Significance
Human primordial germ cell-like cells (hPGCLCs) generated from pluripotent stem cells in vitro hold promise, with broad applications for studies of human germline cells. We show that hPGCLCs generated using several distinct protocols are transcriptomally comparable and that primed pluripotency human iPSCs gain competence to generate hPGCLCs after only 72 hours of reprogramming toward ERK-independent state-naïve pluripotency. hPGCLCs were localized in the outermost surface layer of embryoid bodies and strongly expressed CXCR4. Live cell imaging showed active migratory activity of hPGCLCs, and their exposure to the CXCR4 ligand CXCL12/SDF-1 induced enriched expression of promigratory genes and antiapoptotic genes. These results support the resemblance of hPGCLCs to prechemotaxis human embryonic primordial germ cells migrating in the midline region of embryos.
Keywords: primordial germ cells, induced pluripotent stem cells, embryoid body, PRDM1, TFAP2C
Abstract
Pluripotent stem cell-derived human primordial germ cell-like cells (hPGCLCs) provide important opportunities to study primordial germ cells (PGCs). We robustly produced CD38+ hPGCLCs [∼43% of FACS-sorted embryoid body (EB) cells] from primed-state induced pluripotent stem cells (iPSCs) after a 72-hour transient incubation in the four chemical inhibitors (4i)-naïve reprogramming medium and showed transcriptional consistency of our hPGCLCs with hPGCLCs generated in previous studies using various and distinct protocols. Both CD38+ hPGCLCs and CD38− EB cells significantly expressed PRDM1 and TFAP2C, although PRDM1 mRNA in CD38− cells lacked the 3′-UTR harboring miRNA binding sites regulating mRNA stability. Genes up-regulated in hPGCLCs were enriched for cell migration genes, and their promoters were enriched for the binding motifs of TFAP2 (which was identified in promoters of T, NANOS3, and SOX17) and the RREB-1 cell adhesion regulator. In EBs, hPGCLCs were identified exclusively in the outermost surface monolayer as dispersed cells or cell aggregates with strong and specific expression of POU5F1/OCT4 protein. Time-lapse live cell imaging revealed active migration of hPGCLCs on Matrigel. Whereas all hPGCLCs strongly expressed the CXCR4 chemotaxis receptor, its ligand CXCL12/SDF1 was not significantly expressed in the whole EBs. Exposure of hPGCLCs to CXCL12/SDF1 induced cell migration genes and antiapoptosis genes. Thus, our study shows that transcriptionally consistent hPGCLCs can be readily produced from hiPSCs after transition of their pluripotency from the primed state using various methods and that hPGCLCs resemble the early-stage PGCs randomly migrating in the midline region of human embryos before initiation of the CXCL12/SDF1-guided chemotaxis.
Pluripotency is the cellular capability to differentiate into primordial germ cells (PGCs) and three embryonic germ layers (1). Studies using mouse models have revealed that pluripotency in vivo is only a transient state gained by cells of the inner cell mass (ICM) and epiblast during the early stages of embryonic development. However, mouse ICM cells and epiblast cells can be maintained indefinitely without losing their pluripotency in cell culture as ES cells (ESCs) and epiblast stem cells, respectively, and engineered epigenetic reprogramming of differentiated cells can gain pluripotency to generate induced pluripotent stem cells (iPSCs). Whereas human pluripotent stem cells (hPSCs) maintained in the mTeSR1 medium on Matrigel has a primed-state pluripotency (1), Gafni et al. (2) showed that it can be converted into a naïve state when grown in the naïve human stem cell medium (NHSM). Although transgene-free conversion of primed hPSCs to naïve state was subsequently achieved under several different cell culture conditions, features of the achieved naïve pluripotency varied significantly (1, 3), and long-term maintenance of pluripotent stem cells (PSCs) in a strongly naïve state may increase risk of chromosomal anomalies (1, 4).
Although PSCs can produce cells resembling PGCs [primordial germ cell-like cells (PGCLCs)] in cell culture (5–14), direct conversion of mouse PSCs to PGCLCs is inefficient (6). Hayashi et al. (6) generated cells resembling early epiblast [epiblast-like cells (EpiLCs)] from mouse PSCs and showed their strong ability to produce PGCLCs as cell aggregates in the presence of bone morphogenetic protein 4 (Bmp4). In the absence of Bmp4, robust PGCLC production can also be achieved by overexpression of three transcription factors (TFs)—namely Prdm1, Prdm14, and Tfap2c (7, 14)—or by a single TF Nanog (8) from exogenous vectors. The high competency of EpiLCs for PGCLC production is transient, peaking at day 2 of EpiLC induction but largely lost at day 3 (6, 10). In a previous study (10), we showed robust and global demethylation of genomic DNA (gDNA) involving erasure of DNA methylation at CpG dinucleotides in the imprinting control regions.
In the first report of efficient human primordial germ cell-like cell (hPGCLC) production from PSCs, Irie et al. (15) maintained human induced pluripotent stem cells (hiPSCs) in an ERK-independent naïve pluripotency state for at least 2 wk in a modified NHSM containing LIF, FGF2, and TGF-β1 and four chemical inhibitors (4i) of MEK, p38MAPK, GSK3β, and JNK. Aggregates of hPSCs maintained in the 4i medium robustly differentiated into hPGCLCs, which strongly express NANOS3, TNAP, and CD38 (15). Efficiency of hPGCLC production from the conventional primed pluripotency hiPSCs was very low (<5%) (15), which was later confirmed by Sasaki et al. (16). Including Irie et al. (15), at least four laboratories reported efficient hPGCLC production from PSCs (9, 15–18) but with significant variations in the intermediate cell cultures and marker antigens used for FACS enrichment of hPGCLCs (9, 16, 17) (Table S1).
In this study, we show that a 72-h exposure of the primed pluripotency hiPSCs in the 4i medium is sufficient for a robust production of CD38+ hPGCLCs. In contrast to mouse germ cell development, induction of PRDM1 or TFAP2C did not seem to be the primary determinant of hiPSC differentiation to hPGCLCs, agreeing with observations made by Irie et al. (15). hiPSC differentiation to hPGCLCs in embryoid bodies (EBs) was associated with enriched induction of genes involved in cell migration, and most hPGCLCs were observed at the outermost surface monolayer of EBs. Live cell imaging revealed actively migrating hPGCLCs forming cellular protrusions. All hPGCLCs expressed the CXCR4 chemotaxis receptor, whereas its ligand CXCL12/SDF1 was not significantly expressed in any cells in EBs. Exposure of hPGCLCs to CXCL12/SDF1 induced genes involved in cell migration or antiapoptosis. These results suggest that hPGCLCs in EBs resemble early-stage PGCs randomly migrating in the midline region of human embryos before initiation of their directional migration (i.e., chemotaxis) toward genital ridges under the CXCR4-CXCL12 signaling.
Results
Production of CD38+ hPGCLCs from Short-Term 4i Reprogrammed hiPSCs.
