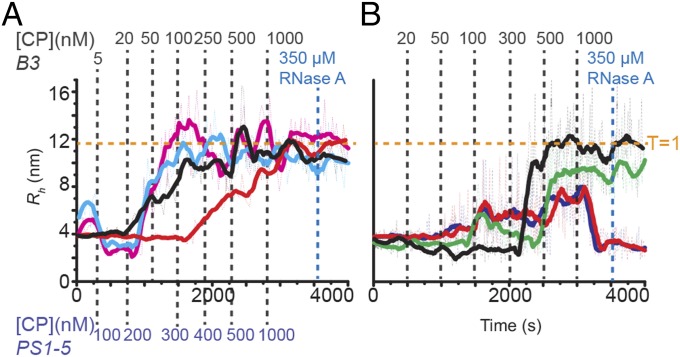
Fig. 3.
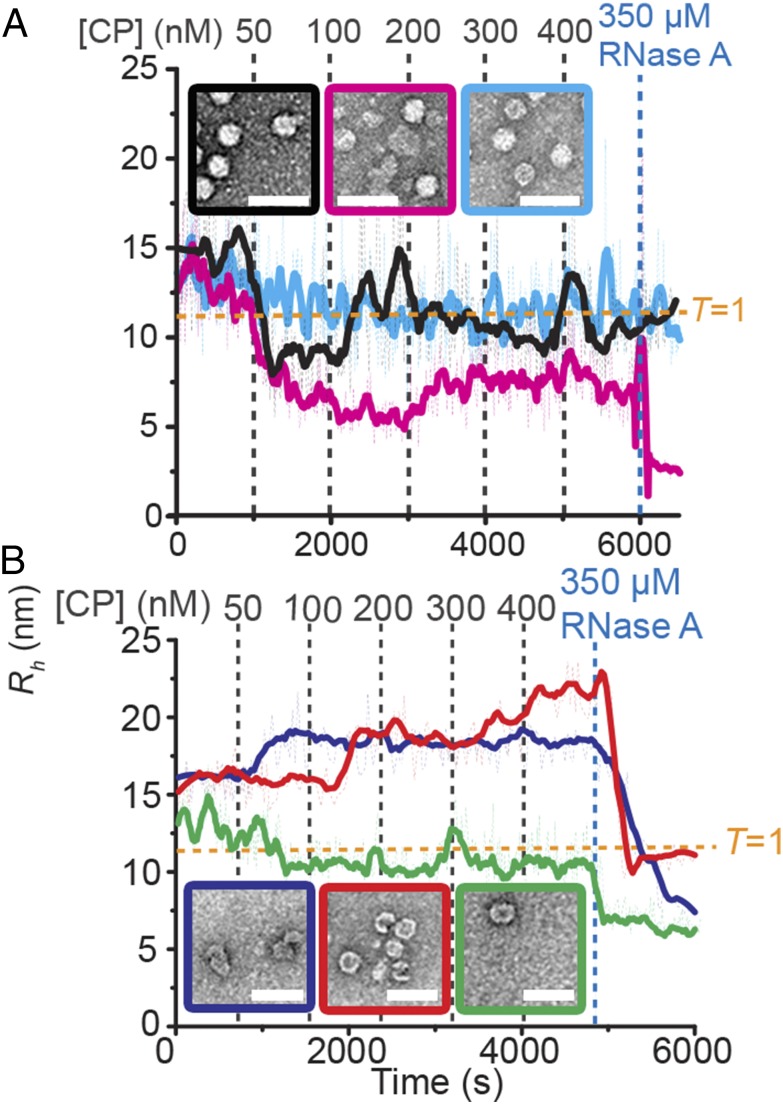
Electrostatic interactions and cooperativity of assembly. (A) Wild-type or R8A CPs were titrated into B3 (1 nM, black or red, respectively) or PS1–PS5 (10 nM, cyan or magenta, respectively), and Rh changes were monitored. Titrations points are shown above (B3 in gray) and below (PS1–PS5 in light blue), respectively. (B) Wild-type STNV CP was titrated into 10 nM of each of PS1–PS5 (black), PS1–PS3 (red), PS3–PS5 (blue), or PS2–PS4 (green) (Fig. 4).
