Significance
CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) is an E3 ubiquitin ligase acting as a central repressor of seedling photomorphogenesis in plants. Many nuclear-localized COP1 substrates have been identified in the last two decades; however, whether COP1 targets cytoplasmic factors for ubiquitination and degradation remains largely unknown. In this study, we show that COP1 interacts with a microtubule-associated protein, WAVE-DAMPENED 2-LIKE 3 (WDL3), in a dark-dependent manner at cortical microtubules. Thus, COP1 targets WDL3 for 26S proteasome-mediated degradation to control hypocotyl elongation in etiolated Arabidopsis seedlings. Collectively, our study uncovers a cytoplasmic substrate of COP1 that functions as a microtubule-associated protein in mediating hypocotyl cell elongation.
Keywords: COP1, WDL3, cortical microtubule, hypocotyl elongation, Arabidopsis
Abstract
CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a well-known E3 ubiquitin ligase, functions as a central regulator of plant growth and photomorphogenic development in plants, including hypocotyl elongation. It has been well-established that, in darkness, COP1 targets many photomorphogenesis-promoting factors for ubiquitination and degradation in the nucleus. However, increasing evidence has shown that a proportion of COP1 is also localized outside the nucleus in dark-grown seedlings, but the physiological function of this localization remains largely unclear. In this study, we demonstrate that COP1 directly targets and mediates the degradation of WAVE-DAMPENED 2-LIKE 3 (WDL3) protein, a member of the microtubule-associated protein (MAP) WVD2/WDL family involved in regulating hypocotyl cell elongation of Arabidopsis seedlings. We show that COP1 interacts with WDL3 in vivo in a dark-dependent manner at cortical microtubules. Moreover, our data indicate that COP1 directly ubiquitinates WDL3 in vitro and that WDL3 protein is degraded in WT seedlings but is abundant in the cop1 mutant in the dark. Consistently, introduction of the wdl3 mutation weakened, whereas overexpression of WDL3 enhanced, the short-hypocotyl phenotype of cop1 mutant in darkness. Together, this study reveals a function of COP1 in regulating the protein turnover of a cytosol-localized MAP in etiolated hypocotyls, thus providing insights into COP1-mediated degradation of downstream factors to control seedling photomorphogenesis.
The ubiquitin/26S proteasome system (UPS) regulates many fundamental cellular processes, such as plant cell growth, cell signaling, protein trafficking, and abiotic stress responses, by controlling the degradation rates of numerous proteins (1, 2). For the majority of UPS substrates, the concerted action of E1 Ub-activating enzymes, E2 Ub-conjugating enzymes, and E3 Ub ligases leads to the covalent attachment of a multiubiquitin chain to proteins destined for degradation (3, 4). The polyubiquitinated target protein is then degraded by the 26S proteasome, an evolutionarily conserved multicatalytic protease (5). Numerous studies have shown that E3 ligases determine the specificity of target proteins and facilitate the diverse functions of the ubiquitination system in many cellular processes (2, 4).
Arabidopsis seedlings display distinct morphologies when grown in the light compared with those grown in the dark. Light-grown seedlings undergo photomorphogenesis (also known as deetiolation), typified by short hypocotyls and open green cotyledons; by contrast, dark-grown seedlings undergo skotomorphogenesis (also known as etiolation), characterized by long hypocotyls, closed cotyledons, and apical hooks (6). CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a well-known E3 ubiquitin ligase, functions as a central repressor of seedling photomorphogenesis (7, 8). COP1 exhibits a light-mediated nucleocytoplasmic repartitioning, being enriched in the nucleus in darkness and depleted from the nucleus in the light (9). Numerous studies have demonstrated that COP1 targets a subset of nuclear-localized photomorphogenesis-promoting factors for degradation in darkness, including transcription factors such as ELONGATED HYPOCOTYL 5 (HY5) (10), HY5 HOMOLOG (HYH) (11), LONG AFTER FAR-RED LIGHT 1 (LAF1) (12), LONG HYPOCOTYL IN FAR-RED 1 (HFR1) (13–15), and B-BOX21 (BBX21) (16), and photoreceptors, such as phytochromes (17, 18). COP1 interacts with SUPPRESSOR OF PHYA-105 (SPA) proteins to form E3 ligase complexes (12, 19, 20). In the light, photo-activated photoreceptors (such as phytochromes and cryptochromes) inactivate the COP1/SPA complex largely by disrupting the direct interaction of COP1 and SPA proteins (21–24), thus allowing the accumulation of photomorphogenesis-promoting transcription factors in the nucleus to initiate photomorphogenesis. In darkness, COP1 protein is mostly localized in the nucleus; however, some COP1 was also detectable outside the nucleus (25, 26). In addition, a recent study showed that the interaction of COP1 with a resistance (R) protein HRT was observed in the periphery of pavement cells in the dark (27). Thus, these observations indicate that a proportion of COP1 may be present outside the nucleus in the dark although the biological significance of this distribution remains largely unclear.
