Significance
Chronic wasting disease (CWD) is a fatal neurodegenerative disease that can cause declines in deer and elk populations. Theoretical work suggests elk will adapt to the disease by favoring a naturally occurring but underrepresented prion protein variant (leucine allele) that enables individuals to live and reproduce for several years after infection with CWD. Using genetic and disease samples from different populations, we found that elk herds infected with CWD for 30–50 y exhibited leucine allele frequencies that were twice as great as those in unexposed populations. Our results are consistent with the hypothesis that genetic selection has occurred due to CWD, but it remains unknown if this will allow elk to mitigate the negative population impacts of CWD.
Keywords: prion protein, chronic wasting disease, Cervus elaphus, elk, pathogen-mediated selection
Abstract
Pathogens can exert a large influence on the evolution of hosts via selection for alleles or genotypes that moderate pathogen virulence. Inconsistent interactions between parasites and the host genome, such as those resulting from genetic linkages and environmental stochasticity, have largely prevented observation of this process in wildlife species. We examined the prion protein gene (PRNP) in North American elk (Cervus elaphus nelsoni) populations that have been infected with chronic wasting disease (CWD), a contagious, fatal prion disease, and compared allele frequency to populations with no history of exposure to CWD. The PRNP in elk is highly conserved and a single polymorphism at codon 132 can markedly extend CWD latency when the minor leucine allele (132L) is present. We determined population exposure to CWD, genotyped 1,018 elk from five populations, and developed a hierarchical Bayesian model to examine the relationship between CWD prevalence and PRNP 132L allele frequency. Populations infected with CWD for at least 30–50 y exhibited 132L allele frequencies that were on average twice as great (range = 0.23–0.29) as those from uninfected populations (range = 0.04–0.17). Despite numerous differences between the elk populations in this study, the consistency of increase in 132L allele frequency suggests pathogen-mediated selection has occurred due to CWD. Although prior modeling work predicted that selection will continue, the potential for fitness costs of the 132L allele or new prion protein strains to arise suggest that it is prudent to assume balancing selection may prevent fixation of the 132L allele in populations with CWD.
Demonstrating selection via molecular adaptation in free-ranging animals is difficult and relatively uncommon due to the large number of abiotic and biotic factors that interact to affect individual fitness. Pathogen-mediated selection in vertebrates is considered to be one of the most likely areas to observe selection due to the close association between parasites and loci that shape immune responses (1). There is increasing evidence of correlations between host susceptibility to parasitism and neutral genetic diversity (2) as well as alleles of the MHC, a highly polymorphic region of the genome that is closely tied to immune response (3). However, the impact of specific alleles, MHC or otherwise, is largely unknown within wildlife populations, preventing testable predictions or an understanding of the trajectory that pathogen-mediated selection may take, particularly in the presence of an emerging disease.
Chronic wasting disease (CWD) is a contagious transmissible spongiform encephalopathy affecting cervids. First observed in 1967, CWD is attributed to conversion of normal cellular prion protein (PrPC) to a misfolded, protease-resistant prion protein (PrPres) which accumulates in lymphoid and central nervous system tissues (4–6). There is no evidence that infected individuals can develop immunity or survive infection (7). Studies on free-ranging deer (Odocoileus spp.) and elk (Cervus elaphus nelsoni) have found that survival and population growth rates can be negatively affected by CWD (8–11). In elk, PRNP is highly conserved with only one of three single nucleotide polymorphisms in the open reading frame (ORF) giving rise to a nonsynonymous change. The resulting alleles encode either methionine (M) or leucine (L) at codon 132 (12, 13). Oral inoculation experiments in captivity have found that incubation time is allele-associated; MM elk developed disease 23 mo postinoculation, ML elk in 40 mo, and LL elk in 59–63 mo (14, 15). Within naturally occurring populations, the methionine allele appears to be overrepresented, particularly in those that have not been exposed to CWD. Colorado elk populations with a low prevalence of CWD (proportion infected <0.02) consisted of 67% MM, 31% ML, and 2% LL elk (n = 259), whereas unexposed elk from South Dakota and Wyoming consisted of 81% MM, 19% ML, and 0% LL (n = 97) (refs. 12 and 16; note that none of these data are included in our analyses).