Given that multiple RNA PSs can overcome lower RNA-CP affinity, as expected for a process in which PSs act collectively, we examined how many PSs are required to generate cooperative assembly. Given the importance of PS3 and the effects seen for fragments containing five PSs, three subfragments of the 127-mer each containing PS3 were tested (Fig. 3B and SI Appendix, Figs. S4 and S5). These are PS1–PS3, PS2–PS4, and PS3–PS5. Each could bind CP at PS3 but differs in the numbers of flanking PS sites, from two 5′ or 3′ of PS3 to just one on each flank. Only the fragment with PS3 centrally located assembles RNase-resistant T = 1 shells, although it does not show a collapse, and the overall yield is lower than for the 127-mer. The other fragments form nonspecific aggregates that eventually spontaneously dissociate.
The interpretation of these results is nontrivial. The effects are clearly not purely electrostatic in nature since the PS2–PS4 fragment (66 nt) which assembles is shorter than PS1-3 (76 nt) and 1 nt shorter than PS3–PS5. To understand the specificity of these reactions we need to consider the folding propensity of each of the PS-encompassing sites. The secondary structure of the 127-mer shown (Fig. 4) was arrived at by constraining its folds to capture the maximum number of SLs with AXXA loop motifs present. In this fragment only PS1 and PS3 are predicted to have a favorable folding free energy (Mfold; ref. 32) in isolation. This is consistent with our previous assays, in which replacement of the CP recognition motifs within each PS with UUUU, and variations in their relative spacing with respect to PS3, resulted in markedly different assembly behavior (12). In solution these RNA molecules will exist as an ensemble of differing conformations. Interaction with the STNV CP displaces this equilibrium, preferentially selecting a single or a few assembly-competent conformations in which the PSs are present. The assembly efficiency seen may therefore be related to the relative populations of such conformers in the ensemble and thus to the free-energy costs of imposing this conformation. Assessing the extent of a conformational ensemble is difficult. A sense of the likelihood of alternate structures can be obtained based on the first 100 secondary structures returned by the Sfold algorithm (33).
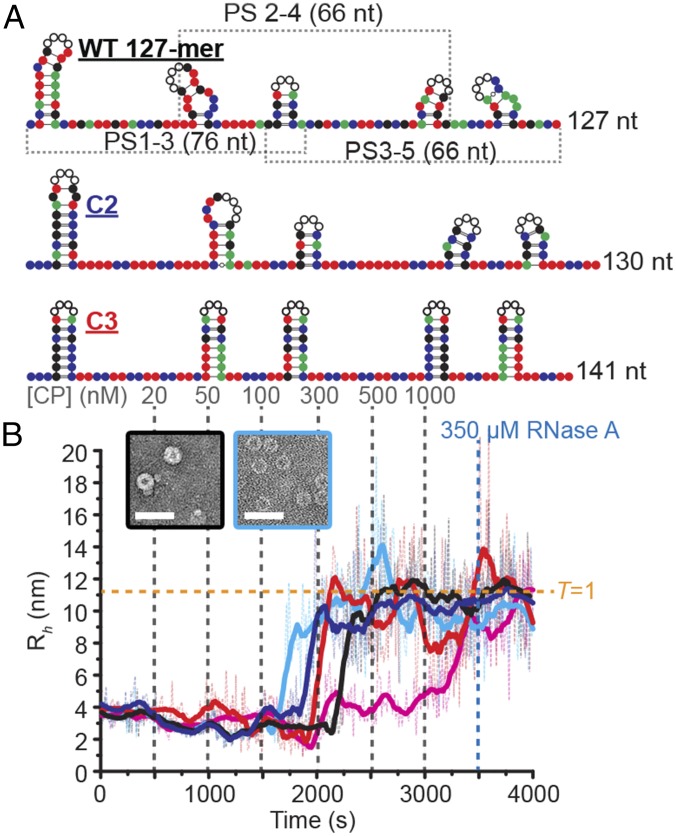
Fig. 4.
Assembly of synthetic cassettes. (A) Sequences and putative secondary structures of the wild-type 127-mer, the C2 and C3 cassettes (SI Appendix, Fig. S6). (B) STNV CP titration of all variant PS1–PS5 constructs; conditions as in Fig. 3B. (Inset) EM images of the products with the wild-type 127-mer (black) and C4 (cyan) cassettes.