Microtubule-associated proteins (MAPs) control the organization, stability, and dynamics of microtubules functioning in plant growth and plant cell morphogenesis (28, 29). Hypocotyl cell elongation is strongly influenced by developmental and environmental cues and is regulated by many factors, including light, phytohormones, and microtubules (30–33). Increasing evidence has shown that some MAPs participate in the regulation of hypocotyl elongation and are regulated by transcription factors of various signaling pathways. For example, MICROTUBULE-DESTABILIZING PROTEIN 40 is involved in brassinosteroid (BR)-promoted hypocotyl elongation of etiolated seedlings and is directly targeted by BRASSINAZOLE-RESISTANT 1, a key transcription factor of the BR signaling pathway (33). Thus, investigation of how MAPs are regulated by upstream factors is essential to understand the regulatory mechanisms of plant growth and development. A recent study showed that COP1 activity is crucial for microtubule destabilization, which is involved in darkness- and abscisic acid-induced stomatal closure (34), but the molecular mechanisms underlying COP1-regulated microtubules remain largely unknown. Because cop1 mutants display a short hypocotyl and open cotyledon phenotype in both light and dark conditions (referred to as cop phenotype) (35), it will be interesting to investigate whether COP1 is involved in posttranscriptional regulation of MAPs to control hypocotyl cell elongation.
Our previous study showed that a microtubule-associated protein from the WAVE-DAMPENED 2 (WVD2)/WVD2-LIKE (WDL) family, WDL3, participated in the control of hypocotyl cell elongation (36).Overexpression of WDL3 resulted in shorter hypocotyl cells and stabilization of cortical microtubules in the light whereas WDL3 RNA interference Arabidopsis seedlings grown in the light developed longer hypocotyls (36). It was shown that WDL3 protein was abundant in the light but was degraded through the 26S proteasome pathway in the dark (36); however, the molecular mechanism of this degradation was unclear. In this study, we show that the E3 ubiquitin ligase, COP1, interacts with and directly ubiquitinates WDL3. Decreasing WDL3 expression partially suppresses the short hypocotyl phenotype of etiolated cop1-6 seedlings. Our study thus reveals a function of COP1 in the cytoplasm to mediate hypocotyl cell elongation by regulating the abundance of a microtubule-associated protein.
Results and Discussion
COP1 Interacts with WDL3.
Our previous study showed that a microtubule-associated protein, WDL3, was degraded by the 26S proteasome pathway in the dark to facilitate hypocotyl elongation of etiolated seedlings, but the underlying mechanism was unknown (36). Due to the central role of COP1 as an E3 ubiquitin ligase in mediating etiolated hypocotyl elongation, we thus hypothesized that COP1 may be involved in targeted degradation of WDL3 in darkness. To test this hypothesis, we first examined whether COP1 could physically interact with WDL3. Our yeast two-hybrid assay data indicated that WDL3 interacted with full-length COP1 (Fig. 1 A, Upper). The COP1 protein contains three conserved structural domains: a RING-finger motif, followed by a coiled-coil domain and a carboxyl-terminal WD40 repeat domain, all of which have been implicated in mediating the interactions of COP1 with other proteins and/or its self-dimerization (35, 37). Deletion experiments showed that WDL3 interacted with the COP1 WD40 domain, but not with the deletion mutant containing the RING and the coiled-coil domains of COP1 (Fig. 1 A, Lower), indicating that the WD40 domain of COP1 mediates the interaction with WDL3. To further investigate the COP1–WDL3 interaction, we carried out in vitro pull-down assays. GST pull-down assay data showed that GST-WDL3, but not GST-tagged MAP65-1 from Arabidopsis, could successfully pull down COP1 in vitro (Fig. 1B), confirming the results obtained by yeast two-hybrid assays.
Fig. 1.
COP1 interacts with WDL3 in the dark. (A) Interactions between WDL3 and COP1, or COP1 domain-deletion mutant forms in the yeast two-hybrid system. (B) In vitro pull-down assays showing that GST-WDL3 interacts with MBP-COP1. MBP-COP1 pulled down by GST-WDL3 was detected by anti-MBP antibody. GST and GST-MAP65-1 were used as controls and did not interact with MBP-COP1. (C) Interaction of COP1 with WDL3 was revealed via firefly LCI assay in tobacco leaves in the dark. Negative controls used were nLuc+cLuc-COP1, WDL3-nLuc+cLuc, and nLuc+cLuc. (Scale bar: 1 cm.) (D) Coimmunoprecipitation of COP1 and WDL3. Total protein was extracted from Arabidopsis protoplasts expressing 35S:COP1-MYC and 35S:WDL3-GFP. Anti-MYC-Agarose Affinity Gel was used for immunoprecipitation, and the precipitated proteins were analyzed by immunoblots using an anti-GFP antibody.
The in vivo interaction of COP1 and WDL3 was validated by a firefly luciferase complementation imaging (LCI) assay in tobacco leaves (38). We found that COP1 interacted with WDL3 in the dark (Fig. 1C), but not in the light (Fig. S1). To further validate the interaction between COP1 and WDL3, a coimmunoprecipitation (co-IP) assay was performed using Arabidopsis protoplasts transiently expressing 35S:COP1-MYC and 35S:WDL3-GFP cultured in the dark. We found that green fluorescent protein (GFP)-tagged WDL3 coimmunoprecipitated with MYC-tagged COP1 (Fig. 1D), further confirming that COP1 interacts with WDL3 in vivo.