Since previous studies showed robust production of hPGCLCs via various precursor cell cultures (Table S1), we speculated that the primed pluripotency state may specifically prevent hPSCs from germline differentiation, while various degrees of deviation from it might be more permissive. To examine this possibility, we exposed primed pluripotency hiPSCs (clone A4; 46 + XY diploid) to the 4i medium for total 72 h (48-h exposure as monolayer cultures followed by 24-h exposure as EBs) and attempted to produce hPGCLCs using the protocol described by Irie et al. (15) (Fig. 1A). Colonies of A4 iPSCs grown in the mTeSR1 medium on feeder cells showed typical morphology of primed pluripotency hPSCs (Fig. 1B, Left), whereas colonies grown for 48 h in the 4i medium already showed a densely packed, sharp-edged appearance (Fig. 1B, Center), which agrees with the reported characteristics of naïve hPSCs (2). After 72 h of growth in the 4i medium, expression of DNMT3A and DNMT3B in A4 iPSCs dramatically decreased, whereas expression of TET1 or DNMT1 was unchanged or only modestly suppressed, respectively (Fig. 1C, Left). Strong suppression of DNMT3B expression was obvious after only 24-h incubation in the 4i medium (Fig. 1C, Right). The rapid changes in expression of the DNMT genes agreed with the characteristics of hiPSCs grown in the NHSM observed by Gafni et al. (2). These results support that a 72-h incubation in the 4i medium was sufficient to significantly deviate the pluripotency of hiPSCs out of the standard primed state maintained in the mTeSR1 medium. Because both the NHSM and 4i media commonly contain 1 μM PD0325901 ERK1/2 inhibitor (2, 15), we speculate that the pluripotency of hiPSCs after a 72-h 4i culture may be shifted toward the ERK-independent naïve state, although exact characteristics are unknown.
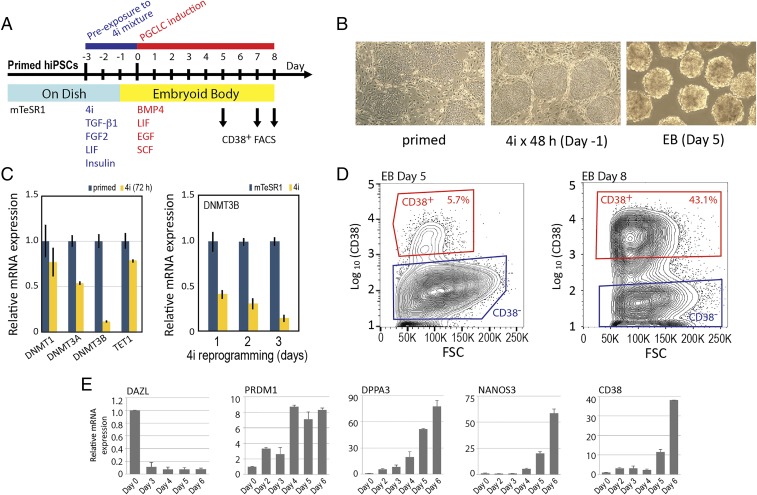
Fig. 1.
Production of hPGCLCs from primed pluripotency hiPSCs via 72-h reprogramming toward naïve pluripotency. (A) Diagram of the 11-d schedule of hPGCLC production from primed pluripotency hiPSCs. (B) Phase contrast images of hiPSCs before and after reprograming toward naïve pluripotency in the 4i medium and day 5 EBs. (C) mRNA expression of TET1 and DNA methyltransferase genes in the primed and 4i reprogrammed hiPSCs. TaqMan real-time qPCR measurements (n = 3, mean ± SD). (D) FACS enrichment of CD38+ and CD38− EB cells from days 5 and 8 EBs. FSC, forward scatter. (E) mRNA expression of germline markers in EBs. TaqMan real-time qPCR measurements (n = 6, mean ± SD).
After a 48-h culture in the 4i medium, we casted hiPSCs into the AggreWell microwells for rapid formation of EBs using the spin EB method (19). EBs were formed in the 4i medium within 24 h and then incubated in the PGCLC medium for 5–8 d (Fig. 1B, Right) followed by single-cell dissociation and FACS enrichment of hPGCLCs as CD38+ cells. FACS contour plots showed a readily discerned, distinct cell population of CD38+ cells, which corresponded to ∼6 and ∼43% of FACS-sorted cells after 5 and 8 d of EB culture in the hPGCLC production medium, respectively (Fig. 1D), whereas CD38+ mRNA was scarcely detected in day 0 EB (Fig. 1C). Real-time qPCR of the total EB cell lysates revealed that expression of PRDM1, an early PGC marker gene that encodes a TF required for PGC specification (15, 20, 21), was induced by day 2 and reached maximal strength by day 4 (Fig. 1E). Expression of three other early PGC marker genes, DPPA3, NANOS3, and CD38, strongly increased from day 4 to 5, whereas their expression at day 0 was scarcely detectable (Fig. 1E). Expression of DAZL, a marker of naïve pluripotency as well as late-stage germline development (22), was rapidly suppressed in EBs, agreeing with previously published studies using other methods of hPGCLC production (16, 20). These results indicate that a 72-h reprogramming of primed pluripotency hiPSCs in the 4i medium is sufficient for subsequent robust production of CD38+ hPGCLCs.
Transcriptomal Profiling of CD38+ hPGCLCs Produced via 72-h Pre-EB 4i Reprogramming.
To obtain insight into mechanisms of germline differentiation of hiPSCs, we determined transcriptomes of the CD38+ hPGCLCs and CD38− cells in day 5 EBs and their precursors. Unsupervised hierarchical clustering of transcriptomes (Fig. 2A and Fig. S1 A and B) identified genes significantly expressed in both CD38+ and CD38− day 5 EB cells but only weakly expressed in their precursors (cluster 1; 81 genes), genes expressed predominantly in CD38+ cells over other cell types (cluster 2; 130 genes), and genes suppressed only in CD38− cells (cluster 3; 153 genes). Positions of these clusters are indicated on the right in Fig. 2A, and their enlarged images with full lists of genes are shown in Fig. S1C. A heat map of marker genes selected from Fig. 2A is also shown as Fig. 2B.
Fig. 2.
Transcriptomal profiles of CD38+ hPGCLCs and precursors. (A and B) Unsupervised hierarchical clustering of CD38+ hPGCLCs and their precursors based on transcriptomes (A) or 24 marker genes (B). Details of the three gene clusters 1–3 indicated in A are shown in Fig. S1 C1–C3, respectively. RNA-seq data were obtained from three independently performed experiments. Gray color indicates “zero value” data, reflecting the absence of expression. (C) Transcriptomal PCA. Arrows indicate the path of hiPSC differentiation to CD38+ hPGCLCs and CD38− EB cells. (D) RNA-seq tracks at the 3′-UTR of PRDM1 mRNA. Positions of exons 6–8, 3′-UTR, and miRNA binding sites are indicated. (E) GO analysis of DEGs between CD38+ hPGCLCs and CD38− EB cells.