When such structures are examined for the fragments encompassing three PSs, a possible explanation for their assembly competencies emerges. For PS1–PS3, the dominant folds encompass PS1 with a minority also containing PS3 (SI Appendix, Table S3). In principle, the minor conformer could promote assembly, but the spacing between PS1 and PS3 is too large to facilitate the cooperative effects of multiple PSs. Similar analysis of PS2–PS4 suggests that the dominant secondary structure does not contain any of the PS folds expected for the 127-mer. However, its predicted secondary structure contains two alternative SLs that are almost always present, one of which presents an AXXA sequence (SI Appendix, Fig. S5). Their relative spacing (4 nt) is short enough to see a cooperative effect. The PS3–PS5 fragment forms two SLs within 10–12 nt of one another, one presenting an AXXA motif as PS5. This would suggest an assembly-competent structure. However, in the ensemble of possible structures, this SL is only present in 6% of the potential folds (SI Appendix, Table S3), which may account for its assembly behavior (Fig. 3B).
The conformational scrambling behavior described above for the fragments encompassing three PSs probably reflects events in vivo, where it is known that sequences within the 127-mer participate in formation of a translational enhancer with sequences in the 3′ UTR (34). That complex cannot be present in the assembly-competent conformer. To explore the effects of such secondary structure folding propensity further, we turned to the design of artificial PS-containing sequences.
Assembly of Nonviral Substrates.
To investigate the requirements for an efficient assembly substrate, we produced synthetic cassettes mimicking aspects of the wild-type 127-mer in which most of the natural viral sequence has been replaced (∼80%). Attempts to create these sequences using a simple base substitution scheme all resulted in unstable secondary structures. We therefore chose to modify the existing SLs by conversion of base pairs to G-C, inversion of existing G-C pairs, or adding extra base pairs and then checking that they would likely fold into similar secondary structures to those in the wild-type 127-mer. The natural viral sequences connecting these SLs were then replaced with strings of As and Gs until only one fold was most likely (Fig. 4 and SI Appendix, Fig. S6). The relative separations of the base-paired stems were kept identical to those in the wild-type 127-mer. As a result of these changes, PSs 1, 2, 4, and 5 have been stabilized compared with the wild-type 127-mer, with all SLs having favorable folding propensity.
To assess the importance of the folding propensity of the dominant PS3 site we also created the following synthetic PS1–PS5 cassettes: (i) unstable PS1–PS5, cassette 1 (C1), in which the folding free energy of PS3, the central PS, is positive (0.3 vs. −2.6 kcal/mol), i.e., a scenario in which PS3 is unlikely to fold spontaneously; (ii) stable PS1–PS5, cassette 2 (C2), in which the folding free energy of the central PS is more negative (−3.5 vs. −2.6 kcal/mol for the 127-mer), i.e., where PS3 is more stable; (iii) all PS3, cassette 3 (C3), in which all five PSs mimic PS3, with stems of all PSs extended to the same length (7 bp) and all CP recognition motifs identical to that in wild-type PS3; and (iv) synthetic, stabilized PS1–PS5, cassette 4 (C4), containing the artificial PSs 1, 2, 4, and 5 from stable PS1–PS5 and the artificial extended SL for PS3 from the all-PS3 construct. The latter is hyperstabilized with respect to PS3 in both the wild-type fragment and C2 (−7.6 vs. −2.6 or −3.5 kcal/mol, respectively).
To compare the behaviors of these cassettes we examined their potential secondary structures. SI Appendix, Table S3 lists the frequency of occurrence of each PS in the ensemble of the first 100 secondary structures returned by the Sfold algorithm (33), together with their relative spacings. In addition, we compared their circular dichroism (CD) spectra. CD provides a physical signal (35), the molar ellipticity at 260 nm, that is proportional to the percentage of base-paired residues and/or tertiary structure. The measurements were made in a buffer containing calcium ions since these are required in the reassembly buffer, there being several Ca2+-binding sites within the STNV capsid (36). Titration of the test RNAs up to 2 mM calcium, the concentration in reassembly buffers, results in mild increases (9–17%) in the 260-nm ellipticity, as expected (SI Appendix, Fig. S7A). The only exception is C1, which does not respond to the presence of the cation. The molar ellipticity values of all test RNAs in this buffer decline as expected with temperature (SI Appendix, Fig. S7B). CD ellipticities at 260 nm of all the RNAs differ, illustrating the complexity of comparing RNA conformational ensembles. The C1 is much less structured throughout the temperature range. Perhaps surprisingly given the apparent secondary structures, the wild-type 127-mer has the highest amount of structure at the lower temperatures. At the highest temperature tested all the RNAs except C1 have roughly similar ellipticity values, implying that they had reached similar levels of denaturation.