Previous studies have shown that WVD2/WDL proteins play diverse physiological roles in plant growth and plant cell morphogenesis (36, 39). WDL5 plays a negative role in regulating etiolated hypocotyl elongation in Arabidopsis (40). Using yeast two-hybrid and firefly LCI assays, we found that COP1 did not interact with WDL5 in yeast and plant cells (Fig. S2), which is in agreement with previous studies showing that WDL3, but not other members of the WVD2/WDL family, was regulated by the ubiquitin/26S proteasome-dependent pathway (36, 39, 41). These data suggest a special biochemical property of WDL3 protein in the WVD2/WDL family. In addition, previous studies indicated that COP1 activity was significantly suppressed by photoreceptors when in the light (21–24). In this study, we also found that COP1 did not interact with WDL3 in the light, which is consistent with our previous finding that WDL3 protein is more stable under light conditions (36).
COP1 Ubiquitinates WDL3 in Vitro.
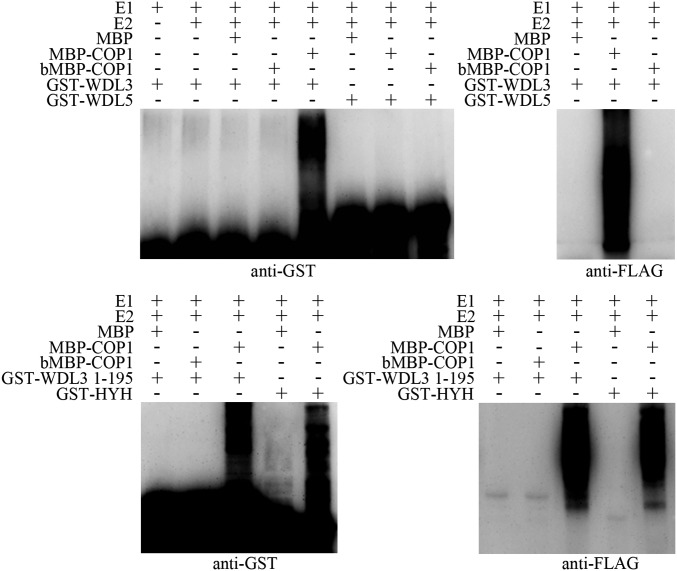
It is well-established that COP1 functions as an E3 ubiquitin ligase that ubiquitinates itself as well as transcription factors such as HY5, LAF1, HFR1, and BBX21 in in vitro ubiquitination assays (12, 15, 16, 19). The physical interaction between COP1 and WDL3 prompted us to test whether WDL3 might be a substrate of the COP1 E3 ligase. We thus conducted an in vitro ubiquitination assay in which we used GST-WDL3 protein purified from Escherichia coli as a substrate and maltose-binding protein (MBP)-COP1 recombinant protein as the E3 ligase. In the presence of E1, E2, and FLAG-tagged ubiquitin, GST-WDL3, but not GST-WDL5, was polyubiquitinated by MBP-COP1 as detected by anti-GST and anti-FLAG antibodies (Fig. 2, Upper). In Arabidopsis, the WVD2/WDL protein family consists of eight members, all of which have a conserved C terminus but a diverse N terminus (39, 41). Using yeast two-hybrid and firefly LCI assays, we showed that the WDL3 N terminus (1-195), but not the C terminus (196-339), interacted with COP1 in vitro and in vivo (Fig. S3), suggesting that the N terminus is important for WDL3 degradation. Consistently, an in vitro ubiquitination assay showed that the WDL3 N terminus (1-195) was polyubiquitinated by MBP-COP1 (Fig. 2, Lower). GST-HYH was used as a positive control for COP1 activity (11). These results illustrated that WDL3 is a substrate of the COP1 E3 ligase and can be directly ubiquitinated by COP1 in vitro.
Fig. 2.
COP1 directly ubiquitinates WDL3 in vitro. In vitro ubiquitination assays showing that WDL3 is ubiquitinated by COP1. Purified MBP, recombinant MBP-COP1, denatured COP1 (bMBP-COP1), and GST-WDL3 were used in the assays. The ubiquitination of WDL3 and WDL3 1-195 was analyzed by immunoblots using anti-GST and anti-FLAG antibodies. GST-HYH was used as a positive control, and GST-WDL5 was used as a negative control.
Interaction of COP1 with WDL3 Occurs at the Cortical Microtubules.