Recent modeling projected 132L allele frequency to increase in populations where CWD occurs due to ML and LL individuals’ surviving longer and successfully reproducing postinfection, which allowed the population to mitigate the negative impacts of the disease when methionine is overrepresented (17). Similar results have been observed in humans (18) and predicted for white-tailed deer (Odocoileus virginianus) (19), but there have been no attempts to evaluate changes in PRNP with empirical data from cervid populations. Here we examine allele frequencies of PRNP from multiple elk populations with and without CWD to determine if pathogen-mediated selection has occurred. Prior work indicates that this selection pressure should be observable after elk populations are exposed to CWD for ∼25 y (17). We predicted that the frequency of the 132L allele should be greater in elk populations where CWD has occurred for at least 30–50 y.
Results
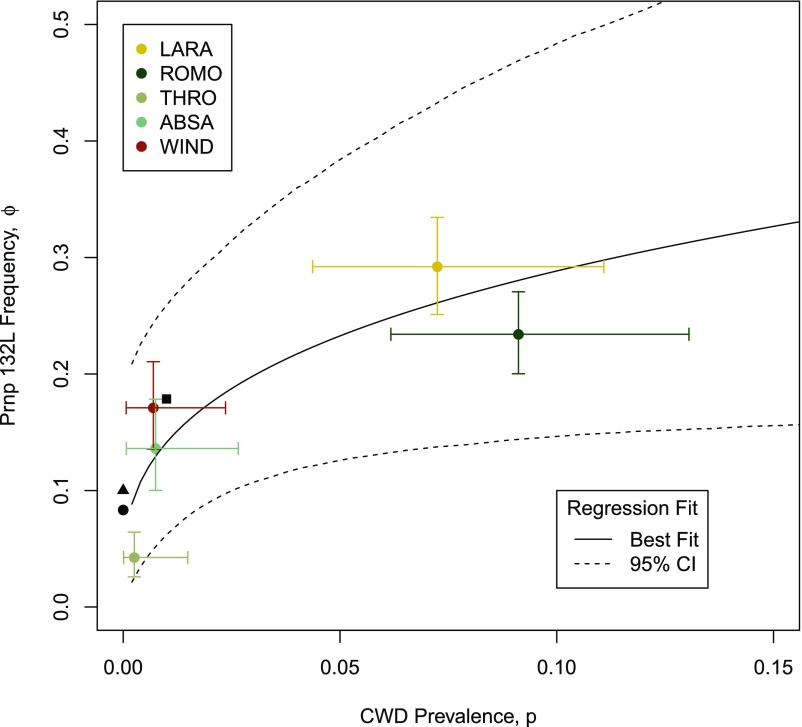
We collected 1,018 samples across the five populations. Diagnostic tests agreed with prior work; infected elk were detected in Laramie Mountains, Wyoming (LARA) and Rocky Mountain National Park, Colorado (ROMO), but not in the Absaroka Range, Wyoming (ABSA), Theodore Roosevelt National Park, North Dakota (THRO), or the northern end of the Wyoming and Wind River Ranges (WIND). The overall proportion of ML and LL genotypes and L allele frequency in LARA and ROMO was two to three times greater than that observed in nonexposed populations. Notably, the 95% credible interval (CI) of L allele frequencies exhibited almost no overlap between elk populations with and without CWD (Table 1).
Table 1.
Total number of individual elk, proportion of individuals in each genotype, observed L allele frequency, and model fit results for CWD prevalence and L allele frequency from five distinct study populations
| Observed | Model fit results | ||||||
| Population | No. of elk | MM | ML | LL | L allele frequency | CWD prevalence (95% CI) | L allele frequency (95% CI) |
| ROMO | 269 | 0.58 | 0.37 | 0.05 | 0.24 | 0.09 (0.06–0.13) | 0.23 (0.20–0.27) |
| LARA | 216 | 0.50 | 0.41 | 0.09 | 0.29 | 0.07 (0.05–0.11) | 0.29 (0.25–0.34) |
| WIND | 186 | 0.69 | 0.27 | 0.04 | 0.17 | 0.01* (0.00–0.02) | 0.17 (0.14–0.21) |
| ABSA | 148 | 0.74 | 0.24 | 0.01 | 0.14 | 0.01* (0.00–0.03) | 0.14 (0.010–0.18) |
| THRO | 199 | 0.93 | 0.07 | 0.01 | 0.04 | 0.00 (0.00–0.01) | 0.04 (0.03–0.06) |
Values in parentheses are the upper and lower 95% CI.