All these cassettes, with the exception of C1, trigger assembly of T = 1 capsids and are able to protect the encapsidated RNA from challenge by nuclease but with very different CP concentration dependences. All but C1 also show similar initial decreases in Rh to the 127-mer (Fig. 4B). The assembly behavior of C1 resembles that of PS3 alone, suggesting that it has lost cooperativity, and its distribution plot and appearance in EM images (SI Appendix, Fig. S8) suggests that it has also lost the ability to regulate capsid formation efficiently. In contrast, the importance of the central PS folding propensity is illustrated by the behavior of C2. Despite the potential issues with a folding ensemble, it shows a similar collapse to the 127-mer and a cooperative assembly to T = 1 particles with an Rh distribution similar to the wild-type fragment. It assembles into VLPs at lower CP concentrations than the wild-type 127-mer, i.e., under these conditions it is a better assembly substrate. Remarkably, C3 also assembles more efficiently than wild type, even though it encompasses PSs that are longer than those found in the 127-mer, suggesting that there is some leeway in the PS secondary structure context in which the recognition motif is presented. This is a little surprising given the critical dependency on PS spacing around PS3 observed previously (12). The efficiency of assembly and the folding propensity of C3 notwithstanding, C4 is by far the best assembly substrate, assembling to VLPs most efficiently (i.e., it assembles more rapidly following the 100-nM CP titration point) (Fig. 4B).
These results suggest that it is possible to abstract the critical assembly features from a viral genomic RNA fragment. Given the alterations in the stem lengths and loop sizes in the synthetic fragments it would also appear that there is considerable scope for engineering templates with improved PS folding propensity.
Transfer of Critical Assembly Features to Genomic-Scale RNAs.
As a test of whether these experiments have successfully identified essential assembly features, we examined how inclusion of this improved RNA “cassette” alters the assembly efficiency of the STNV-1 genome. As a control we created a genome lacking PSs by altering all AXXA motifs within stable SLs to UXXU (ΔAXXA) (SI Appendix, Fig. S9). This has only a modest effect on the total number of SLs that can form in the modified genome. We then created a series of chimeric genomes fusing C1, C4, and a 127-mer PS1–PS5U, in which all the loop motifs are substituted by Us, which we have shown previously are unable to support assembly in isolation (12), onto the wild-type genomic fragment from 128 to 1239 nt. In addition, we fused C4 to the equivalent ΔAXXA 3′ fragment (SI Appendix, Table S4). The resultant smFCS assembly curves, Rh distributions, and EM images are shown in Fig. 5 and SI Appendix, Fig. S9. The ΔAXXA genome aggregates, the RNA remains RNase-sensitive, and it is clearly a very poor assembly substrate, as expected for an RNA lacking PSs. C4, which is the best assembly substrate in isolation, only partially reverses these properties, creating an RNA that collapses but remains RNase-sensitive, forming only a few misshapen VLPs. This result confirms that genome assembly relies on PSs outside the 127-mer. Indeed, PS1–PS5U 127-mer lacking CP-binding motifs fused to a wild-type 3′ fragment has improved assembly properties, although it fails to collapse and appears to form aggregates of T = 1 capsids that resolve into discrete particles and malformed structures on RNase treatment. Clearly, well-regulated assembly requires both sets of PSs to be functional, and this is confirmed with the C1 chimera, which collapses but struggles to form T = 1 capsids, with the RNA remaining accessible to nuclease. The proof of these ideas is seen with C4 fused to a wild-type 3′ fragment. It collapses rapidly to T = 1, forming nuclease-resistant T = 1 shells in high yield. Thus, an artificial sequence encompassing improved PS characteristics is able to regulate the assembly pathway of a fragment that is over 10 times its own size.
Fig. 5.
Assembly assays with genomic chimeras: (A) STNV CP was titrated into 1 nM of STNV-1 (black), C1-WT (magenta), or C4-WT (cyan) genomes, and the resulting Rh was monitored using smFCS. The Rh of the recombinant T = 1 particle is indicated in orange. (Inset) Color-coded EM images of the resultant products. (B) STNV CP was titrated into 1 nM of ∆AXXA (blue), PS1–PS5U-WT (red), or C4-∆AXXA (green) genomes, and the resulting Rh was monitored using smFCS. The Rh of the recombinant T = 1 particle is indicated in orange. (Inset) Color-coded EM images of the resultant products.