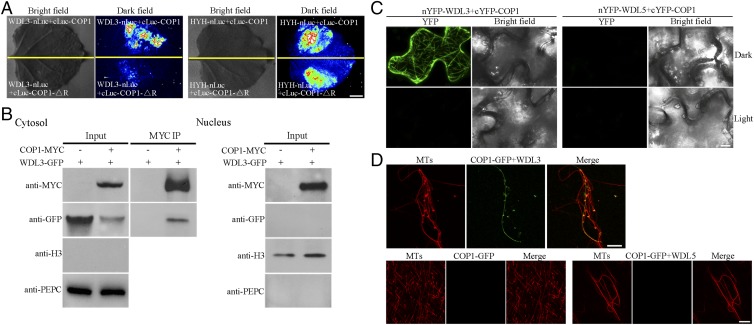
It is well established that, in darkness, COP1 functions in the nucleus (9), but recent studies also reported that some COP1 was also detectable outside the nucleus (26, 27). Since WDL3 acts as a microtubule-associated protein in the cytoplasm, it is intriguing to investigate where the interaction of COP1 and WDL3 occurs within cells. Our data showed that WDL3 is capable of interacting with COP1-△R in yeast (Fig. 1A); however, the interaction of WDL3 with COP1-△R was unable to be detected in dark-grown plants (Fig. 3 A, Left). A previous study showed that deletion of the RING domain resulted in COP1 localization only in the nucleus (42). A nuclear-localized transcription factor, HYH, was found to interact with both COP1 and COP1-△R in darkness (Fig. 3 A, Right), indicating that COP1-△R is indeed localized in the nucleus in darkness. Thus, our observations indicate that the interaction of COP1 and WDL3 may occur in the cytoplasm in darkness in living plant cells. We then isolated the nuclear and soluble fractions from dark-grown plants expressing 35S:COP1-MYC and 35S:WDL3-GFP. The subsequent coimmunoprecipitation assay showed that both the tagged COP1 and WDL3 proteins were detected in the soluble fraction and that their interaction did occur in this fraction (Fig. 3 B, Left). In contrast, only tagged COP1 was detected in the nuclear fraction (Fig. 3 B, Right). Anti-Histone 3 (H3) (a nuclear marker) (43) and anti-phosphoenolpyruvate carboxylase (PEPC) (a soluble marker) (44) blots were performed to show that H3 was only detected in the nuclear fraction and PEPC was only detected in the soluble fraction (Fig. 3B). Collectively, these data demonstrate that COP1 interacts with WDL3 outside the nucleus of the plant cell.
Fig. 3.
Interaction of COP1 with WDL3 at microtubules. (A) WDL3 did not interact with COP1-△R revealed by the firefly LCI assays in tobacco leaves in the dark. HYH was used as a positive control and exhibited interaction with COP1 or COP1-△R under the same conditions. (Scale bar: 1 cm.) (B) Coimmunoprecipitation of cytoplasmic COP1 and WDL3 extracted from tobacco leaves expressing 35S:COP1-MYC and 35S:WDL3-GFP. Anti-MYC-Agarose Affinity Gel was used for immunoprecipitation, and the precipitated proteins were analyzed by immunoblots using an anti-GFP antibody. PEPC and H3 were used as cytosolic and nuclear markers, respectively. (C) BiFC assays showing interaction between COP1 and WDL3 in tobacco leaves in the dark. Negative controls used were nYFP-WDL5+cYFP-COP1. (Scale bar: 20 μm.) (D) Confocal microscopy images of microtubules polymerized from rhodamine-labeled tubulin (20 μM) incubated in the presence or absence of 3 μM GST-WDL3 or GST-WDL5 with COP1-GFP fusion proteins for 20 min. (Scale bar: 10 μm.)
To further verify the interaction of COP1 and WDL3 in living plant cells, we performed bimolecular fluorescence complementation (BiFC) assays. The fluorescence signal from yellow fluorescent protein (YFP) formed filamentous structures only in pavement cells of tobacco leaves when 35S:nYFP-WDL3 and 35S:cYFP-COP1 were coexpressed in the dark, but not in the light or in tobacco leaves coexpressing 35S:nYFP-WDL5 and 35S:cYFP-COP1 (Fig. 3C). These observations suggest that the interaction of COP1 and WDL3 occurs at cortical microtubules. To further confirm their localization, we performed an in vitro assay using paclitaxel-stabilized microtubules polymerized from rhodamine-labeled tubulin and His-COP1-GFP and GST-WDL3 fusion proteins purified from E. coli. Confocal microscopy showed that microtubules exhibited single filamentous patterns. The colocalization of microtubules with COP1-GFP was not found in the absence of GST-WDL3 whereas filamentous structures of COP1-GFP were detected when GST-WDL3, but not GST-WDL5, was added into the system (Fig. 3D). These data demonstrate that the interaction of COP1 and WDL3 occurs at cortical microtubules.
It was previously shown that the interaction of COP1 with a resistance (R) protein HRT from Arabidopsis was observed in the periphery of the cell when exposed to the dark (27). Together with this report, we hypothesized that COP1 may associate with target proteins localized in different cellular compartments.
WDL3 Protein Accumulates in the cop1-6 Mutant in the Dark.
