Significance
UvrD helicase plays essential roles in multiple DNA metabolic processes. Although UvrD monomers can translocate along single-stranded DNA, processive DNA unwinding in vitro requires self-assembly to form at least a UvrD dimer. However, the mechanism of activation by dimerization is not known. Using single-molecule fluorescence approaches, we show that the 2B subdomain of UvrD can freely rotate among at least four substates. Binding of a DNA substrate to a UvrD monomer induces an open conformation that does not activate its helicase activity. UvrD dimerization on DNA is accompanied by closing of the 2B subdomain conformation and helicase activation. The results emphasize the important role of the 2B subdomain in regulating helicase activity.
Keywords: single molecule, conformational heterogeneity, DNA motors, DNA repair
Abstract
Escherichia coli UvrD DNA helicase functions in several DNA repair processes. As a monomer, UvrD can translocate rapidly and processively along ssDNA; however, the monomer is a poor helicase. To unwind duplex DNA in vitro, UvrD needs to be activated either by self-assembly to form a dimer or by interaction with an accessory protein. However, the mechanism of activation is not understood. UvrD can exist in multiple conformations associated with the rotational conformational state of its 2B subdomain, and its helicase activity has been correlated with a closed 2B conformation. Using single-molecule total internal reflection fluorescence microscopy, we examined the rotational conformational states of the 2B subdomain of fluorescently labeled UvrD and their rates of interconversion. We find that the 2B subdomain of the UvrD monomer can rotate between an open and closed conformation as well as two highly populated intermediate states. The binding of a DNA substrate shifts the 2B conformation of a labeled UvrD monomer to a more open state that shows no helicase activity. The binding of a second unlabeled UvrD shifts the 2B conformation of the labeled UvrD to a more closed state resulting in activation of helicase activity. Binding of a monomer of the structurally similar Escherichia coli Rep helicase does not elicit this effect. This indicates that the helicase activity of a UvrD dimer is promoted via direct interactions between UvrD subunits that affect the rotational conformational state of its 2B subdomain.
UvrD from Escherichia coli is a nonhexameric SF1A DNA helicase/translocase, structurally similar to E. coli Rep and Bacillus stearothermophilus PcrA, which is involved in many aspects of genome maintenance (1, 2), including DNA repair (3, 4), replication (5–8), and recombination (7, 9–11). Its ATP-dependent activities include translocation along single-stranded (ss) DNA (12–16), duplex DNA unwinding (17–22), displacement of RecA filaments from ssDNA (10, 11), and pushing of proteins along ssDNA (23).
The activities of UvrD can be modulated in vitro by its assembly state and/or by binding partners (1). UvrD monomers can translocate directionally (3′ to 5′) along ssDNA [∼190 nucleotides (nts) s−1] (12, 13, 15, 16), in a reaction that is tightly coupled to ATP hydrolysis (14). However, UvrD monomers show little helicase activity (12, 16, 21, 24, 25). In the absence of accessory proteins, helicase activity in vitro requires formation of a UvrD dimer (16, 21, 25–27). The dimer unwinds dsDNA with rates of ∼70 base pair (bp) s−1 (20, 21, 24, 27). Rep and PcrA monomers also show no helicase activity and require oligomerization or an accessory protein for helicase activity in vitro (28–31).
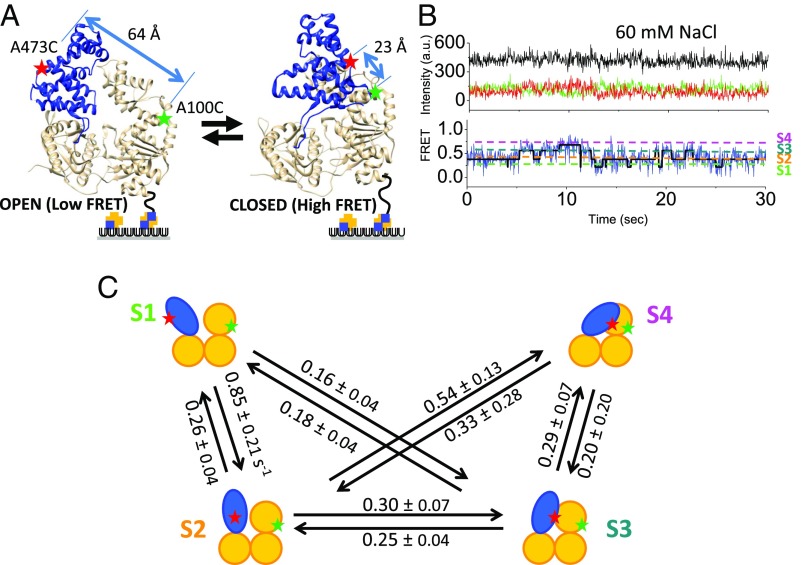
UvrD, Rep, and PcrA all contain four subdomains (1A, 2A, 1B, and 2B) (2, 32–35), and the 2B subdomain can undergo a substantial rotation about a hinge region connecting it to the 2A subdomain (2, 22, 32–37). A structure of an apo UvrD monomer shows the 2B subdomain in an open conformation (35), whereas a structure of a UvrD monomer bound to an ss/ds DNA junction shows the 2B subdomain in a closed conformation (Fig. 1A) in which the 2B domain contacts the duplex DNA (2). The positions of the 2B subdomain in its open and closed structures differ by an ∼160° rotation (2, 35). The rotational state of the 2B subdomain in apo UvrD is influenced by salt concentration and type, with a closed conformation favored at low salt and an open conformation favored at high salt (35).