CWD has never been documented in elk from WIND or ABSA; prevalence represents model predictions that were limited to sample sizes in this study, resulting in greater uncertainty and a prediction of 0.01 for mean prevalence in these areas.
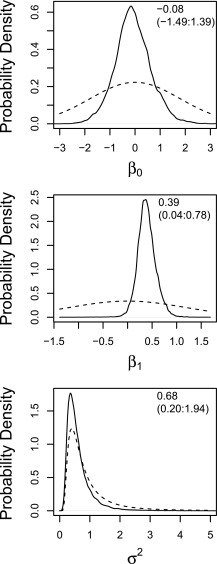
We found a strong correlation between CWD prevalence and the frequency of the L allele. There was a clear trend for higher prevalence of CWD to cooccur with greater frequency of the 132L allele (Fig. 1). The marginal posterior probability that the regression coefficient β1 > 0 was 0.982 with the specified model, representing a significant shift away from the prior (Fig. 2).
Fig. 1.
Regression of derived predicted mean allele frequency based on CWD prevalence (solid line) and model results for the five elk populations included in analyses (whisker plots showing 95% CI). Two of the elk populations have been exposed to CWD for at least 30–50 y (LARA and ROMO), while the disease has never been detected in the other three populations (WIND, THRO, and ABSA). Data extracted from other elk populations are shown for comparison but not included in analyses (■ = Colorado, ▲ = Wyoming, ● = South Dakota; refs. 12 and 16).
Fig. 2.
Prior (dashed line) and marginal posterior (solid line) distributions of the regression intercept (β0), regression coefficient between allele frequency and CWD prevalence (β1), and associated variance (σ2). Model fit results for each parameter is included in the upper right of each graph as posterior mean (95% CI).
Discussion
The positive relationship between frequency of the L allele in the PRNP and prevalence of CWD is consistent with the prediction that pathogen-mediated selection has occurred in free-ranging elk infected with PrPres. Model simulations have previously predicted this outcome (17), but this study demonstrates evidence for selection with empirical data from multiple free-ranging cervid populations. Research on captive elk indicates that the mechanism for this selection is delayed onset of clinical CWD postinfection among individuals with an L allele (14, 15, 17). Although a relationship between the delayed onset of CWD and PRNP has yet to be demonstrated in free-ranging elk, it has been observed in wild deer (20). Additionally, greater sensitivity of antemortem diagnostic tests in free-ranging elk without an L allele (i.e., the MM genotype) has been attributed to faster accumulation of PrPres in the lymphoid tissues (21–23). There is currently no evidence that the L allele prevents prion infection or CWD in elk (16), suggesting the delayed onset of clinical disease and associated increase in survival time allowing additional reproduction may be the sole cause of PRNP selection observed in this study.
We found a clear but moderate increase in L allele frequency when average prevalence of CWD was 0.07–0.09 after 30–50 y of exposure to PrPres. These findings do not negate or support the conclusion that the frequency of the L allele will continue to increase in the future and mitigate the negative survival impacts of CWD. Recent work projected that L allele frequency will exhibit further increases beyond those observed in our study, eventually surpassing 0.50 in populations infected with CWD and allowing elk populations to sustain positive population growth even in the presence of relatively high transmission rates (17). However, this outcome is contingent on the assumption that the L allele does not have any negative fitness-related impacts. This topic has not been sufficiently investigated in free-ranging cervids; some evidence of PRNP-related impacts on fitness has been observed in domestic sheep, a species affected by the transmissible spongiform encephalopathy scrapie (24–26). Our results indicate that the M allele is consistently overrepresented in populations without CWD; the majority of individuals were MM homozygotes (≥0.69) and M allele frequency ranged from 0.83 to 0.96. Similar findings have been observed in populations not included in this study from South Dakota (M allele frequency = 0.90) (12) and in areas to the east and west of ROMO (M allele frequency = 0.82) (16); CWD in ROMO neighboring areas may be a more recent occurrence as prevalence was ≤0.02 at the time of sampling (16). These consistent observations are unlikely due to genetic drift, random effects, or geographic location, as we would expect a much more variable L allele frequency in the absence or relatively low prevalence of CWD. Given this, we suggest it remains just as likely that there is a fitness-associated cost associated with the L allele which could lead to balancing selection that will limit the future increase of this allele in elk populations with CWD.