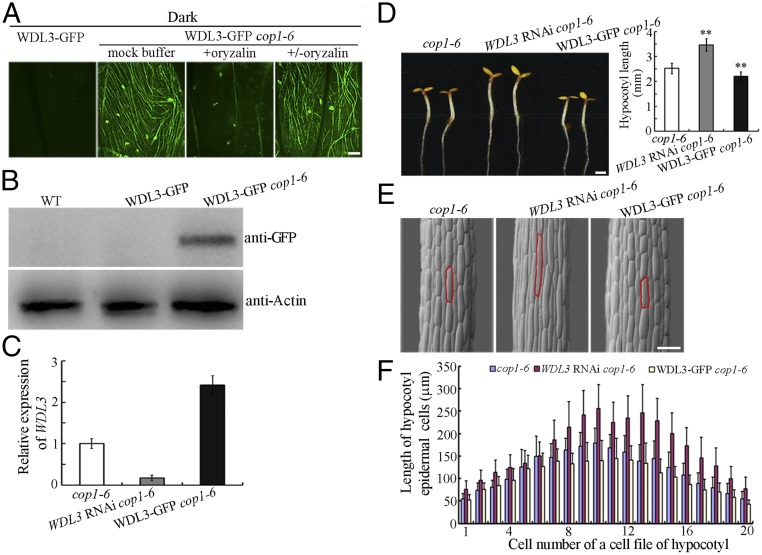
Since COP1 interacts with and ubiquitinates WDL3, we next asked whether the abundance of WDL3 protein is regulated by COP1. Dark-grown cop1 mutant seedlings display characteristics of light-grown seedlings, including short hypocotyls (35). We crossed a WDL3-GFP transgenic plant with a weak cop1 mutant allele (cop1-6) (35). Confocal microscopy revealed that WDL3-GFP signals were visible in filamentous structures in hypocotyl epidermal cells of etiolated WDL3-GFP cop1-6 seedlings, but not in those of WDL3-GFP transgenic seedlings. These filamentous structures containing WDL3-GFP could be disrupted by treatments with the microtubule (MT)-disrupting drug, oryzalin, and recovered after oryzalin washout (Fig. 4A), demonstrating that WDL3 protein is stable in the cop1-6 mutant and is capable of binding cortical microtubules in the dark. Western blots using an anti-GFP antibody further confirmed that the WDL3 protein level was negatively regulated by COP1 in the dark (Fig. 4B and Fig. S4).
Fig. 4.
Overexpression of WDL3 partially suppressed hypocotyl growth of etiolated cop1 mutants. (A) Confocal images of epidermal cells from etiolated hypocotyls of WDL3-GFP and WDL3-GFP cop1-6 seedlings. The filamentous pattern of WDL3-GFP was disrupted when cells of WDL3-GFP cop1-6 seedlings were treated with oryzalin whereas filamentous structures of WDL3-GFP were partially recovered after oryzalin washout. (B) Western blots using proteins extracted from WDL3-GFP and WDL3-GFP cop1-6 seedlings. Anti-Actin was used as a loading control. (C) WDL3 expression was determined using real-time quantitative RT-PCR with RNAs extracted from the cop1-6, WDL3 RNAi cop1-6, and WDL3-GFP cop1-6 seedlings. The expression of WDL3 in the cop1-6 mutants was set to 1. The data represent the mean ± SD of three independent experiments. (D) WDL3 RNAi cop1-6 seedlings developed longer hypocotyls whereas WDL3-GFP cop1-6 seedlings developed shorter hypocotyls (t test, **P < 0.01) when grown on half-strength Murashige and Skoog growth medium in the dark for 4 d. The graph shows the average hypocotyl length measured from a minimum of 32 seedlings. Error bars represent the mean ± SD. (Scale bar: 1 mm.) (E) Scanning electron microscopy images of hypocotyl epidermal cells from cop1-6, WDL3 RNAi cop1-6, and WDL3-GFP cop1-6 seedlings grown in the dark for 4 d. (F) Lengths of hypocotyl epidermal cells of cop1-6, WDL3 RNAi cop1-6, and WDL3-GFP cop1-6 seedlings grown in the dark for 4 d. Error bars represent the mean ± SD. (Scale bars: A, 10 μm; E, 100 μm.)
Since WDL3 protein is more stable in the cop1-6 mutant in the dark, we examined whether increasing or decreasing WDL3 expression could alter the hypocotyl phenotype of dark-grown cop1-6 seedlings. We crossed WDL3 RNAi with cop1-6 to generate the WDL3 RNAi cop1-6 double mutant. Real-time quantitative RT-PCR results showed that the level of WDL3 transcript was dramatically reduced in the WDL3 RNAi cop1-6 but was considerably enhanced in the WDL3-GFP cop1-6 seedlings (Fig. 4C). Phenotypes of 4-d-old dark-grown seedlings of the WDL3-GFP cop1-6 and WDL3 RNAi cop1-6 double mutants showed that increasing WDL3 expression could significantly suppress hypocotyl elongation of etiolated cop1-6 seedlings whereas decreasing WDL3 expression in cop1-6 mutants resulted in obviously longer hypocotyls (Fig. 4D). Because the organization of cortical microtubules is related to the elongation rate of etiolated hypocotyl epidermal cells (45, 46), we found that parallel arrays of cortical microtubules were generally transversely oriented to the longitudinal hypocotyl growth axis in the etiolated hypocotyls of WDL3 RNAi cop1-6 seedlings. By contrast, random, oblique, or longitudinal cortical microtubules were observed in most of the cop1-6 hypocotyl cells (Fig. S5). These observations are consistent with the longer hypocotyl phenotype of etiolated WDL3 RNAi cop1-6 mutant seedlings. Previous studies indicated that microtubules were very stable in cop1 mutants and that WDL3 is a microtubule stabilizer (34, 36). Consistent with these reports, we found that decreasing WDL3 expression enhanced the sensitivity of the cortical microtubules in cop1-6 cells to oryzalin treatment (Fig. S6). These data further confirmed that COP1 is responsible for the degradation of WDL3 with regard to microtubule organization and stability.