Fig. 1.
The 2B subdomain of UvrD populates at least four discrete rotational conformational states. (A) The 2B subdomain (blue) in the open and closed states in crystal structures. Movement of the 2B domain is monitored by the change in FRET efficiency between a donor (Cy3) and an acceptor (Cy5) on the 1B (A100C) and the 2B (A473C) subdomains. (B) A single-molecule time trace [Cy3 (green), Cy5 (red), and FRET efficiency (blue)] in imaging buffer plus 60 mM NaCl (25.0 °C) shows anticorrelated changes in Cy3 and Cy5 with a constant total intensity (black). Transitions are observed among four FRET states (S1 to S4) as shown by the hidden Markov fit (solid black line) to the trajectory. (C) Schematic of the four FRET substates and their interconversion rates obtained from hidden Markov analyses.
Based on crystal structures, it has been proposed that the 2B subdomain plays a catalytic role in DNA unwinding for both UvrD (2) and PcrA (33) by binding to the duplex DNA. However, removal of the 2B subdomain in Rep to form RepΔ2B activates the helicase activity of the monomer (38, 39). Therefore, the 2B subdomain is autoinhibitory for Rep monomer helicase activity and thus plays a regulatory, rather than a catalytic role (39). Constraining the 2B subdomain of Rep in a closed conformation by chemical cross-linking also activates its monomer helicase activity (40). Other studies indicate that a closed conformational state of the 2B subdomain in UvrD correlates with DNA unwinding activity. When a DNA substrate is under tension in an optical tweezers experiment, a UvrD monomer displays limited helicase activity but with low processivity (22). During DNA unwinding, the 2B subdomain of UvrD is predominantly in a closed conformation, whereas upon switching to an open conformation, DNA unwinding activity is lost (22). These studies also showed that a UvrD dimer unwinds DNA with higher processivity than a monomer (22), but the basis for the higher processivity of the dimer remains unknown. One proposal is that dimerization activates helicase activity by influencing the rotational conformational state of the 2B subdomain (1, 22, 35, 38, 39).
These studies point to the importance of understanding the dynamics of the 2B subdomain and its effect on the enzyme activities of SF1A helicases/translocases. Here we use single-molecule Förster resonance energy transfer (FRET) to examine the rotational dynamics of the 2B subdomain in UvrD and show that activation of helicase activity by UvrD dimerization is accompanied by formation of a closed 2B subdomain conformational state.
Results
The 2B Subdomain Can Populate at Least Four Discrete Rotational Conformational States.
We investigated the rotational dynamics of the 2B subdomain using single-molecule total internal reflection fluorescence microscopy (smTIRF). We used a previously characterized UvrD variant, UvrD[A100C, A473C] (referred to as DM-1B/2B), containing two Cys residues at positions 100 on the 1B subdomain and 473 on the 2B subdomain within an otherwise Cys-less UvrD, UvrD∆Cys (35), which also was biotinylated on its N terminus. The two Cys residues were labeled stochastically with Cy3 (donor) and Cy5 (acceptor) dyes that undergo FRET. These positions are highlighted in the closed and open conformations of UvrD in Fig. 1. As shown previously, UvrD DM-1B/2B yields a high FRET signal when the 2B subdomain is in a closed conformation and a low FRET signal when the 2B subdomain is in an open conformation (35). Previous ensemble studies of Cy3/Cy5-labeled DM-1B/2B showed that the Cy3 and Cy5 fluorescence changes are due entirely to FRET and that increasing the NaCl concentration from 20 to 600 mM induces a transition from a closed to an open 2B subdomain, with a transition midpoint of ∼60 mM NaCl at 25 °C (35).