There are two additional factors that could affect selection and the long-term trajectory of elk populations with CWD. First, different PrP strains have been found to exist in domestic sheep (27, 28) and new conformations can arise, particularly when transmission occurs between hosts with different prion genotypes (29). For example, passage of a single deer CWD isolate through deer of varying PRNP genotypes followed by bioassay using transgenic mouse models have helped identify novel strains that may be associated with certain PRNP alleles (29). This suggests that greater PrP variability could interact with and overcome selection for alleles that reduce susceptibility to prion infection or are associated with prolonged incubation times of prion diseases. Second, recent captive work has found that elk with an L allele can shed PrPres well before the signs of disease, suggesting that prolonged survival and selection for such alleles may lead to greater transmission potential and environmental contamination (30). Together, these factors emphasize that managers should be cautious when trying to assess the future long-term impacts of prion genotype frequency on elk survival and population growth, particularly given that CWD has been found to decrease elk survival and population growth even in the presence of greater L allele frequency (10).
There was a strong relationship between allele frequency and prevalence of CWD (posterior probability that was 0.982). This is notable because multiple factors could have confounded this correlation. In particular, we used prevalence as a proxy for CWD impact, but that did not consider time since introduction of PrPres, reintroduction history of each elk population, or regional factors such as winter severity that may have influenced selection. For example, elk were reintroduced into ROMO in 1914 and THRO in 1985, elk in the WIND population are supplemented with food during winter, and elk in ABSA and WIND inhabit ranges occupied by gray wolves (Canis lupus). This suggests that selective pressures acting on PRNP in elk are strong enough, even in the absence of disease, to overcome the potential impacts of genetic drift, founder effects, and random effects. The one exception to this may be THRO, where 47 elk from Wind Cave National Park, South Dakota were reintroduced in 1985 (31), which may have influenced the observed lower frequency of the L allele.
Pathogen-mediated selection has been described under experimental conditions (32), but studies of wild populations have found no evidence for selection (33, 34) due to complex interactions between parasite species and MHC alleles (35) and linked alleles that confound results (3). Despite a relatively short time for selection to occur in a long-lived species, the proportion of elk with genotypes that delay the onset of clinical disease is higher in populations that have only been exposed to PrPres for ∼50 y. Future work on this topic will offer a rare opportunity to demonstrate the potential for balancing versus directional selection in a free-ranging mammal.
Materials and Methods
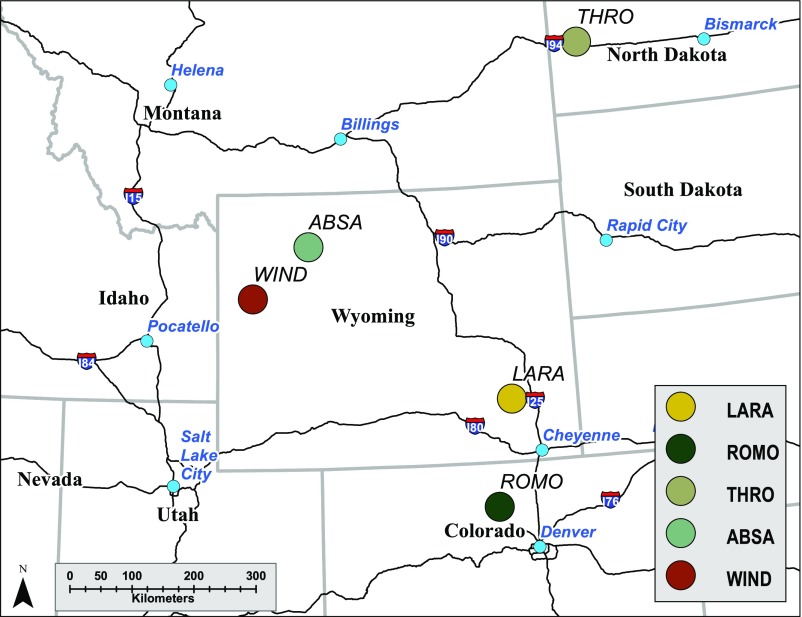
We collected genetic and diagnostic samples from adult female elk in two populations with CWD and three populations where surveillance activities have never detected a case of CWD (Fig. 3). Samples from populations with CWD were from ROMO, where CWD in elk was first detected in 1981 (36), and LARA, where CWD was first detected in 1985 (37). Epidemic models estimated that CWD has likely been in these areas since the 1960s (37). Samples from nonexposed populations were from ABSA, WIND, and THRO. Samples from WIND were collected from feedgrounds that can increase both food availability and elk densities in winter. All other samples from exposed and nonexposed populations were from areas that were not food-supplemented.