Scanning electronic microscopy revealed no major differences in hypocotyl cell profiles among cop1-6, WDL3-GFP cop1-6, and WDL3 RNAi cop1-6 seedlings. The cell numbers in individual hypocotyl epidermal cell files of different seedlings were similar (∼18 to ∼20). However, the hypocotyl cell length of etiolated cop1-6 mutant seedlings was decreased when WDL3 was overexpressed but increased when WDL3 was underexpressed (Fig. 4 E and F), consistent with the negative role of WDL3 in regulating hypocotyl cell elongation. In addition, we used the CRISPR/Cas9 technique (47) to generate two wdl3 mutants (wdl3-1 and wdl3-2), and the observations showed that hypocotyl cell lengths of 7-d-old light-grown wdl3-1 and wdl3-2 seedlings were significantly longer (Fig. S7), consistent with previously reported phenotypes of WDL3 RNAi seedlings (36). Moreover, hypocotyl cell lengths of 4-d-old dark-grown wdl3-1 cop1-6 and wdl3-2 cop1-6 seedlings were much longer than those of cop1-6 mutants (Fig. S8). Taken together, our data demonstrate that WDL3 is a downstream factor of COP1-mediated hypocotyl growth and is involved in inhibiting hypocotyl cell elongation.
In conclusion, our results provide definitive evidence for a physiological role of COP1 outside the nucleus in regulating hypocotyl elongation of etiolated seedlings. Combined with previous studies involving transcription factors including HY5, HYH, and BBX21, our study provides a molecular framework of COP1-mediated hypocotyl cell elongation (Fig. S9). In the dark, photomorphogenesis-promoting transcription factors (including HY5, HYH, and BBX21) are degraded by COP1 in the nucleus, and the microtubule-associated protein WDL3 is degraded by COP1 in the cytoplasm; thus, faster etiolated hypocotyl cell elongation is facilitated. In the light, COP1 function in the nucleus is impeded by light, which consequently allows the photomorphogenesis-promoting transcription factors to accumulate and suppress hypocotyl cell elongation; additionally, WDL3 is stabilized in the cytosol, which consequently allows WDL3 to inhibit hypocotyl cell elongation by binding and regulating cortical microtubules. In the light, the majority of COP1 is localized in the cytosol of hypocotyl cells (9); however, COP1-targeted WDL3 degradation is inactivated, possibly because COP1 is unable to interact with WDL3 in the light (Fig. S1). The underlying molecular mechanism remains currently unknown. It was recently shown that phytochromes inactivate the COP1/SPA complex in the nucleus by disrupting the direct interaction of COP1 and SPA proteins (24). Phytochrome-mediated inactivation of COP1 activity may also occur in the cytosol, because it was observed long ago that a red light pulse promotes the rapid formation of phyA-containing cytosolic spots, referred to as sequestered areas of phytochromes (SAPs), which appear before the nuclear transport of phyA (6, 48, 49, 50). However, further investigation is needed to verify this hypothesis. Collectively, our data indicate that COP1 controls photomorphogenesis not only by degrading nuclear-localized factors (such as transcription factors), but also by degrading cytosol-localized effectors (such as microtubule-associated protein WDL3).
Materials and Methods
Plant Materials and Growth Conditions.
The WT, cop1-6, and WDL3-GFP Arabidopsis seedlings were of the Columbia (Col) ecotype. WDL3-GFP cop1-6 and WDL3 RNAi cop1-6 were generated by genetic crossing. After surface sterilization, seeds were sown on Murashige and Skoog growth medium (1/2× Murashige and Skoog basal salts, 1% sucrose, and 0.8% agar). After 2 d of incubation at 4 °C in darkness, the seeds were exposed to 8 h of white light to induce uniform germination, as described previously (51), and then transferred to grow in the dark.
Yeast Two-Hybrid Assay.
To test the interactions between COP1 and WDL3, and between COP1 and WDL5, the coding sequences of full-length, or the indicated fragments of, WDL3, WDL5, or COP1 were separately fused into the pGBKT7 (for expressing binding domain-fused protein) or pGADT7 (for expressing activation domain-fused protein) vectors. The respective combinations of plasmids were cotransformed into the yeast strain AH109. Transformed yeast cells were separately spread onto 2D synthetic dropout medium (-Trp/-Leu) and 4D selective medium (-Trp/-Leu/-His/-Ade) and incubated at 28 °C for ∼4 to 5 d. To determine the intensity of protein interaction, saturated yeast cultures were diluted to 10−1 and 10−2 and then spotted onto selection medium.
In Vitro Pull-Down Assay.
Recombinant proteins were expressed and purified from E. coli according to the manufacturer’s (GE Healthcare) protocol. Prey proteins of interest were incubated with the immobilized bait protein for 2 h at 4 °C. After binding in vitro, unbound prey proteins were washed with cold PBS buffer five times, and the eluted proteins were boiled in 5× SDS loading dye. The protein samples were then separated by 10% SDS/PAGE gels and detected by immunoblots.
Firefly Luciferase Complementation Imaging Assay.
The coding sequences of WDL3, WDL3 1-195, WDL3 196-339, and WDL5 were fused to the N terminus of nLUC (35S:nLuc vector) (38), respectively, and the coding sequences of COP1 and COP1-△R were fused to the C terminus of cLUC (35S:cLuc vector) (38), respectively. The respective combinations of vectors were transformed into the Agrobacterium strain GV3101, which was then infiltrated into tobacco leaves. After infiltration, the tobacco leaves were grown in light for 3 d and then incubated in darkness for 2 h. The abaxial sides of leaves were sprayed with 1 mM luciferin and then kept in the dark for 10 min. A charge-coupled device (CCD) (1300B; Roper Scientific) camera was used to capture the LUC signal at −110 °C. The exposure time was 20 min for cLUC-COP1+WDL3-nLuc.