UvrD monomers, labeled with Cy3 and Cy5 and biotinylated on its N terminus, were immobilized onto a NeutrAvidin surface (Fig. 1) as described in Materials and Methods, and FRET signals of individual UvrD molecules were monitored by exciting Cy3 fluorescence with a 532-nm laser using smTIRF. An example smFRET trajectory in imaging buffer plus 60 mM NaCl is shown in Fig. 1B. This [NaCl] represents the midpoint in the FRET change as determined in previous ensemble studies (35) (see also Fig. 2D). These smFRET trajectories indicate that the 2B subdomain undergoes dynamic movements relative to the 1B subdomain consistent with rotation of the 2B subdomain about the hinge region connecting it to the 2A subdomain. The FRET trajectories indicate that the 2B subdomain can populate at least four discrete substates, rather than only two states as seen in the two crystal structures (2, 35). We refer to these four substates as S1, S2, S3, and S4, with S1 being the most open state with the lowest FRET efficiency (Fig. 1B). At 60 mM NaCl the four FRET states interconvert freely in solution reflecting a dynamic heterogeneity in the 2B rotational conformational state. Estimates of the rates of interconversion among the four substates were obtained using hidden Markov analyses (41), and these are shown in Fig. 1C. Direct transitions between the S1 and S4 states are observed only rarely. Because any transitions from S1 to S4 would need to go through the intermediate states S2 and S3, a direct transition between S1 and S4 would only be observed when the dwell times of the two intermediate states (S2 and S3) are very short.
Fig. 2.
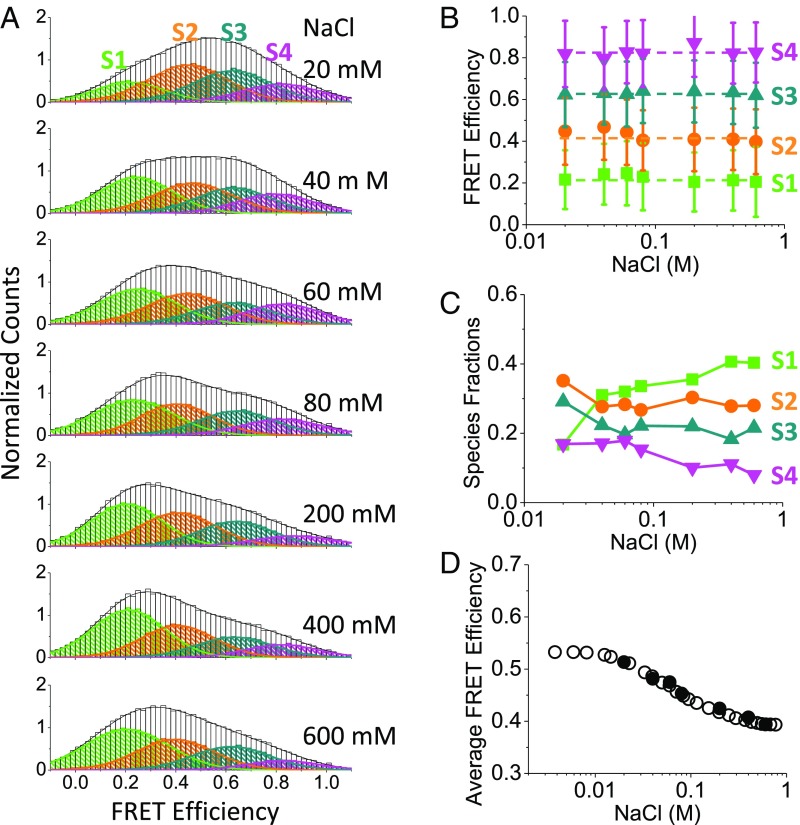
Population distribution of conformational states shifts to the more open states with increasing NaCl concentration. (A) Normalized histograms (imaging buffer) showing FRET efficiencies and species fractions of the 2B substates at [NaCl] of 20 mM (n = 145), 40 mM (n = 102), 60 mM (n = 165), 80 mM (n = 73), 200 mM (n = 94), 400 mM (n = 76), and 600 mM (n = 89). The high FRET states (S3 and S4) are more populated at low [NaCl], and the low FRET states (S1 and S2) are more populated at high [NaCl]. (B) FRET efficiencies of the four substates are independent of [NaCl]. (C) Relative populations of the four FRET substates change with [NaCl]. (D) Average FRET efficiencies from single-molecule experiments (filled circles) overlaid with the normalized FRET efficiencies from ensemble studies (35) show a net change from a closed to an open state with increasing [NaCl] (60 mM NaCl midpoint).