Fig. 3.
Sample locations for elk populations with CWD (ROMO and LARA) and without CWD (THRO, ABSA, and WIND).
Samples were collected from 2008 to 2015 from harvested and agency-culled elk, or elk in demographic studies. Diagnostic samples were collected from LARA, ROMO, THRO, and ABSA and consisted primarily of brainstem (medulla oblongata at the level of the obex) and retropharyngeal lymph nodes, as well as antemortem rectal biopsy samples for 118 of the 269 ROMO samples (21). Each tissue was either fixed in neutral buffered formalin and assayed for PrPres by immunohistochemistry or held fresh/frozen and assayed by ELISA (38, 39). Diagnostic samples were not available for the 186 elk from WIND included in this study, which we assumed were not infected with CWD. We tested this assumption by assaying an additional 2,057 postmortem samples for PrPres from adult female elk in WIND that were collected between 2008–2015.
DNA was extracted from frozen tissue or blood using commercial kits (FastDNA Spin Kit; MP Biomedicals or iPrep PureLink gDNA Blood Kit; Invitrogen/Life Technologies) following the manufacturer’s instructions. Tissue utilized for extraction was primarily brain- or lymph node-derived but DNA was isolated for some animals from heart, kidney, liver, lung, or spleen. Blood utilized for DNA extraction was collected into EDTA anticoagulant. PCR amplification and sequencing of the PRNP ORF was performed as previously described (6). All samples were sequenced using reverse primer 12; additional sequence reactions were performed for some samples using forward primer 245 (6). PRNP alleles were identified by the deduced amino acid at codon 132 (M or L).
We modeled the relationship between prevalence and allele frequencies as follows. There was no evidence of deviation from Hardy–Weinberg equilibrium within populations so we used allele counts to avoid the loss of power which would result from a multinomial model of genotype categories. Let be the CWD test result for animal in population , for , where is the total number of populations observed and is the observed 132L allele count for animal in population where = 0 for a 132M homozygote, 1 for a heterozygote, and 2 for the 132L homozygote. These data are modeled from binomial distributions with appropriate probabilities:
where is the probability of testing positive for CWD in population and is the 132L allele frequency in the population. Normal regression links the logit-transformed frequency of positive CWD tests to logit-transformed frequency of the 132L allele across populations:
Priors are assigned for the regression coefficients, and , with means equal to zero and uninformative variances given the logit transformation
represents the value of at the intercept, or after transformation. We set the prior variance on to result in 95% of the prior probability density for lying between 0.1 and 0.9 when . represents the predicted change in for a single unit change in with 95% of the prior probability density between .
A vague informative prior is used to keep reasonably small and to aid in model convergence:
This model was fit to the available data with a Metropolis–Hastings algorithm using Markov chain Monte Carlo methods in program R over 100,000 iterations with 10% discarded for burn-in. Model fit was evaluated using posterior predictive checks as previously described (40). There was no evidence of deviation between the observed and replicated data with Bayesian P values ranging from 0.5 to 0.7.