Cluster 1 genes included PRDM1 and naïve pluripotency/ICM markers TCL1B and TFCP2L1 (16, 20). TFAP2C and another ICM marker KLF4 (16, 20) were also expressed in both CD38+ and CD38− EB cells relatively strongly, although varying degrees of weaker expression of TFAP2C were observed with some of the precursor hPSC cells (Fig. 2 A and B). The weak expression of TFCP2L1 and KLF4 in both the primed hiPSCs and the short-term 4i reprogrammed hiPSCs was consistent with the previously reported characteristics of the primed pluripotency hPSCs and the ERK-independent naïve pluripotency hiPSCs, respectively (2, 3, 15), indicating that robust production of CD38+ hPGCLCs does not require strong expression of these markers of naïve pluripotency in the pre-EB precursor cultures. Instead, a deviation from the primed pluripotency achieved after a 72-h incubation in the 4i medium seems sufficient. However, the ICM markers TCL1B, TFCP2L1, and KLF4 were strongly expressed in CD38− cells in day 5 EBs, suggesting a certain degree of commonality in the gene regulation network between ICM and EB cells incubated in the hPGCLCs. Cluster 2 genes included known markers of human primordial germ cells (hPGCs)/hPGCLCs, such as NANOS3, SOX15, SOX17, and CD38 (15, 22, 23), as well as mesodermal markers T, HOXD1, and WNT3 and a naïve pluripotency marker TCL1A (16). Cluster 3 genes were enriched with markers shared by hPSCs and hPGCs/hPGCLCs, such as ALPL, KIT, NANOG, UTF1, and POU5F1 (15, 16). Many pluripotency markers (but not hPGC/hPGCLC markers), such as LIN28A, SALL4, DPPA4, PRDM14, ZIC3, and SOX2, were strongly expressed in the pluripotent precursor cells (including day 0 EB cells) but only weakly expressed in day 5 EBs. Transcriptomal principal component analysis (PCA) (Fig. 2C) showed reproducibility of our protocol, although EB cell differentiation to nongermline lineages seemed somewhat stochastic.
The marker gene expression profile of our CD38+ hPGCLCs was nearly identical to those of the CD38+/TNAP+ hPGCLCs produced by Irie et al. (15) from hiPSCs grown in the 4i medium for at least 2 wk, confirming that the CD38+ cells produced in this study via a 72-h 4i reprogramming were bona fide hPGCLCs. Choi et al. (4) recently reported (together with J.O. and T.S.) that there is a significant risk of chromosomal aneuploidy and anomalous DNA demethylation in mouse PSCs after prolonged maintenance (≥20 passages) in the ERK1/2-independent naïve pluripotency state; our modification of hPGCLC production protocol using the ERK1/2-inhibiting 4i medium for only a limited period of time may be beneficial to reduce the risk of genetic mutations and epimutations, although the effect of prolonged ERK1/2-repressing culture on hPSCs remains to be shown.
Not only CD38+ hPGCLCs but also CD38− cells in day 5 EBs significantly expressed PRDM1 and TFAP2C, which are early-stage hPGC/hPGCLC markers (15), although expression of PRDM1 was weaker in CD38− cells than CD38+ cells (Fig. 2 A and B and Fig. S1 C1 and D). However, the mRNA transcripts for PRDM1 expressed in CD38− cells lacked most of the 3′-UTR, whereas CD38+ hPGCLCs expressed transcripts with the full-length 3′-UTR (24, 25) (Fig. 2D and Fig. S2B). Such 3′-UTR length polymorphism seemed to be specific to PRDM1 among genes shown in Fig. 2B (Fig. S2B). The 3′-UTR of PRDM1 mRNA missing in CD38− cells contained binding sites for miRNA let-7, miR-9, and miR-BHRF1-2 (Fig. 2D and Fig. S2B), which play significant roles in PRDM1 protein degradation in normal or malignant human B lymphocytes (26–28). West et al. (21) reported that in vitro differentiation of mouse ESCs to PGCLCs is dependent on suppression of let-7 miRNA by Lin28 and that overexpression of Prdm1 rescued differentiation of lin28-deficient ESCs to PGCLCs. In our hPGCLC production system, both LIN28A and LIN28B were significantly expressed in CD38+ and CD38− cells (Fig. S1D). The possible importance of the 3′-UTR polymorphism of PRDM1 mRNA and/or let-7 miRNA in hiPSC differentiation to hPGCLCs remains to be determined.
Gene Ontology (GO) analysis of the differentially expressed genes (DEGs) between CD38+ and CD38− EB cells [1,445 genes; false discovery rate (FDR) < 5%; more than fourfold change] revealed very strong enrichment of genes involved in cell migration (Fig. 2E, Left) and plasma membrane activities (Fig. 2E, Right), both of which included formation of cell projections. Most of the DEGs supporting these enrichments were expressed more strongly in CD38+ hPGCLCs than CD38− EB cells. These results suggest that the germline differentiation of EB cells may affect their ability to change their shape for migration, agreeing with the reported highly migratory feature of hPGCs (29).
We attempted to determine whether CD38+ hPGCLCs in day 8 EBs reflect a more advanced stage of germline development than day 5 hPGCLCs (Fig. S3). Transcriptomal PCA (Fig. S3C) and hierarchical clustering of 3,500 DEGs with the lowest FDRs (Fig. S3D) revealed a clear distinction between days 5 and 8 CD38+ hPGCLCs. However, GO analysis of these DEGs did not reveal any meaningful enrichment. Expression of marker genes for pluripotency or early-stage hPGCs/hPGCLCs was unchanged between days 5 and 8, except for a slight decrease in LIN28A expression (Fig. S3E), and expression of late-stage hPGC marker genes, such as DAZL or DDX4, was not significant. These results suggest that hPGCLCs at day 8 EB culture did not reflect more advanced germline differentiation than at day 5. Interestingly, 152 DEGs that were expressed more strongly in day 8 hPGCLCs than day 5 hPGCLCs (Fig. S3 D, bar F and F) were significantly enriched with processed pseudogenes derived from ribosomal protein (RP) genes (eight RPS and three RPL pseudogenes; P < 0.0001 by permutation test) (Fig. S3F, asterisks). Since expression of processed RP pseudogenes shows strong tissue specificity, presumably reflecting distinct epigenetic organizations (30), their differential expression in hPGCLCs might reflect epigenetic changes between days 5 and 8 EB cultures.