Bimolecular Fluorescence Complementation Assays.
To determine where the interaction of COP1 and WDL3 occurred within the plant cell, the full-length coding sequences of WDL3, WDL5, or COP1 were separately fused into the pSPYNE or pSPYCE vectors (52). The plasmids of nYFP-WDL3 and cYFP-COP1 or of nYFP-WDL5 and cYFP-COP1 were introduced into Agrobacterium strain GV3101, which was then infiltrated into young tobacco leaves. After infiltration, the tobacco leaves were grown in light for 3 d and then incubated in darkness for 2 h. The YFP fluorescence signal was then detected using a Zeiss LSM 710 META confocal microscope.
Co-IP Assay.
For co-IP experiments, the 35S:WDL3-GFP and 35S:COP1-MYC constructs were transformed into Arabidopsis mesophyll protoplasts. After overnight incubation in light, the protoplasts were incubated in darkness for 3 h and then lysed. COP1-MYC proteins were immunoprecipitated by anti-MYC-Agarose Affinity Gel (Cat. No. A7470; Sigma-Aldrich). The mixtures were kept at 4 °C with gentle shaking for 3 h. The agarose beads were recovered by centrifugation at 14,000 × g for 5 s and washed three times with cold PBS buffer. The coimmunoprecipitation products were detected by immunoblot analysis with an anti-GFP antibody (Sigma-Aldrich).
To obtain enough proteins from the nuclear and soluble fractions, a tobacco expression system was used. After infiltration, the tobacco leaves were grown in light for 3 d and then incubated in darkness for 2 h. Soluble and nuclear proteins from tobacco leaves were isolated using a Plant Nuclei Isolation/Extraction Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The soluble proteins from tobacco leaves expressing 35S:WDL3-GFP alone, or expressing 35S:COP1-MYC and 35S:WDL3-GFP together, were immunoprecipitated with anti-MYC-Agarose Affinity Gel (Cat. No. A7470; Sigma-Aldrich). The mixtures were kept at 4 °C with gentle shaking for 3 h, and then the agarose beads were recovered by centrifugation at 14,000 × g for 5 s and washed with cold PBS three times. The samples were detected by anti-MYC and anti-GFP antibodies (Sigma-Aldrich). PEPC and H3 were used as cytosolic and nuclear markers, respectively.
In Vitro Ubiquitination Assays.
In vitro ubiquitination assays were performed as described previously (19, 53). Ubiquitination reaction mixtures (60 μL) contained 30 ng of UBE1 (E1; Boston Biochem), 30 ng of UbcH5b (E2; Boston Biochem), 10 μg of FLAG-tagged ubiquitin (FLAG-Ub; Boston Biochem), 200 ng of GST-WDL3, GST-WDL3 1-195, GST-HYH, or GST-WDL5, and 0.5 μg of MBP-COP1 (incubated with 20 μM zinc acetate) or bMBP-COP1 (denatured MBP-COP1 by being boiled for 10 min at 95 °C before addition to the ubiquitination reaction system) in a reaction buffer containing 50 mM Tris, pH 7.5, 10 mM MgCl2, and 10 mM ATP. After 2 h of incubation at 30 °C, reactions were stopped by adding 5× sample buffer. Thirty microliters of each mixture was then separated in 8% SDS/PAGE gels. Ubiquitinated GST-WDL3, GST-WDL3 1-195, and GST-HYH were detected using anti-GST and anti-FLAG antibodies (Sigma-Aldrich).
Generation of wdl3 Mutants Using CRISPR/Cas9 Technique.
The egg cell-specific promoter-controlled CRISPR/Cas9 system has been described previously (47). WT (Col) Arabidopsis plants were transformed with the CRISPR/Cas9 constructs by floral dipping. The homozygous wdl3 mutants were identified by sequencing. The wdl3-1 cop1-6 and wdl3-2 cop1-6 double mutants were generated by genetic crossing.
Measurement of Hypocotyl Lengths.
To measure the hypocotyl lengths of seedlings, seeds were cold treated at 4 °C for 2 d and then transferred to continuous white light for 8 h to induce uniform germination. The seeds were then transferred to darkness and incubated at 22 °C for 4 d. The images of seedlings, with a ruler aside, were taken by a camera, and the hypocotyl lengths of seedlings were calculated using ImageJ software. The lengths of 1 cm on the ruler and of the respective hypocotyls in the same picture were measured by the software, and the real lengths of the respective hypocotyls were calculated according to their ratios.
Quantification of Cortical Microtubules in the Cell.
The quantification of cortical microtubules was performed as previously described (36) using ImageJ software. Three repeated measurements were performed for each cell, and at least 32 cells from each treatment were used. The values were recorded, and the significance of difference was analyzed using a paired Student’s t test.