We next examined the effect of [NaCl] on the distributions of the 2B conformational states. The same four FRET states are observed at each [NaCl], with the relative populations of the four FRET states shifting with [NaCl] (Fig. 2 A and B) (see Fig. S1 for examples of the smFRET trajectories at each [NaCl]). As observed in previous ensemble studies (35), the average FRET efficiency for Cy3/Cy5-labeled DM-1B/2B decreases upon increasing the [NaCl] indicating a gradual opening of the 2B subdomain. The population distributions of the four FRET states redistribute with increasing [NaCl] so that the average FRET value decreases, consistent with previous ensemble FRET results (35) (Fig. 2D). The major changes associated with the increase in [NaCl] from 20 to 600 mM NaCl are an increase in S1 population and a decrease in the S3 and S4 populations (Fig. 2C). The S2 state shows relatively little change.
We compared the FRET efficiencies of the four substates to the expected FRET efficiencies of the two 2B conformational states observed in the two UvrD crystal structures (Fig. S2) using EFRET = [1 + (R/R0)6]−1, with R = 23 Å for the closed state (2), R = 64 Å for the open state (35), and R0 = 54 Å for the Cy3/Cy5 pair (42). The expected FRET efficiency of the open state in the apo UvrD crystal structure (35) agrees well with the observed S1 state FRET efficiency. However, the expected FRET efficiency estimated for the closed conformation in the UvrD–ss/dsDNA crystal structure (2) is ∼1, much higher than observed for the most closed S4 state (∼0.79). This suggests that the most closed conformation (S4) that we observe for apo UvrD at low [NaCl] is more open than observed in the crystal structure.
The 2B Rotational Conformation State Distribution Changes upon DNA Binding.
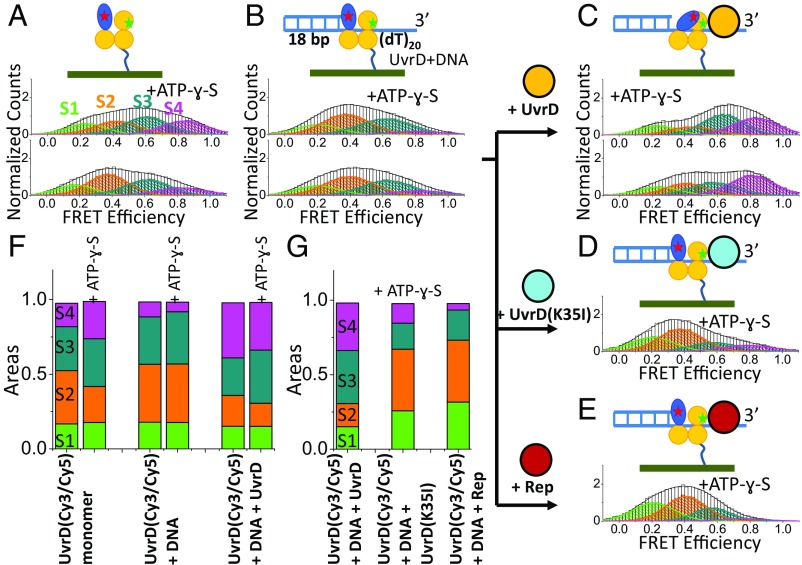
We next examined the population distributions of the four FRET states of the UvrD monomer upon binding of a DNA unwinding substrate, 3′-(dT)20 ssDNA with an 18 base pair duplex [3′-(dT)20ds18 bp], in imaging buffer plus 2 mM MgCl2 and 20 mM NaCl with or without 0.5 mM ATP-ɣ-S. As shown in Fig. 3 A and B, upon binding the DNA substrate the population of the S2 state increases, whereas that of the S4 state decreases resulting in a more open 2B conformation on average (Fig. 3 B and F). Note that the most closed S4 state is the least populated. This agrees with previous ensemble studies indicating that the 2B conformation is shifted to a more open state when a UvrD monomer is bound to a ss/dsDNA substrate (35). However, it is not consistent with the crystal structure of a UvrD monomer–ss/dsDNA complex that shows the 2B subdomain in a much more closed state (2). The addition of ATP-ɣ-S changes the population distribution only slightly, increasing the S3 population, while decreasing the S4 population (Fig. 3 B and F). The average conformation is still more open than observed in the crystal structure. When bound to the DNA substrate, UvrD monomers still exhibit dynamic interconversion among the four rotational conformational states (Fig. S3).
Fig. 3.