Confirmation of this interpretation of the results was obtained by directly comparing assembly efficiency of wild-type genome versus the C4–wild-type chimera, each carrying a different dye, under conditions where there was only enough CP to package one of these RNAs fully. QELS and EM images of the products following elution from a gel filtration column identifies two types of STNV VLPs. Fluorescence emission measurements (SI Appendix, Table S5) for each VLP suggest that the chimera constitutes up to 70% of the resultant VLPs (SI Appendix, Fig. S9), i.e., the C4 cassette chimera outcompetes the wild-type genome for CP binding.
Discussion
We have shown that the dual codes inherent in RNA PS-mediated virus assembly, i.e., that genomic RNAs simultaneously encode a genetic message as well as instructions for efficient capsid assembly, are separable. An important question is why do the codes not separate during the course of viral evolution, especially as replication in ssRNA viruses occurs via error-prone processes that lead to creation of a quasi-species of genome variants. There are now three examples of viruses using RNA PS-mediated virus assembly where we have structural information that partially answers this question. In bacteriophage MS2 (12), human parechovirus-1 (19) and STNV (12), at least one of the PS sites in the genome also encodes amino acid residues forming part of the PS-binding site. This intimate embedding of both codes has the consequence of favoring assembly only of progeny RNAs in which PS-mediated assembly persists. Similarly, the density of functions encoded within such RNAs is well-known. The natural 5′ 127-mer in the STNV genome also forms an essential transcriptional/translational enhancer contact with the 3′ end sequence. Since that structure and assembly are mutually excluding functions, the natural sequence has evolved to balance their relative propensities of formation to enable both functions at appropriate stages in the viral lifecycle.
The focus here is the assembly code liberated from the wild-type viral RNA sequence. Indeed, by sequentially investigating each aspect of the STNV assembly sequence in its natural context we have been able to reproduce its effects in triggering in vitro assembly of STNV CPs using a synthetic nonviral RNA cassette. Additional refinements allowed us to produce sequences that are either less or more efficient than the wild-type STNV 127-mer, and we demonstrated that these effects can be transferred to genome-length RNAs. These results confirm the nature of PS-mediated assembly for STNV. Assembly in vitro initiates within the 127-mer by CP recognition of the PS3 stem-loop. Higher-order CP binding is dependent on the correct positioning and folding of the neighboring PSs (PS2 and PS4), each presenting a consensus CP recognition motif in the loop. The 127-mer potentially encompasses five PSs that make the initial binding cooperative with respect to protein concentration, leading to a collapse in the hydrodynamic radius of the RNA, a necessary precursor to encapsidation. Thereafter additional PSs 3′ to the 127-mer ensure accurate completion of the viral capsid. Electrostatic interactions contribute to these protein-RNA contacts but are not the major driving force, which instead is a high-affinity sequence-specific interaction of the stem and loop regions of the PSs with the inner surface of the protein capsid.
Previously, Wilson and colleagues (37, 38) showed they could direct assembly of nonviral RNAs into rods of tobacco mosaic virus (TMV) CP by creating RNA chimeras encompassing the TMV assembly initiation site. This was successful, with the length of the protein-coated rods formed being determined by the length of the RNA being packaged, as expected from the known assembly mechanism. This approach was less successful when applied to spherical ssRNA viruses, where the highest-affinity MS2 PS has positive effects on in vitro encapsidation of short RNAs, but is less important on longer ones (39). Note, all these experiments were done at micromolar concentrations, where the effects of PS-mediated assembly are obscured by the tendency of the CP-CP to form spontaneously (40). The results described above suggest an efficient route for encapsidation of bespoke, nonviral RNAs in shells of viral CPs. In vitro assembly may be possible for a large number of CP-RNA combinations, but it differs from in vivo assembly where, in many viruses, there is good evidence suggesting that only nascent genomic transcripts emerging from the viral polymerase complex are packaged into progeny virions. In such reactions, the RNA is very likely to fold kinetically, avoiding some of the issues with RNA conformational ensembles in the in vitro reactions such as those described here.
Viruses and VLPs are finding increasing potential in medical applications as gene therapy or drug-delivery vectors (28–30), as well as acting as nonreplicating synthetic vaccines. Viral protein shells are also of interest for nanotechnology applications. The results described here offer an important insight into ways to create such structures with high efficiency and potentially carrying nonviral RNAs with advantageous properties. This will be essential for the production of designer synthetic virions.