Supplementary Material
Acknowledgments
We thank Dr. Xiaoli Lin and Dr. Jingbo Jin (Chinese Academy of Sciences) for the comments on the article. This work was supported by National Science Fund for Distinguished Young Scholars Grant 31625005 (to T.M.) and National Natural Science Foundation of China Grant 31471272 (to T.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708087114/-/DCSupplemental.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 3.Guerra DD, Callis J. Ubiquitin on the move: The ubiquitin modification system plays diverse roles in the regulation of endoplasmic reticulum- and plasma membrane-localized proteins. Plant Physiol. 2012;160:56–64. doi: 10.1104/pp.112.199869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012;196:13–28. doi: 10.1111/j.1469-8137.2012.04266.x. [DOI] [PubMed] [Google Scholar]
- 5.Glickman MH. Getting in and out of the proteasome. Semin Cell Dev Biol. 2000;11:149–158. doi: 10.1006/scdb.2000.0161. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Li G, Wang H, Wang Deng X. Phytochrome signaling mechanisms. Arabidopsis Book. 2011;9:e0148. doi: 10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng XW, et al. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 8.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 9.von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 10.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 11.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo HS, et al. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;423:995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- 13.Duek PD, Elmer MV, van Oosten VR, Fankhauser C. The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol. 2004;14:2296–2301. doi: 10.1016/j.cub.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 14.Jang IC, Yang JY, Seo HS, Chua NH. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, et al. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu D, et al. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA. 2016;113:7655–7660. doi: 10.1073/pnas.1607687113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010;22:2370–2383. doi: 10.1105/tpc.109.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saijo Y, et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20:2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lian HL, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011;25:1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011;21:841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheerin DJ, et al. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell. 2015;27:189–201. doi: 10.1105/tpc.114.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, et al. Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet. 2013;9:e1004025. doi: 10.1371/journal.pgen.1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, et al. Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol. 2016;170:2340–2350. doi: 10.1104/pp.15.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong RD, et al. Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2010;107:13538–13543. doi: 10.1073/pnas.1004529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buschmann H, Lloyd CW. Arabidopsis mutants and the network of microtubule-associated functions. Mol Plant. 2008;1:888–898. doi: 10.1093/mp/ssn060. [DOI] [PubMed] [Google Scholar]
- 29.Sedbrook JC, Kaloriti D. Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 2008;13:303–310. doi: 10.1016/j.tplants.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Fan XY, et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant. 2012;5:591–600. doi: 10.1093/mp/sss041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo XM, et al. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niwa Y, Yamashino T, Mizuno T. The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:838–854. doi: 10.1093/pcp/pcp028. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, et al. Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell. 2012;24:4012–4025. doi: 10.1105/tpc.112.103838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khanna R, et al. COP1 jointly modulates cytoskeletal processes and electrophysiological responses required for stomatal closure. Mol Plant. 2014;7:1441–1454. doi: 10.1093/mp/ssu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNellis TW, et al. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, et al. Light-regulated hypocotyl elongation involves proteasome-dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis. Plant Cell. 2013;25:1740–1755. doi: 10.1105/tpc.113.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi C, Deng XW. COP1: From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15:618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen CY, et al. WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis. Plant Physiol. 2003;131:493–506. doi: 10.1104/pp.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J, Ma Q, Mao T. Ethylene regulates the Arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiol. 2015;169:325–337. doi: 10.1104/pp.15.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrin RM, Wang Y, Yuen CY, Will J, Masson PH. WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 2007;49:961–971. doi: 10.1111/j.1365-313X.2006.03015.x. [DOI] [PubMed] [Google Scholar]
- 42.Stacey MG, Hicks SN, von Arnim AG. Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell. 1999;11:349–364. doi: 10.1105/tpc.11.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, et al. The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell. 2010;22:1313–1332. doi: 10.1105/tpc.109.069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z, et al. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell. 2017;66:117–128.e5. doi: 10.1016/j.molcel.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Le J, Vandenbussche F, De Cnodder T, Van Der Straeten D, Verbelen JP. Cell elongation and microtubule behaviour in the Arabidopsis hypocotyl: Responses to ethylene and auxin. J Plant Growth Regul. 2005;24:166–178. [Google Scholar]
- 46.Crowell EF, et al. Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. Plant Cell. 2011;23:2592–2605. doi: 10.1105/tpc.111.087338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang ZP, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speth V, Otto V, Schäfer E. Intracellular localisation of phytochrome in oat coleoptiles by electron microscopy. Planta. 1986;168:299–304. doi: 10.1007/BF00392353. [DOI] [PubMed] [Google Scholar]
- 49.Nagatani A. Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol. 2004;7:708–711. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 50.Kevei E, Schafer E, Nagy F. Light-regulated nucleo-cytoplasmic partitioning of phytochromes. J Exp Bot. 2007;58:3113–3124. doi: 10.1093/jxb/erm145. [DOI] [PubMed] [Google Scholar]
- 51.Xu D, et al. Arabidopsis COP1 SUPPRESSOR 2 represses COP1 E3 ubiquitin ligase activity through their coiled-coil domains association. PLoS Genet. 2015;11:e1005747. doi: 10.1371/journal.pgen.1005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waadt R, et al. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008;56:505–516. doi: 10.1111/j.1365-313X.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- 53.Xu D, et al. The RING-finger E3 ubiquitin ligase COP1 SUPPRESSOR1 negatively regulates COP1 abundance in maintaining COP1 homeostasis in dark-grown Arabidopsis seedlings. Plant Cell. 2014;26:1981–1991. doi: 10.1105/tpc.114.124024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.