Effects of binding of DNA, ATP-ɣ-S, and a second UvrD molecule on the 2B conformational state distribution. (A) FRET histogram of labeled UvrD(A100C, A473C) with 0.5 mM ATP-ɣ-S (n = 72) or without (n = 55). (B) FRET histogram upon addition of 300 nM 3′(dT)20ds18 bp to A with (n = 48) or without ATP-ɣ-S (n = 89). (C) FRET histogram upon addition of 0.5 µM wtUvrD to B greatly enhances the S4 population in the presence (n = 54) or absence (n = 69) of ATP-ɣ-S. (D) FRET histogram upon addition of 0.5 µM UvrD(K35I) to B (n = 43). (E) FRET histogram upon addition of 1.0 µM Rep to B (n = 45). (F) Populations of the four substates for the experiments in A–C with or without ATP-ɣ-S. (G) Populations of the four substates for the experiments in C–E in the presence of ATP-ɣ-S.
UvrD Dimerization on DNA Shifts the 2B Conformation to a More Closed State.
The DNA unwinding (helicase) activity of UvrD is stimulated greatly in vitro by UvrD dimerization (12, 16, 21, 24, 27), although the molecular basis for this activation is not known. Based on our observation that the 2B subdomain of the E. coli Rep protein is autoinhibitory for Rep monomer helicase activity and that removal of the 2B subdomain activates the helicase activity of a Rep monomer (38, 39), we hypothesized that UvrD dimerization might relieve a similar autoinhibition by affecting the 2B subdomain rotational conformational state. Indeed, upon addition of unlabeled UvrD to the surface bound Cy3/Cy5 labeled UvrD monomer that is bound to a 3′-(dT20)-18 bp DNA substrate, we observe a shift in the population distribution of the 2B subdomain to the more closed conformations, with the most significant increase in the population of the most closed S4 state (Fig. 3 B, C, and F). Therefore, binding of a second UvrD molecule induces a more closed 2B subdomain of the Cy3/Cy5 labeled UvrD bound to the DNA. Inclusion of ATP-ɣ-S with the UvrD dimer–DNA complex results in a further increase in the S3 population at the expense of the S4 and S2 states (Fig. 3 C and F). Overall, inclusion of ATP-ɣ-S increases the population of the more closed S3 and S4 states relative to the more open S1 and S2 states. Whereas we observe dynamic transitions among the four conformational states for apo UvrD monomer and UvrD monomer bound to a DNA substrate as discussed above, we no longer observe these transitions upon formation of a UvrD dimer on the DNA. That is, although all four states are still observed, this 2B conformational state heterogeneity appears static on the time scale of our experiments (minutes) (Fig. S3).
Maluf et al. (21) previously showed in ensemble studies that binding of a second wild-type UvrD to a UvrD monomer–DNA complex stimulated helicase activity, whereas binding of either a heterologous wt Rep monomer or an ATPase deficient UvrD mutant (K35I) did not stimulate helicase activity. In fact, the binding of neither UvrD(K35I) nor Rep to the UvrD(Cy3/Cy5)–DNA complex induces a more closed 2B conformation of the Cy3/Cy5 labeled UvrD; the population of the S4 state remains very low (Fig. 3 D, E, and G), even at 1 μM Rep.
DNA Unwinding Activity Correlates with the Closed S4 Conformational State.
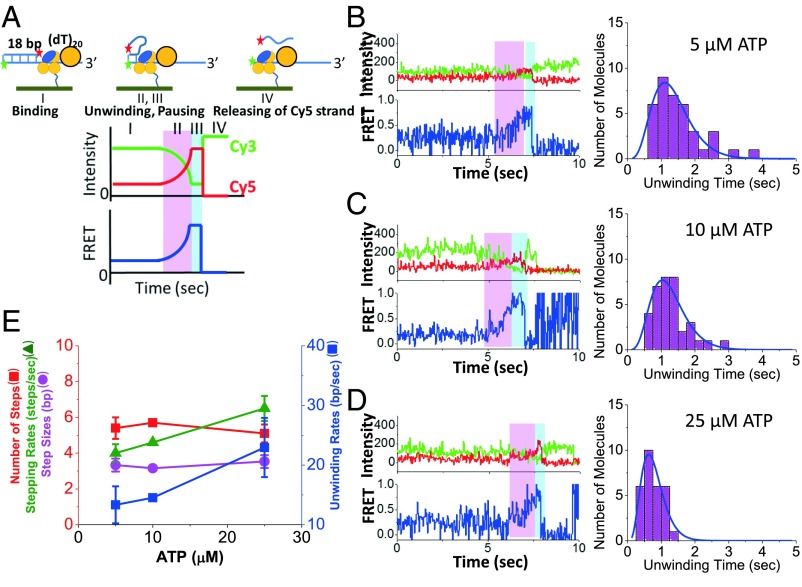
We next examined the DNA unwinding (helicase) activity of a UvrD monomer as well as a UvrD dimer to determine if there is any correlation with the 2B subdomain conformational states in imaging buffer plus 2 mM MgCl2, 20 mM NaCl. To monitor DNA unwinding, we used a short ss/dsDNA substrate described by Arslan et al. (40) containing a 3′-(dT)20 binding site for UvrD attached to an 18-bp duplex DNA labeled with Cy3 and Cy5 at opposite ends of the duplex (Fig. 4A) so that DNA unwinding results in an increase in FRET (increase in Cy5 fluorescence) and thus can be easily distinguished from