To examine the reproducibility of the transcriptomal profiles across independent hiPSC clones, we generated two more hiPSC clones “A5” and “B6” from human neonatal foreskin fibroblasts using the same method by which clone A4 was generated. All three hiPSC clones had normal diploid karyotype (46, XY) (Fig. S4 A and C) and strongly expressed TRA-1–60, TRA-1–81, POU5F1, and NANOG. These hiPSC clones were subjected to hPGCLC production [RNA-sequencing (RNA-seq) data normalization (Fig. S4 D and E), hierarchical clustering (Fig. S4 F, all DEGs and G, selected marker genes), and transcriptomal PCA (Fig. S4H)], and CD38+ hPGCLCs were isolated from day 7 EBs. Unsupervised clustering (Fig. S4 F and G) and PCA (Fig. S4H) showed strong reproducibility of transcriptomal profiles for all three hiPSC clones and all four cell types examined. We then compared CD38+ hPGCLC and CD38− EB cell transcriptomes at days 5 (Fig. 2B), 7 (Fig. S4G), and 8 (Fig. S3E) and observed a decrease in PRDM1 and TFAP2C expression in CD38− cells over time, whereas the expression of these genes was maintained in CD38+ hPGCLCs. These results suggest that sustained expression of PRDM1 and TFAP2C in EB cells may be necessary for hiPSC differentiation to hPGCLCs.
Transcriptomal Consistency of hPGCLCs Produced Using Various Protocols and FACS Markers.
To obtain insight into biological consistency of hPGCLCs produced using different protocols (Table S1), we compared transcriptomes of our hPGCLCs with those reported by Irie et al. (15) and Sasaki et al. (16) as well as human fetal gonocytes (23), gestation week 7 hPGCs, gonadal somatic cells, and the TCam-2 human seminoma cell line (15). Hierarchical clustering involving 46 selected pluripotency and germline marker genes clearly separated the subjects into six clusters corresponding to cell types—namely PSCs, CD38− EB cells, hPGCLCs, advanced germline cells (AGCs), gonadal somatic cells, and TCam-2 (Fig. 3A; color-coded protocols and cell types are shown in Fig. 3B). The PSC cluster cells strongly expressed pluripotency markers (Fig. 3A, rectangle 1). Our CD38− EB cells strongly expressed ICM/germline markers LEF1, DPPA3, KLF2, KLF4, TFC2L1, TCL1B, and TFAP2C (Fig. 3A, rectangle 2), whereas expression of other germline markers, such as CD38, NANOS3, or SOX17, was low. PRDM1 was significantly expressed in CD38− EB cells, although less strongly than hPGCLCs. The hPGCLC cluster cells strongly expressed known hPGC/hPGCLC markers (Fig. 3A, rectangle 3). The especially strong expression of five markers—namely T, CD38, PRDM1, NANOS3, and SOX17—in all hPGCLCs produced by three independent laboratories suggests their practical usefulness as an hPGCLC signature, whereas expression of other hPGC/hPGCLC markers, such as TCL1A, KIT, DND1, SOX15, or GATA4, was either heterogeneous or relatively low. Expression of late-stage germline markers, including DAZL or SYCP3, was generally weak in hPGCLCs but strong in week 7 embryonic hPGCs, AGCs, and TCam-2 cells (Fig. 3A, rectangle 4). PCA with these 46 marker genes also separated the subjects into five clusters along cell types (Fig. 3C and Fig. S3G). These results show significant similarity of the CD38+ hPGCLCs produced in this study with those generated in previous reports.
Fig. 3.
Marker gene expression profiles of human germ cells, hPGCLCs, and precursors across various protocols. (A) Unsupervised hierarchical clustering of human embryonic and in vitro germline cells generated in four independent laboratories using different protocols. Clustering was performed based on 46 marker genes of pluripotency (blue), PGC (red), and AGCs (green). Color-coded cell types shown at the top of the heat map are indicated in B. Groups of genes expressed in the (1) pluripotent precursor cells, (2) nongermline EB cells (CD38−), (3) hPGCLCs, and (4) AGCs are shown with yellow rectangles. Gray color indicates zero value data, reflecting the absence of expression. GS, gonadal somatic cells; TC, TCam-2 human seminoma cells. (B) Color identifications of transcriptomal data. Data from this study are indicated by red caps in B and circles in C. Data reported by Sasaki et al. (16), Irie et al. (15), and Gkountela et al. (23) are indicated in green, blue, and orange, respectively. (C) PCA of human embryonic and in vitro germline cells with data shown in A. PC, principal component.
Hierarchical clustering with 131 genes most strongly expressed in our CD38+ hPGCLCs (Fig. S1C2) also roughly separated the subjects along with the cell types (Fig. S5). In this heat map, a set of 34 genes was strongly expressed in hPGCLCs (Fig. S5 A, blue bar and B, enlargement), and 9 of them—namely HOXD1, ALOX5, WNT2, VSTM2B, PLDB1, IRX6, KANK3, TMEM123, and TMPRSS11E (RP11-387A1.6 is identical to the second exon of HOXD1)—were specific to hPGCLC. Other genes, including germline markers NANOS3, SOX17, TCL1A, CD38, T, and WNT3, were also expressed in 7-wk hPGCs, week 16–16.5 gonocytes, or TCam-2. Several known hPGC markers, such as PRDM1, TFAP2C, or KIT, were not included in this heat map, because they were also significantly expressed in CD38− EB cells and/or pluripotent precursors. Strong expression of NANOS3, SOX17, TCL1A, T, and CD38 was also observed in hPGCLCs produced and enriched as KIT+ cells by von Meyenn et al. (9) (Fig. S6 and Table S1). Expression of UTF1, DND1, or SOX15 was weak in their hPGCLCs (Fig. S6B) as well as hPGCLCs produced by other laboratories (Fig. 3A, rectangle 3). Significant induction of NANOS3 and SOX17 was also observed by Sugawa et al. (17) with their hPGCLCs enriched as KIT+/TRA-1–81+ cells (Table S1). Taken together, these results suggest that hPGCLCs produced via different pre-EB precursors and/or FACS enrichment marker antigens have comparable gene expression characteristics.
Enrichment of TF Binding Motifs in Promoters of DEGs Between CD38+ hPGCLCs and CD38− EB Cells.
To obtain insight into the mechanisms of hiPSC differentiation to CD38+ hPGCLCs, we examined enrichment of TF binding motifs in promoter sequences of DEGs between CD38+ and CD38− cells in day 5 EBs using the Transcription Factor (TRANSFAC) database and the F-match algorithm (31). We identified 13 TF binding motifs that were significantly enriched in promoters of 537 genes expressed more strongly in CD38+ hPGCLCs than CD38− EB cells (Fig. 4), whereas no enriched motifs were found with genes down-regulated in CD38+ hPGCLCs. Strikingly, 1 of these 13 enriched TF motifs was the optimal 9-nt TFAP2C binding sequence GCCTGAGGC (32). Although other members of the TFAP2 TFs can also bind to this sequence, expression of TFAP2C was strongest among them in CD38+ hPGCLCs and their precursors (Fig. S1E). The list of 44 genes with promoters that harbor predicted TFAP2 binding consensus motifs (Table S2) included SOX17, NANOS3, and T, which are robustly expressed in hPGCLC (Figs. 2B and 4B, locations of TFAP2 binding motifs), suggesting the functional significance of TFAP2 for hPGCLC induction. However, our list showed only a marginal overlap with a previously reported list of predicted Tfap2c-regulated genes in mouse PGCs (33), sharing as few as three genes—namely NANOS3, PRUNE2, and GSTM1. Among other TFs shown in Fig. 4, RREB-1 is also noteworthy, because it is required for formation of cell protrusions and lamellipodia as well as the dynamic cell–cell interactions (34).