Supplementary Material
Acknowledgments
We thank M. Hooten for assistance with model specification and S. Ratchford, H. Killion, J. Jennings-Gaines, J. Spaak, E. Wheeler, T. Spraker, V. Jameson, T. Gidlewski, M. Graham, D. Lesiak, A. Wexler, D. Zhuang, D. Baker, S. Kichman, C. Sexton, M. Oehler, and numerous field personnel from the Wyoming Game and Fish Department for assistance in sample collection or laboratory analysis. This work was funded by the National Park Service and US Department of Agriculture Research Service Grant CRIS 2090-32000-030-00. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the US Department of Agriculture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gompper ME, Monello RJ, Eggert LS. Genetic variability and viral seroconversion in an outcrossing vertebrate population. Proc Biol Sci. 2011;278:204–210. doi: 10.1098/rspb.2010.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Froeschke G, Sommer S. Role of selection versus neutral processes determining genetic variation in a small mammal along a climatic gradient in southern Africa. Evol Ecol. 2014;28:1169–1190. [Google Scholar]
- 4.Williams ES, Young S. Chronic wasting disease of captive mule deer: A spongiform encephalopathy. J Wildl Dis. 1980;16:89–98. doi: 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 5.Williams ES, Miller MW. Chronic wasting disease in deer and elk in North America. Rev Sci Tech. 2002;21:305–316. doi: 10.20506/rst.21.2.1340. [DOI] [PubMed] [Google Scholar]
- 6.Spraker TR, Balachandran A, Zhuang D, O’Rourke KI. Variable patterns of distribution of PrP(CWD) in the obex and cranial lymphoid tissues of Rocky Mountain elk (Cervus elaphus nelsoni) with subclinical chronic wasting disease. Vet Rec. 2004;155:295–302. doi: 10.1136/vr.155.10.295. [DOI] [PubMed] [Google Scholar]
- 7.Williams ES, Kirkwood JK, Miller MW. Transmissible spongiform encephalopathies. In: Williams ES, Barker IK, editors. Infectious Diseases of Wild Mammals. Iowa State Univ Press; Ames, IA: 2001. pp. 292–301. [Google Scholar]
- 8.Miller MW, et al. Lions and prions and deer demise. PLoS One. 2008;3:e4019. doi: 10.1371/journal.pone.0004019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sargeant GA, Weber DC, Roddy DE. Implications of chronic wasting disease, cougar predation, and reduced recruitment for elk management. J Wildl Manage. 2011;75:171–177. [Google Scholar]
- 10.Monello RJ, et al. Survival and population growth of a free-ranging elk population with a long history of exposure to chronic wasting disease. J Wildl Manage. 2014;78:214–223. [Google Scholar]
- 11.Edmunds DR, et al. Chronic wasting disease drives population decline of white-tailed deer. PLoS One. 2016;11:e0161127. doi: 10.1371/journal.pone.0161127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Rourke KI, et al. PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 1999;80:2765–2769. doi: 10.1099/0022-1317-80-10-2765. [DOI] [PubMed] [Google Scholar]
- 13.White SN, Spraker TR, Reynolds JO, O’Rourke KI. Association analysis of PRNP gene region with chronic wasting disease in Rocky Mountain elk. BMC Res Notes. 2010;3:314. doi: 10.1186/1756-0500-3-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamir AN, et al. Preliminary observations of genetic susceptibility of elk (Cervus elaphus nelsoni) to chronic wasting disease by experimental oral inoculation. J Vet Diagn Invest. 2006;18:110–114. doi: 10.1177/104063870601800118. [DOI] [PubMed] [Google Scholar]
- 15.O’Rourke KI, et al. Elk with a long incubation prion disease phenotype have a unique PrPd profile. Neuroreport. 2007;18:1935–1938. doi: 10.1097/WNR.0b013e3282f1ca2f. [DOI] [PubMed] [Google Scholar]
- 16.Perucchini M, Griffin K, Miller MW, Goldmann W. PrP genotypes of free-ranging wapiti (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 2008;89:1324–1328. doi: 10.1099/vir.0.83424-0. [DOI] [PubMed] [Google Scholar]
- 17.Williams AL, Kreeger TJ, Schumaker BA. Chronic wasting disease model of genetic selection favoring prolonged survival in Rocky Mountain elk (Cervus elaphus) Ecosphere. 2014;5:1–10. [Google Scholar]
- 18.Mead S, et al. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science. 2003;300:640–643. doi: 10.1126/science.1083320. [DOI] [PubMed] [Google Scholar]
- 19.Robinson SJ, Samuel MD, Johnson CJ, Adams M, McKenzie DI. Emerging prion disease drives host selection in a wildlife population. Ecol Appl. 2012;22:1050–1059. doi: 10.1890/11-0907.1. [DOI] [PubMed] [Google Scholar]
- 20.Johnson C, et al. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol. 2006;87:2109–2114. doi: 10.1099/vir.0.81615-0. [DOI] [PubMed] [Google Scholar]