photobleaching events. The expected unwinding time course for this DNA is depicted schematically in Fig. 4A and shows a gradual increase in FRET due to the fact that the Cy5 label moves closer to the Cy3 label as the DNA is unwound (stage II). The unwinding time is defined as the time difference between the start of the increase in FRET efficiency and when it reaches its maximum value. This is followed by a FRET plateau defined by a pause time (stage III), during which some conformational change may occur before release of the unwound DNA strand. The final release of the Cy5 labeled DNA strand (stage IV) leads to the disappearance of the Cy5 fluorescence and thus an increase in the Cy3 fluorescence resulting from the loss of FRET. In these experiments, a single unlabeled, biotinylated UvrD monomer was immobilized on the surface, and the Cy3/Cy5 labeled DNA was added to form a UvrD-DNA(Cy3/Cy5) complex as depicted in Fig. 4A. At 25 °C, there is little unwinding of the DNA by UvrD monomers (<1%) because the FRET efficiency remains at ∼0.3 over a wide range of ATP concentrations. As shown in Fig. 3, the UvrD 2B subdomain is observed to be mostly in the S2 and S3 states under this condition. However, upon addition of nonbiotinylated, unlabeled UvrD to form a UvrD dimer, plus ATP (5, 10, or 25 µM), significant DNA unwinding is observed (Fig. 4 B–D). The percentage of DNA molecules unwound was 28% (37/132) at 5 µM ATP, 34% (35/103) at 10 µM ATP, and 35% (29/83) at 25 µM ATP. Under these same conditions, in separate experiments (Fig. 3 C, F, and G), 33% of the Cy3/Cy5 labeled lead UvrD molecules are in the S4 conformational state. Hence, DNA helicase activity correlates with population of the most closed S4 state of the 2B subdomain.
Fig. 4.
Unwinding of DNA by a UvrD dimer. (A) Schematic of the expected DNA unwinding time course phases. Phase I is binding of Cy3/Cy5-DNA to UvrD on the surface. Phase II (magenta) is binding of a second UvrD plus ATP results in DNA unwinding with anticorrelated changes in Cy3 and Cy5 resulting in a FRET increase. Phase III (cyan) is a brief plateau in FRET efficiency. Phase IV is release of the Cy5 DNA strand. Experimental DNA unwinding trajectories and histograms of unwinding times at (B) 5 µM (n = 37), (C) 10 µM (n = 35), and (D) 25 µM ATP (n = 29). (E) Histograms were fit to a gamma distribution to obtain the DNA unwinding parameters as a function of [ATP].
The observed unwinding rates show the expected dependence on ATP concentration, with smaller average unwinding times at higher ATP concentrations (Fig. 4 B–D). The histograms for each ATP concentration were fit to a gamma distribution that describes an n-step DNA unwinding model (43) (Materials and Methods). These distributions were then analyzed to obtain DNA unwinding rates, kinetic step sizes, number of steps, and stepping rates (Fig. 4E), using the method of Neuman et al. (44). The stepping rates (k = 4.0 ± 0.5, 4.6 ± 0.1, and 6.5 ± 0.7 steps/s) and the DNA unwinding rates (r = 13 ± 3, 15 ± 1, and 23 ± 5 bp/s) increase with ATP concentration (5, 10, and 25 µM ATP), whereas the kinetic step size (d = 3.3 ± 0.2 base pairs) and the number of steps (n = 5.4 ± 0.3 steps) are independent of [ATP]. These results are in agreement with previous ensemble DNA unwinding studies that reported a kinetic step size of 4 ± 1 bp (20) and a maximum DNA unwinding rate of ∼70 bp/s at saturating ATP concentrations (≥0.25 mM) (20, 21). Similar results were obtained when the stepping rates and the step sizes were computed using the mean and variance of the unwinding times (44) (Fig. S4). The average pausing time (Fig. S5) or the total unwinding time (Fig. S6) is also dependent on ATP concentrations suggesting that some additional enzymatic steps occur without measurable FRET changes.