Fig. 4.
TF binding motifs enriched in the regulatory sequences of 537 DEGs expressed more strongly in CD38+ hPGCLCs than in CD38− EB cells. (A) List of significantly enriched TFs (P < 0.01). The P value was defined by the F-match algorithm and calculated by the TRANSFAC server. (B) Locations of the TFAP2 binding motifs in promoters of NANOS3, SOX17, and T/BRACHYURY. Matches to the core motif (GCCNNNGGC) are shown in red, and matches to the additional contributing sequences are shown in orange.
Localization of CD38+ hPGCLCs at Surface of EBs.
Since iPSC differentiation to hPGCLCs was associated with enriched expression of cell migration genes (Fig. 2E), we determined locations of hPGCLCs in EBs by immunohistochemistry. Because an anti-CD38 antibody suitable for identification of hPGCLCs in formaldehyde-fixed, paraffin-embedded (FFPE) slides was not available, we used POU5F1 as an alternative hPGCLC marker antigen (35). Although POU5F1 was expressed in hPSCs, its expression in day 7 EBs was specific to CD38+ hPGCLCs (Fig. 5A). Unexpectedly, we observed POU5F1+ hPGCLCs almost exclusively in the outermost surface monolayer of EB cells (Fig. 5B and Fig. S7B). The majority of the POU5F1+ cells (>90%) were distributed at the EB surface without aggregation (Fig. 5B, Left), although aggregated clusters of POU5F1+ cells were occasionally observed (Fig. 5B, Right). Significantly biased localization of POU5F1+ hPGCLCs in the outermost surface monolayer of EBs was statistically confirmed (Fig. 5C). A weak POU5F1 signal, readily suppressed by low concentrations of a POU5F1 blocking peptide (Fig. S7A), was also detected predominantly at the subsurface area of EBs (Fig. S7B). We also attempted to determine quantitative aspects of expression of PRDM1 protein by immunofluorescence and immunohistochemistry but were unsuccessful because of technical difficulties.
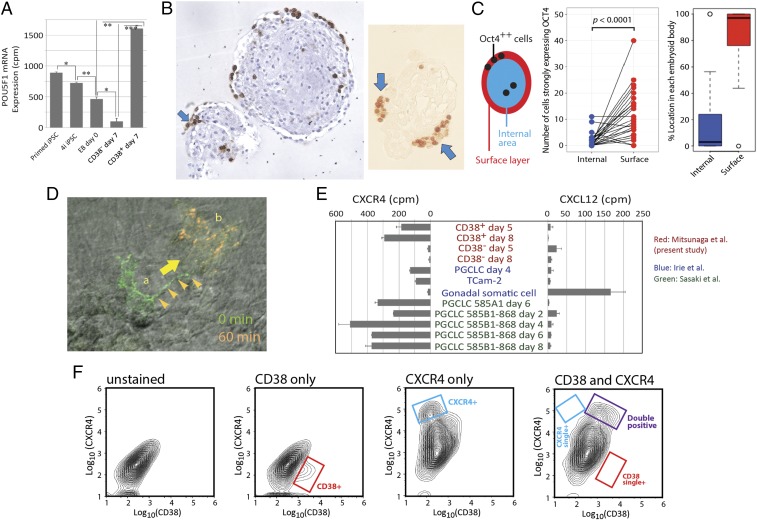
Fig. 5.
Localization of CD38+ hPGCLCs at the outermost surface layer of EBs. (A) Expression of POU5F1 mRNA transcripts: RNA-seq (n = 3, mean ± SEM). Statistical significance (t test) is indicated by asterisks. *P < 0.05; **P < 0.01; ***P < 0.001. (B) Localization of POU5F1+ hPGCLCs at the outermost surface layer of day 8 EBs. hPGCLCs were observed in formaldehyde-fixed, paraffin-embedded EBs by immunohistochemistry. (Left) hPGCLCs distributed at the outermost surface monolayer of EBs (POU5F1 protein is stained brown with hematoxylin counterstaining). (Right) hPGCLCs are found as aggregates at EB surface (no counterstaining). Arrows indicate aggregated cells strongly expressing POU5F1. (C) Statistical evaluation of surface enrichment of POU5F1++ (strongly positive) hPGCLCs. (Left) Definition of the “surface layer,” which is the outermost surface monolayer of EBs, and the “internal area,” which is the remaining region. (Center) Number of POU5F1++ cells in the internal or surface regions of each EB (n = 23; P value was calculated by paired t test). (Right) Box plot representation of percentage localization of POU5F1++ cells in the internal or surface regions of EBs. (D) Time-lapse live imaging of a CD38+ hPGCLC showing active cell migration. A live EB was immobilized on Matrigel overnight, and an hPGCLC that emerged from the EB mass was stained with a fluorescence dye-conjugated anti-CD38 antibody. Fluorescence images were taken before and after a 60-min time lapse and digitally converted to green and orange pseudocolors, respectively. The CD38+ hPGCLC marked as a showed a well-developed cellular protrusion (arrowheads) and moved to the location marked as b within 60 min. (E) Expression of the CXCR4 chemotaxis receptor and its ligand CXCL12 in human germline cells and gonadal somatic cells. RNA-seq data generated in four independent laboratories were conormalized to obtain relative expression profiles (of CXCR4 and CXCL12 mRNA across cell types. cpm, Counts per million. (F) FACS analysis of total day 5 EB cells for cell surface expression of CD38 and CXCR4. The double-stained FACS chart indicates gates for single- and double-positive cells.
Because the DEGs up-regulated in CD38+ hPGCLCs rather than in CD38− EB cells were enriched for cell migration genes and because the majority of hPGCLCs at the EB surface were observed without forming cell aggregates, we speculated that hPGCLCs may have highly migratory characteristics just as human embryonic PGCs do (29). To obtain evidence of hPGCLC migration, we attempted to obtain time-lapse images of CD38+ cells by immunofluorescence staining of live cells. Whole EBs at day 6 were unsuitable for live fluorescence imaging because of strong autofluorescence. However, when EBs were inoculated on Matrigel-coated dishes to spread overnight, cells that emerged from the EB mass and formed a monolayer were largely free of autofluorescence. Taking advantage of this observation, we stained live cells surrounding the immobilized EB masses with a fluorescence dye-conjugated anti-CD38 antibody and obtained antibody-dependent fluorescence images matching cell contours and cellular protrusions (Fig. S7C). Two live fluorescence images taken at a 60-min time interval showed significant changes in CD38+ cell shape and location (in Fig. 5D, fluorescence colors were digitally adjusted to green and orange for images taken before and after the interval, respectively), agreeing with the highly migratory nature of human embryonic PGCs (29). Our attempts to obtain video microscopic tracings of live-stained CD38+ cells were unsuccessful, apparently because of the apparent sensitivity of CD38+ cells to changes in the cell culture environment.