Materials and Methods
Wild-type and mutant STNV CP was prepared by dissociation of recombinant VLPs produced in Escherichia coli. RNAs used in assembly assays were either transcribed as described previously (12), or gene blocks were purchased (IDT) and cloned into a PACYC184 plasmid for subsequent transcription (SI Appendix). smFCS assays and data analysis were performed as previously described, with any variations described in SI Appendix (11).
Supplementary Material
Acknowledgments
We thank Profs. Peter Prevelige, University of Alabama, Birmingham, and Ian Robinson, University College London, for stimulating our analysis of “synthetic” packaging sequences. This work was supported by grants from the UK Biotechnology and Biological Sciences Research Council (BB/J00667X/1 and BB/L022095/1). R. Twarock acknowledges funding via a Royal Society Leverhulme Trust Senior Research Fellowship (LT130088). P.G.S. and R. Twarock thank The Wellcome Trust for financial support for virus work (Joint Investigator Awards 110145 and 10146).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706951114/-/DCSupplemental.
References
- 1.Stockley PG, et al. Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid. Bacteriophage. 2016;6:e1157666. doi: 10.1080/21597081.2016.1157666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prevelige PE., Jr Follow the yellow brick road: A paradigm shift in virus assembly. J Mol Biol. 2016;428:416–418. doi: 10.1016/j.jmb.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Garmann RF, et al. Role of electrostatics in the assembly pathway of a single-stranded RNA virus. J Virol. 2014;88:10472–10479. doi: 10.1128/JVI.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudnick J, Bruinsma R. Icosahedral packing of RNA viral genomes. Phys Rev Lett. 2005;94:038101. doi: 10.1103/PhysRevLett.94.038101. [DOI] [PubMed] [Google Scholar]
- 5.van der Schoot P, Bruinsma R. Electrostatics and the assembly of an RNA virus. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71:061928. doi: 10.1103/PhysRevE.71.061928. [DOI] [PubMed] [Google Scholar]
- 6.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci USA. 2006;103:17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balint R, Cohen SS. The effects of dicyclohexylamine on polyamine biosynthesis and incorporation into turnip yellow mosaic virus in Chinese cabbage protoplasts infected in vitro. Virology. 1985;144:194–203. doi: 10.1016/0042-6822(85)90317-4. [DOI] [PubMed] [Google Scholar]
- 8.Bruinsma RF. Physics of RNA and viral assembly. Eur Phys J E Soft Matter. 2006;19:303–310. doi: 10.1140/epje/i2005-10071-1. [DOI] [PubMed] [Google Scholar]
- 9.Beren C, Dreesens LL, Liu KN, Knobler CM, Gelbart WM. The effect of RNA secondary structure on the self-assembly of viral capsids. Biophys J. 2017;113:339–347. doi: 10.1016/j.bpj.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qu F, Morris TJ. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J Virol. 1997;71:1428–1435. doi: 10.1128/jvi.71.2.1428-1435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borodavka A, Tuma R, Stockley PG. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc Natl Acad Sci USA. 2012;109:15769–15774. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel N, et al. Revealing the density of encoded functions in a viral RNA. Proc Natl Acad Sci USA. 2015;112:2227–2232. doi: 10.1073/pnas.1420812112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dykeman EC, Stockley PG, Twarock R. Solving a Levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proc Natl Acad Sci USA. 2014;111:5361–5366. doi: 10.1073/pnas.1319479111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basnak G, et al. Viral genomic single-stranded RNA directs the pathway toward a T=3 capsid. J Mol Biol. 2010;395:924–936. doi: 10.1016/j.jmb.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolfsson Ó, Toropova K, Ranson NA, Stockley PG. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J Mol Biol. 2010;401:309–322. doi: 10.1016/j.jmb.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolfsson Ó, et al. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J Mol Biol. 2016;428:431–448. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford RJ, et al. Satellite tobacco necrosis virus (STNV) virus like particle in complex with the B3 aptamer. J Mol Biol. 2013;425:1050–1064. doi: 10.1016/j.jmb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart H, et al. Identification of novel RNA secondary structures within the hepatitis C virus genome reveals a cooperative involvement in genome packaging. Sci Rep. 2016;6:22952. doi: 10.1038/srep22952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shakeel S, et al. Genomic RNA folding mediates assembly of human parechovirus. Nat Commun. 