Recall that under the conditions that result in DNA unwinding, where a second UvrD monomer is bound to the DNA, the 2B subdomain of the lead UvrD monomer is shifted to populate the S4 state (Fig. 3C). This suggests that DNA unwinding requires the 2B subdomain of the lead UvrD subunit of the dimer to be in the more closed S4 conformational state. The fact that the binding of a second UvrD monomer facilitates this conformational shift provides direct evidence for interactions between the two UvrD monomers bound to the DNA substrate. That is, the two UvrD monomers interact to form a functional dimer, rather than functioning as two independent monomers, consistent with our previous conclusions (21, 27).
In contrast to the above experiments showing DNA unwinding by a UvrD dimer, the addition of either Rep or an ATPase deficient UvrD(K35I) to the surface immobilized wtUvrD monomer bound to the Cy3/Cy5 labeled DNA did not stimulate the helicase activity of the wtUvrD monomer, even at 1 µM Rep or 0.5 µM UvrD(K35I). This result is consistent with previous ensemble kinetic studies (21). The inability of Rep or UvrD(K35I) to stimulate DNA unwinding also correlates with their inability to induce the more closed S4 state of the lead UvrD monomer (Fig. 3 D and E).
Discussion
The monomeric forms of E. coli Rep, E. coli UvrD, and B. stearothermophilus PcrA are poor helicases, although they are rapid and processive ssDNA translocases (12, 16, 21, 24, 27–30, 38, 39). Significant helicase activity of Rep (28), UvrD (21, 24, 27, 45), and PcrA (30) in ensemble studies in vitro is only evident with enzyme in excess over DNA indicating that multiple monomers or an oligomeric form is responsible for helicase activity. Even in the absence of DNA, UvrD can self-assemble to form dimers and tetramers (45) and a preformed dimer possesses helicase activity (27).
Crystal structures show that the 2B subdomains of UvrD, Rep, and PcrA can undergo substantial rotation about a hinge region connected to the 2A subdomain (2, 32–35). The helicase activity of a Rep monomer can be activated by removal of its 2B domain (38, 39) or by cross-linking the 2B subdomain in a closed state (40). Recent studies have also shown that interaction of PcrA monomer with an accessory protein, RepD, activates helicase activity (31) and shifts the 2B subdomain to a closed state (40). Finally, although helicase activity has been detected for a UvrD monomer, those experiments were performed with the DNA under tension, which may enhance monomer activity (22). Under these conditions, monomer helicase activity was observed when its 2B subdomain was in a closed state. Under the same conditions, UvrD dimers displayed higher helicase activity with increased processivity (22).
Here we show that the 2B subdomain of UvrD can populate at least four rotational substates, rather than the two open and closed states identified in crystal structures (2, 35). These four states freely interconvert in the apo and DNA-bound UvrD monomer indicating dynamic heterogeneity. The distribution of substates is affected by salt concentration as well as DNA and ATP binding. Formation of a helicase active UvrD dimer shifts the 2B subdomain of the lead UvrD motor to the most closed 2B conformational state, S4. This state is not populated in a UvrD monomer–DNA complex but becomes significantly populated when a second UvrD binds to the DNA to form a dimer. Substitution of the second UvrD with Rep or an ATPase dead UvrD mutant does not activate the helicase activity (21) and also does not significantly populate the S4 state.
Whereas we observe a dynamic heterogeneity of conformational states for UvrD monomer, even when bound to DNA, these transitions are suppressed upon formation of a UvrD dimer on the DNA. That is, on the time scale of our experiments (minutes) the 2B conformational state heterogeneity is static. Therefore, only if one UvrD subunit dissociates from the DNA can the remaining UvrD monomer rapidly redistribute among its various 2B substates. Maluf et al. (27) showed that the pathway for UvrD dimer dissociation from DNA occurs mainly via dissociation of one UvrD with a rate constant of 0.030 ± 0.006 s−1 (i.e., a relaxation time of 34 ± 6 s). This slow dissociation is consistent with the apparent static heterogeneity that we observe for the 2B subdomain within UvrD dimers bound to DNA. The static heterogeneity that we observe here is reminiscent of that proposed by Liu et al. (46) to explain the variation in DNA unwinding rates among single molecules of RecBCD helicase.