Expression of the CXCR4 Chemotactic Cytokine Receptor in CD38+ PGCLCs and Transcriptional Effects of CXCL12.
In mouse embryos, PGCs migrate laterally from the midline regions toward genital ridges following a gradient of chemoattractant ligand CXCL12/SDF1 secreted by gonadal somatic cell precursors and its plasma membrane receptor CXCR4 expressed by PGCs (36). Consistent with these observations, CXCR4 was strongly expressed in hPGCLCs and in the TCam-2 seminoma cells, while CXCL12 expression was strong in human embryonic gonadal somatic cells (Fig. 5E). Neither CD38+ hPGCLCs nor CD38− EB cells expressed significant amounts of CXCL12 mRNA. FACS contour plots (Fig. 5F) showed that the EB cell population stained only with either anti-CD38 or anti-CXCR4 antibody completely disappeared when cells were double-stained with these two antibodies, while a new CD38+/CXCR4+ double-positive cell population emerged. These results indicated that all CD38+ hPGCLCs expressed CXCR4 and that all CXCR4-positive cells in EBs were CD38+ hPGCLCs.
We next attempted to examine the transcriptional effects of exogenous CXCL12 on gene expression in hPGCLCs (based on three replicates of RNA-seq). CXCL12-activated CXCR4 signaling in human CD34+ hematopoietic stem cells (37) or Jurkat T cells (38) requires Rho GTPase-activated kinases 1/2 (ROCK1/2). However, a potent ROCK1/2 inhibitor Y27632 is included in our hPGCLC medium as well as the media used by Irie et al. (15), Sasaki et al. (16), and Sugawa et al. (17) but not that used by von Meyenn et al. (9) (Table S1). When we removed Y27632 from hPGCLC medium starting at day 5 EB culture, CD38+ cells isolated at day 7 still strongly expressed NANOS3, NANOG, and SOX17 as well as CD38 (Fig. 6A), indicating that Y27632 is dispensable for hPGCLC production on and after day 5. Under this Y27362-free condition, addition of CXCL12 to the hPGCLC medium on and after day 5 resulted in suppression of CXCR4 expression (Fig. 6A), consistent with an observation made with human CD34+ hematopoietic stem cells (37) and possibly reflecting a negative feedback regulation. Expression of PTGER3 (encoding prostaglandin E2 receptor isoform 3) and BCL2 (encoding an antiapoptotic protein) was dramatically induced after exposure to CXCL12, whereas expression of CASP8 (encoding caspase 8, the initiator caspase required for death receptors-induced apoptosis) was suppressed (Fig. 6A). These changes in expression of apoptosis-regulating genes suggest that CXCL12 may reduce CD38+ cell sensitivity to cytokine-induced apoptosis.
Fig. 6.
Effects of CXCL12 on mRNA expression in hPGCLCs. (A) Expression of mRNA transcripts in CD38+ hPGCLCs or CD38− EB cells at day 8 EBs. EBs were cultured in the standard condition until day 5 and then incubated for an additional 3 d in the presence or absence of Rho Kinase Inhibitor (ROCKi) and/or CXCL12 in the PGCLC production medium. Amounts of mRNA expression are shown as counts per million (cpm) values of normalized reads of three repeated RNA-seq experiments. (B and C) GO analysis of DEGs in hPGCLCs produced in the ROCKi-deficient medium in the presence or absence of CXCL12. Three RNA-seq experiments determined 1,190 up-regulated (B) and 735 down-regulated (C) DEGs. Statistically significant GO terms (FDR < 5%) are shown. (D) RNA-seq traces of CXCR4, PTGER3, BCL2, and CASP8 mRNA expression. The height of peaks represents relative mRNA expression of a gene in the six cell culture conditions and was normalized for each gene. ZRAMB2 locates adjacent to PTGER3 and is shown as control. KDSR and VPS4B are controls located adjacent to BCL2. ACTB expression shows quantitative reproducible RNA-seq experiments across the six different cell culture conditions. Note that PTGER3 and BCL2 traces are shown only at representative exons.
To predict the transcriptomal impact of CXCL12 on CD38+ hPGCLCs, DEGs were identified from the three replicates of RNA-seq data (1,190 up-regulated and 735 down-regulated in the presence of CXCL12 during days 5–8 of EB culture in the PGCLC medium lacking Y27632) and subjected to GO analysis using the DAVID server. Indeed, GO analysis of CXCL12-induced DEGs in CD38+ hPGCLCs showed significant enrichment of genes involved in negative regulation of apoptosis as well as cell migration (Fig. 6B). However, CXCL12-suppressed DEGs were enriched for genes involved in various aspects of mitotic nuclear division (Fig. 6C). These results suggest that CXCR4 activation in hPGCLCs may promote cell migration coupled with suppression of apoptosis and mitotic nuclear division. Additional studies will be needed to examine in detail the biological functions of CXCR4 in hPGCLCs and their relevance to human embryonic PGCs in vivo.
Discussion
Our study provides evidence that hPGCLCs produced using different protocols (Table S1), including ours involving only transient exposure of primed pluripotency hiPSCs to the 4i medium (Fig. 1), share concordant transcriptomal characteristics, despite a variety of pre-EB precursors and FACS enrichment strategies (Fig. 3). Based on the transcriptomal profiling along with steps of hPGCLC production (Fig. 2 A and B), we speculate that specific induction of cluster 2 genes may be important for germline differentiation of EB cells, while persistent expression of cluster 3 genes may prevent differentiation to somatic lineages. Irie et al. (15) reported that human ES cells (hESCs) maintained in the 4i medium for more than 2 wk showed a slight up-regulation of T and HAND1 compared with the conventional hESCs, speculating that 4i-reprogrammed hESCs might have primitive streak-like features, which could explain the competence to directly give rise to hPGCLCs. Our data also show a slight but significant increase in expression of T in the short-term 4i reprogrammed hiPSCs compared with the primed-state hiPSCs (Fig. 2B). Future studies will be needed to evaluate the relevance of the short-term 4i reprogrammed cells with the primitive streak-like state. KJF4, TFAP2C, and cluster 1 genes, with importance in specification of hPGC/hPGCLCs that has been shown or predicted, were expressed in both CD38+ and CD38− EB cells. Expression of KLF4, TCL1B, and TFCP2L1, which are markers of ICM-like strong naïve pluripotency (16, 20), may imply a certain degree of commonality of gene networks between EB cells exposed to the hPGCLC medium and ICMs.
The strong expression of hPGC/hPGCLC markers, such as NANOS3 (15), indicates that our CD38+ cells isolated from day 5 EBs are hPGCLCs. However, weak but significant expression of NANOS3 and SOX17 in CD38− EB cells at day 5 (Fig. 2B) might reflect their undergoing commitment to germline differentiation. Kobayashi et al. (18) recently showed that high levels of SOX17 are needed for robust induction of hPGCLCs. While SOX17 is essential for hPGCLC induction (15), it is not a specific germline maker but also labels mesendodermal cells. Additional studies will be necessary to determine whether the predicted hPGCLC precursor cells weakly expressing SOX17 and/or NANOS3 in the CD38− cells in day 5 EBs can later differentiate to hPGCLCs. We also observed that the PRDM1 mRNA transcripts expressed in CD38− EB cells lacked the 3′-UTR harboring miRNA binding sites required for regulation of mRNA stability (26–28) (Fig. 2D and Fig. S2). Whether the 3′-UTR polymorphism of PRDM1 mRNA and/or let-7 miRNA have significance in hiPSC differentiation to hPGCLCs remains to be determined.