- 21.Monello RJ, et al. Efficacy of antemortem rectal biopsies to diagnose and estimate prevalence of chronic wasting disease in free-ranging cow elk (Cervus elaphus nelsoni) J Wildl Dis. 2013;49:270–278. doi: 10.7589/2011-12-362. [DOI] [PubMed] [Google Scholar]
- 22.Keane DP, et al. Chronic wasting disease in a Wisconsin white-tailed deer farm. J Vet Diagn Invest. 2008;20:698–703. doi: 10.1177/104063870802000534. [DOI] [PubMed] [Google Scholar]
- 23.Keane D, et al. Validation of use of rectoanal mucosa-associated lymphoid tissue for immunohistochemical diagnosis of chronic wasting disease in white-tailed deer (Odocoileus virginianus) J Clin Microbiol. 2009;47:1412–1417. doi: 10.1128/JCM.02209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawalha RM, Brotherstone S, Conington J, Villanueva B. Lambs with scrapie susceptible genotypes have higher postnatal survival. PLoS One. 2007;2:e1236. doi: 10.1371/journal.pone.0001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawalha RM, Villanueva B, Brotherstone S, Rogers PL, Lewis RM. Prediction of prion protein genotype and association of this genotype with lamb performance traits of Suffolk sheep. J Anim Sci. 2010;88:428–434. doi: 10.2527/jas.2009-2009. [DOI] [PubMed] [Google Scholar]
- 26.Sweeney T, Hanrahan JP. The evidence of associations between prion protein genotype and production, reproduction, and health traits in sheep. Vet Res. 2008;39:28. doi: 10.1051/vetres:2008004. [DOI] [PubMed] [Google Scholar]
- 27.Bruce ME, et al. Strain characterization of natural sheep scrapie and comparison with BSE. J Gen Virol. 2002;83:695–704. doi: 10.1099/0022-1317-83-3-695. [DOI] [PubMed] [Google Scholar]
- 28.Stack MJ, Chaplin MJ, Clark J. Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases and Romney and Cheviot breed sheep experimentally inoculated with BSE using two monoclonal antibodies. Acta Neuropathol. 2002;104:279–286. doi: 10.1007/s00401-002-0556-2. [DOI] [PubMed] [Google Scholar]
- 29.Duque Velásquez C, et al. Deer prion proteins modulate the emergence and adaptation of chronic wasting disease strains. J Virol. 2015;89:12362–12373. doi: 10.1128/JVI.02010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng YC, et al. Early and non-invasive detection of chronic wasting disease prions in elk feces by real-time quaking induced conversion. PLoS One. 2016;11:e0166187. doi: 10.1371/journal.pone.0166187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sargeant GA, Oehler MW., Sr Dynamics of newly established elk populations. J Wildl Manage. 2007;71:1141–1148. [Google Scholar]
- 32.Eizaguirre C, Lenz TL, Kalbe M, Milinski M. Rapid and adaptive evolution of MHC genes under parasite selection in experimental vertebrate populations. Nat Commun. 2012;3:621. doi: 10.1038/ncomms1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyce WM, et al. Genetic variation of major histocompatibility complex and microsatellite loci: A comparison in bighorn sheep. Genetics. 1997;145:421–433. doi: 10.1093/genetics/145.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winternitz JC, Wares JP, Yabsley MJ, Altizer S. Wild cyclic voles maintain high neutral and MHC diversity without strong evidence for parasite-mediated selection. Evol Ecol. 2014;28:957–975. [Google Scholar]
- 35.Loiseau C, et al. Diversifying selection on MHC class I in the house sparrow (Passer domesticus) Mol Ecol. 2009;18:1331–1340. doi: 10.1111/j.1365-294X.2009.04105.x. [DOI] [PubMed] [Google Scholar]
- 36.Spraker TR, et al. Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J Wildl Dis. 1997;33:1–6. doi: 10.7589/0090-3558-33.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Miller MW, et al. Epizootiology of chronic wasting disease in free-ranging cervids in Colorado and Wyoming. J Wildl Dis. 2000;36:676–690. doi: 10.7589/0090-3558-36.4.676. [DOI] [PubMed] [Google Scholar]
- 38.Hibler CP, et al. Field validation and assessment of an enzyme-linked immunosorbent assay for detecting chronic wasting disease in mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus), and Rocky Mountain elk (Cervus elaphus nelsoni) J Vet Diagn Invest. 2003;15:311–319. doi: 10.1177/104063870301500402. [DOI] [PubMed] [Google Scholar]
- 39.Spraker TR, et al. Detection of PrP(CWD) in postmortem rectal lymphoid tissues in Rocky Mountain elk (Cervus elaphus nelsoni) infected with chronic wasting disease. J Vet Diagn Invest. 2006;18:553–557. doi: 10.1177/104063870601800605. [DOI] [PubMed] [Google Scholar]
- 40.Gelman A, Meng XL, Stern H. Posterior predictive assessment of model fitness via realized discrepancies. Stat Sin. 1996;1:733–760. [Google Scholar]