2017;8:5. doi: 10.1038/s41467-016-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel N, et al. HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Nat Microbiol. 2017;2:17098. doi: 10.1038/nmicrobiol.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dykeman EC, et al. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J Mol Biol. 2011;408:399–407. doi: 10.1016/j.jmb.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 22.Toropova K, Stockley PG, Ranson NA. Visualising a viral RNA genome poised for release from its receptor complex. J Mol Biol. 2011;408:408–419. doi: 10.1016/j.jmb.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 23.Dent KC, et al. The asymmetric structure of an icosahedral virus bound to its receptor suggests a mechanism for genome release. Structure. 2013;21:1225–1234. doi: 10.1016/j.str.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koning RI, et al. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat Commun. 2016;7:12524. doi: 10.1038/ncomms12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorzelnik KV, et al. Asymmetric cryo-EM structure of the canonical Allolevivirus Qβ reveals a single maturation protein and the genomic ssRNA in situ. Proc Natl Acad Sci USA. 2016;113:11519–11524. doi: 10.1073/pnas.1609482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai X, et al. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature. 2017;541:112–116. doi: 10.1038/nature20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Q, et al. Genetic, structural, and phenotypic properties of MS2 coliphage with resistance to ClO2 disinfection. Environ Sci Technol. 2016;50:13520–13528. doi: 10.1021/acs.est.6b04170. [DOI] [PubMed] [Google Scholar]
- 28.Galaway FA, Stockley PG. MS2 viruslike particles: A robust, semisynthetic targeted drug delivery platform. Mol Pharm. 2013;10:59–68. doi: 10.1021/mp3003368. [DOI] [PubMed] [Google Scholar]
- 29.Wu M, Brown WL, Stockley PG. Cell-specific delivery of bacteriophage-encapsidated ricin A chain. Bioconjug Chem. 1995;6:587–595. doi: 10.1021/bc00035a013. [DOI] [PubMed] [Google Scholar]
- 30.Li J, et al. Messenger RNA vaccine based on recombinant MS2 virus-like particles against prostate cancer. Int J Cancer. 2014;134:1683–1694. doi: 10.1002/ijc.28482. [DOI] [PubMed] [Google Scholar]
- 31.Bunka DHJ, et al. Degenerate RNA packaging signals in the genome of satellite tobacco necrosis virus: Implications for the assembly of a T=1 capsid. J Mol Biol. 2011;413:51–65. doi: 10.1016/j.jmb.2011.07.063. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaempfer R, van Emmelo J, Fiers W. Specific binding of eukaryotic initiation factor 2 to satellite tobacco necrosis virus RNA at a 5′-terminal sequence comprising the ribosome binding site. Proc Natl Acad Sci USA. 1981;78:1542–1546. doi: 10.1073/pnas.78.3.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sosnick TR, Fang X, Shelton VM. Application of circular dichroism to study RNA folding transitions. Methods Enzymol. 2000;317:393–409. doi: 10.1016/s0076-6879(00)17026-0. [DOI] [PubMed] [Google Scholar]
- 36.Unge T, et al. Satellite tobacco necrosis virus structure at 4.0 Å resolution. Nature. 1980;285:373–377. [Google Scholar]
- 37.Sleat DE, Turner PC, Finch JT, Butler PJ, Wilson TM. Packaging of recombinant RNA molecules into pseudovirus particles directed by the origin-of-assembly sequence from tobacco mosaic virus RNA. Virology. 1986;155:299–308. doi: 10.1016/0042-6822(86)90194-7. [DOI] [PubMed] [Google Scholar]
- 38.Gallie DR, Plaskitt KA, Wilson TM. The effect of multiple dispersed copies of the origin-of-assembly sequence from TMV RNA on the morphology of pseudovirus particles assembled in vitro. Virology. 1987;158:473–476. doi: 10.1016/0042-6822(87)90225-x. [DOI] [PubMed] [Google Scholar]
- 39.Witherell GW, Wu HN, Uhlenbeck OC. Cooperative binding of R17 coat protein to RNA. Biochemistry. 1990;29:11051–11057. doi: 10.1021/bi00502a006. [DOI] [PubMed] [Google Scholar]
- 40.Beckett D, Uhlenbeck OC. Ribonucleoprotein complexes of R17 coat protein and a translational operator analog. J Mol Biol. 1988;204:927–938. doi: 10.1016/0022-2836(88)90052-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.