Specification of mouse PGCs requires coordinated actions of three TFs—namely Prdm1, Prdm14, and Tfap2C—which have been described as a tripartite TF network (5, 13, 14). In contrast, specification of hPGCLCs is dependent on SOX17, which induces PRDM1 (15) and endoderm genes. PRDM1 plays critical roles in specification of hPGCs/hPGCLCs by suppressing SOX17-induced expression of endodermal genes as well as BMP- or WNT-induced expression of mesodermal genes (13, 18). Whereas TFAP2C mRNA was strongly expressed in both CD38+ hPGCLCs and CD38− EB cells (Figs. 2 A and B and 3 and Figs. S1, S3, and S4), promoter sequences of DEGs up-regulated in hPGCLCs were significantly enriched with the TFAP2 binding motif (Fig. 4A). Interestingly, these possible TFAP2-regulated DEGs up-regulated in hPGCLCs (Table S2) included SOX17, NANOS3, and T (Fig. 4B). These results support the importance of TFAP2C in hiPSC differentiation to hPGCLCs, although its expression is not restricted to hPGCLCs.
GO analysis of DEGs up-regulated in CD38+ hPGCLCs compared with CD38− EB cells revealed significant enrichment of genes involved in cellular activities relevant to migration, including cell projection morphogenesis, or regulation of locomotion (Fig. 2E). Promoter sequences of these DEGs were significantly enriched with the binding motif of RREB-1 (Fig. 4A), a TF required for migration and spreading of human MCF-10A cells (34). Defects of its Drosophila homolog gene hindsignt caused excess accumulation of intercellular adhesion molecules and suppression of cell spreading in ovarian follicular epithelium, indicating its importance in reducing cell–cell adhesion when interconnected groups of cells undergo dynamic changes in cell shape (34). Based on these roles of hindgut/RREB-1, we speculate that human RREB-1 may be important during migration of PGCs, involving transient and reversible adhesion of PGCs to other types of cells as well as to marked changes in cell shape (Fig. 5D).
We detected hPGCLCs in EBs almost exclusively at their outermost surface monolayer (Fig. 5 A and C and Fig. S7 A and B). Live cell imaging revealed active migration of hPGCLCs (Fig. 5D). In mouse EBs it has been shown that only the outermost surface monolayer will differentiate to primitive endoderm (PrE) and noteworthy visceral endoderm (VE) (39, 40). This is because in vivo specification of mouse PGCs from epiblast requires BMP2 secreted from VE to activate the tripartite TF network (5). Before implantation, mouse PrE deposits basement membrane (BM) to its boundary with ICM, and this BM is apparently required for PrE differentiation to VE (40). Similarly, several BM proteins are deposited beneath the outermost monolayer of mouse EBs (41, 42). Mouse PSCs partially differentiated toward PrE emerge in the standard naïve pluripotency culture, and on EB formation, such PrE-primed PSCs move toward the outside because of differential adhesion (39, 40). Exclusive localization of hPGCLCs at the outermost monolayer of EBs might occur for similar reasons. The distribution of hPGCLCs in wide areas of EB surface without aggregation may reflect their random migration on the subsurface BM. Compared with the conventional methods for forming loose cell aggregates using low-attachment U- or V-shaped wells (Table S1), our use of microwells for the spin method EB formation (19) may increase initial density of hiPSCs, resulting in tighter intercellular adhesion, which in turn, affects directed differentiation of EBs (40).
Our GO analysis of DEGs between CD38+ hPGCLCs and CD38− EB cells in day 5 EBs revealed significant enrichment of cell migration genes (Fig. 2E). CXCR4 was strongly expressed in all hPGCLCs (Figs. 5 E and F and 6A), whereas expression of CXCL12 in whole EB cells was weak (Fig. 5E). Expression of c-KIT, another plasma membrane receptor involved in PGC migration and survival, was also strong in CD38+ hPGCLCs (Fig. 2 A and B and Figs. S3 D and E and S4 F and G). Importantly, our hPGCLC production medium contained stem cell factor (SCF), the c-KIT ligand, which is also the case for most other hPGCLC production protocols (Table S1). Removal of SCF from the hPGCLC production medium resulted in significant dismantling of EBs with no yield of hPGCLCs. In mouse embryos, CXCL12 is produced at the genital ridges and diffused into the surrounding mesenchyme, guiding chemotactic migration of CXCR4+ PGCs (36, 43) (Fig. S8). In contrast, SCF is expressed by somatic cells throughout the path of PGC migration to support survival and the random migratory activities of PGCs (43–45) (Fig. S8). Supported by SCF in the medium, hPGCLCs may survive and randomly migrate on the surface of EBs. We, therefore, speculate that hPGCLCs at the surface of EBs may resemble the prechemotactic embryonic hPGCs in the midline regions (Fig. S8). The reported low degree of gDNA demethylation in hPGCLCs agrees with the notion that hPGCLCs resemble midline hPGCs (9, 15–17, 20).
Exposure of hPGCLCs at the surface of EBs to CXCL12 in the absence of ROCK1/2 inhibitor induced cell migration genes and antiapoptotic genes (Fig. 6 A and B), while expression of CASP8 was suppressed (Fig. 6A). These results may imply that the midline hPGCs may be prone to the extrinsic pathway apoptosis until the exposure to CXCL12 (Fig. S8). Exposure of hPGCLCs to CSCL12 also induced PTGER3 (Fig. 6A). Notably, Ptger3 activation has been reported to be required for expression of both Cxcr4 and Cxcl12 by dendritic cells in mouse lymph nodes (46), and a nine-gene biomarker predicting metastatic prognosis of human head and neck squamous cell carcinomas with 90% accuracy included CXCR4, CXCL12, and PTGER3 (47). It is thus interesting to speculate that there may be functional interactions between prostaglandin E2 signaling and CXCL12 signaling in hPGCs/hPGCLCs.
In summary, we have shown robust production of hPGCLCs after a 72-h culture of primed pluripotency hiPSCs in the 4i medium as well as transcriptomal consistency of hPGCLCs using different protocols and/or FACS enrichment antigens. Most hPGCLCs strongly expressed CXCR4 and were localized at the outermost surface monolayer of EBs. Exposure of hPGCLCs to CXCL12 induced migratory and antiapoptotic genes. Thus, our results provide evidence that hPGCLCs produced using different protocols have comparable biological characteristics resembling prechemotactic, embryonic midline hPGCs.
Materials and Methods
Footprint-free hiPSC clones were generated from commercially available, deidentified neonatal foreskin dermal fibroblasts and maintained in mTeSR1 on Matrigel. hPGCLCs were generated from hiPSCs using a protocol based on the study of Irie et al. (15), with modifications involving 72-h exposure of primed pluripotency hiPSCs to the 4i medium and the use of AggreWell microwells for the spin EB technique of EB formation (19). CD38+ and CD38− cells were FACS-enriched from single-cell suspension using FACS. Total RNA was subjected to TaqMan qPCR or RNA-seq using Illumina NextSeq 500, and deep sequencing data were analyzed using the STAR aligner and R/Bioconductor packages Rsubread, edgeR, prcomp, and plot3D. Promoter enrichment analysis of TF binding motifs was performed using the TRANSFAC database. Immunohistochemistry of FFPE serial sections was performed using standard methods. Time-lapse immunofluorescence live imaging was performed on EBs spread on Matrigel using fluorescence dye-conjugated anti-CD38 antibodies and an inverted fluorescence microscope.
Detailed methods are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank Ayaka Abe for technical contributions. This study was supported by Flight Attendant Medical Research Institute (FAMRI) (J.H.H.) and NIH Grants R01ES023316 (to T.S.) and R21ES024861 (to T.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE102943).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707779114/-/DCSupplemental.
References
- 1.Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- 2.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 3.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi J, et al. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnúsdóttir E, Surani MA. How to make a primordial germ cell. Development. 2014;141:245–252. doi: 10.1242/dev.098269. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 7.Nakaki F, et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 8.Murakami K, et al. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature. 2016;529:403–407. doi: 10.1038/nature16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Meyenn F, et al. Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Dev Cell. 2016;39:104–115. doi: 10.1016/j.devcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi N, et al. Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells. Proc Natl Acad Sci USA. 2016;113:9545–9550. doi: 10.1073/pnas.1610259113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent JJ, et al. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 14.Magnúsdóttir E, et al. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irie N, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki K, et al. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Sugawa F, et al. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34:1009–1024. doi: 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi T, et al. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546:416–420. doi: 10.1038/nature22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonchuk J. Formation of embryoid bodies from human pluripotent stem cells using AggreWell™ plates. Methods Mol Biol. 2013;946:523–533. doi: 10.1007/978-1-62703-128-8_32. [DOI] [PubMed] [Google Scholar]
- 20.Tang WWC, et al. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West JA, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo F, et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Gkountela S, et al. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol. 2013;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang DH, Angelin-Duclos C, Calame K. BLIMP-1: Trigger for differentiation of myeloid lineage. Nat Immunol. 2000;1:169–176. doi: 10.1038/77861. [DOI] [PubMed] [Google Scholar]
- 25.Tunyaplin C, Shapiro MA, Calame KL. Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter. Nucleic Acids Res. 2000;28:4846–4855. doi: 10.1093/nar/28.24.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie K, et al. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: A potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J, et al. Follicular dendritic cell-induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin’s B-cell lymphomas. Leukemia. 2011;25:145–152. doi: 10.1038/leu.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, et al. EBV-miR-BHRF1-2 targets PRDM1/Blimp1: Potential role in EBV lymphomagenesis. Leukemia. 2016;30:594–604. doi: 10.1038/leu.2015.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Felici M. Oogenesis. Springer; London: 2012. Origin, migration, and proliferation of human primordial germ cells; pp. 19–37. [Google Scholar]
- 30.Tonner P, Srinivasasainagendra V, Zhang S, Zhi D. Detecting transcription of ribosomal protein pseudogenes in diverse human tissues from RNA-seq data. BMC Genomics. 2012;13:412. doi: 10.1186/1471-2164-13-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matys V, et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ. Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer. 2010;49:948–962. doi: 10.1002/gcc.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schemmer J, et al. Transcription factor TFAP2C regulates major programs required for murine fetal germ cell maintenance and haploinsufficiency predisposes to teratomas in male mice. PLoS One. 2013;8:e71113. doi: 10.1371/journal.pone.0071113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melani M, Simpson KJ, Brugge JS, Montell D. Regulation of cell adhesion and collective cell migration by hindsight and its human homolog RREB1. Curr Biol. 2008;18:532–537. doi: 10.1016/j.cub.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 35.Mamsen LS, Brøchner CB, Byskov AG, Møllgard K. The migration and loss of human primordial germ stem cells from the hind gut epithelium towards the gonadal ridge. Int J Dev Biol. 2012;56:771–778. doi: 10.1387/ijdb.120202lm. [DOI] [PubMed] [Google Scholar]
- 36.Molyneaux KA, et al. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- 37.Rossi L, et al. The extracellular nucleotide UTP is a potent inducer of hematopoietic stem cell migration. Blood. 2007;109:533–542. doi: 10.1182/blood-2006-01-035634. [DOI] [PubMed] [Google Scholar]
- 38.Tan W, Martin D, Gutkind JS. The Galpha13-Rho signaling axis is required for SDF-1-induced migration through CXCR4. J Biol Chem. 2006;281:39542–39549. doi: 10.1074/jbc.M609062200. [DOI] [PubMed] [Google Scholar]
- 39.Canham MA, Sharov AA, Ko MSH, Brickman JM. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brickman JM, Serup P. Properties of embryoid bodies. Wiley Interdiscip Rev Dev Biol. 2017;6:e259. doi: 10.1002/wdev.259. [DOI] [PubMed] [Google Scholar]
- 41.Li S, Edgar D, Fässler R, Wadsworth W, Yurchenco PD. The role of laminin in embryonic cell polarization and tissue organization. Dev Cell. 2003;4:613–624. doi: 10.1016/s1534-5807(03)00128-x. [DOI] [PubMed] [Google Scholar]
- 42.Li X, et al. Fibroblast growth factor signaling and basement membrane assembly are connected during epithelial morphogenesis of the embryoid body. J Cell Biol. 2001;153:811–822. doi: 10.1083/jcb.153.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: Strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Y, Runyan C, Shoemaker A, Surani A, Wylie C. Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development. 2009;136:1295–1303. doi: 10.1242/dev.030619. [DOI] [PubMed] [Google Scholar]
- 45.Runyan C, et al. Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development. 2006;133:4861–4869. doi: 10.1242/dev.02688. [DOI] [PubMed] [Google Scholar]
- 46.Ogawa F, et al. Prostanoid induces premetastatic niche in regional lymph nodes. J Clin Invest. 2014;124:4882–4894. doi: 10.1172/JCI73530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clatot F, et al. The gene expression profile of inflammatory, hypoxic and metabolic genes predicts the metastatic spread of human head and neck squamous cell carcinoma. Oral Oncol. 2014;50:200–207. doi: 10.1016/j.oraloncology.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Cheng L, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 50.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez F, et al. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 57.Dennis G, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 58.Kel A, Voss N, Jauregui R, Kel-Margoulis O, Wingender E. Beyond microarrays: Finding key transcription factors controlling signal transduction pathways. BMC Bioinformatics. 2006;7:S13. doi: 10.1186/1471-2105-7-S2-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.