It is not clear why a closed 2B subdomain is associated with helicase activity of UvrD, Rep, and PcrA. A UvrD monomer bound to a 3′-ss-dsDNA crystallizes with its 2B subdomain in a highly closed state (2). Based on this structure a model was proposed for how a UvrD monomer might unwind DNA, and this model presumes that interactions between the duplex DNA and the 2B subdomain in its closed state are functional for unwinding (2). Our studies show that the 2B subdomain of a UvrD monomer bound to a 3′-ss/dsDNA substrate in solution populates the more open S1, S2, and S3 states but not the most closed S4 state. This suggests that conformational states observed in crystal structures are not necessarily representative of those in solution. The S1 state, which is most populated at high [NaCl], corresponds most closely to the conformational state exhibited in the apo UvrD crystal structure (35). Even when UvrD is bound as a dimer to a 3′-ss/ds DNA substrate, promoting the more closed S4 state, this S4 state is still more open than the conformation observed in a UvrD–DNA substrate crystal structure (2). It is not clear whether a closed state of the 2B subdomain is needed to promote interactions with the duplex DNA or to alleviate an inhibitory effect. However, the fact that removal of the 2B subdomain activates the helicase activity of the structurally similar Rep monomer (38, 39) suggests that interactions of the 2B subdomain with the duplex are not generally important for SF1A helicase activity.
High ratios of UvrD to DNA are needed to observe robust helicase activity in vitro. However, complete unwinding of an ensemble population of DNA duplexes, even short ones, has never been observed in single round experiments even at saturating UvrD concentrations where UvrD dimers are populated (20, 21). Our results provide an explanation for this observation. Even at UvrD concentrations that promote dimer formation, the 2B subdomain of the lead UvrD still exists in a distribution of conformational states that interconvert slowly. If only the subset of DNA molecules with the lead UvrD motor in the S4 state can initiate DNA unwinding, then complete unwinding of the DNA population would never be observed in a single round experiment (27). In addition, a much slower second phase of DNA unwinding is always observed in such experiments (20, 21). This slow phase must reflect a slow transition from an inactive to an active UvrD dimer, consistent with the static heterogeneity that we observe for UvrD dimers bound to a DNA substrate.
Our previous ensemble studies indicated that DNA unwinding by UvrD requires at least a dimeric form (21, 27). However, evidence was lacking for direct interactions between two monomers (subunits). We suggested that dimerization might result in movement of the 2B domain to eliminate autoinhibition by this domain (1, 39). Indeed, we show here that formation of an active UvrD dimer induces a rotation of the 2B domain to a closed (S4) state, and this correlates with DNA helicase activity. The inability of Rep to activate helicase activity of a UvrD monomer indicates a specific interaction between the two UvrD subunits as suggested previously (21, 27).
Materials and Methods
Imaging buffer is 10 mM Tris⋅HCl, pH 8.3, at 25 °C, 20% (vol/vol) glycerol, 3 mM Trolox, 0.1 mg/mL BSA, 0.8% (wt/vol) dextrose, 20 units/mL glucose oxidase, and 20 units/mL catalase. DNA was synthesized and purified as described (47). ATP (Sigma Aldrich) and ATP-γ-S (Enzo) solutions were prepared as described (35). UvrD proteins were purified and labeled as described (35). Single-molecule FRET efficiencies were calculated after correction for Cy3 leakage into the Cy5 channel and instrument detection efficiencies (47, 48) (SI Materials and Methods).
SI Materials and Methods
DNA.
The ss-dsDNA, referred to as 3′-(dT)20ds18, was formed by annealing 5′-GCC TCG CTG CCG TCG CCA-3′ and 5′-TGG CGA CGG CAG CGA GGC TTT TTT TTT TTT TTT TTT TT-3′. The DNA used to monitor unwinding was formed by annealing 5′-Cy5-GCC TCG CTG CCG TCG CCA-3′ (100% Cy5) and 5′-Cy3-TGG CGA CGG CAG CGA GGC TTT TTT TTT TTT TTT TTT TT-3′ (95% Cy3). DNA duplexes were prepared by annealing two complimentary single-stranded DNA. The DNA duplex stocks (10 µM) were prepared by mixing (1–1) two single-stranded DNAs (in 10 mM Tris, pH 8.1, 0.1 M NaCl) and heating to 100 °C. The solution was slowly cooled in a boiling 1-L water bath to room temperature for 4 h. The duplex formation was confirmed by resolving on a 10% acrylamide gel.
UvrD Protein.
The Avidity Avitag sequence GLNDIFEAQKIEWHE was added to the N terminus of UvrD with the Agilent Technologies Lightning Site-Directed Mutagenesis Kit. For UvrDΔCys(A100C, A473C) Avitag, the primers used were CGA AGC CCA GAA GAT CGA GTG GCA CGA GAG CAG CCA TCA TCA TCA TCA TCA CAG CAG CGG and CGA TCT TCT GGG CTT CGA AGA TGT CGT TTA GGC CCA TGG TAT ATC TCC TTC TTA. For UvrD Avitag, the primers used were CGA AGC CCA GAA GAT CGA GTG GCA CGA GAG CAG CCA TCA TCA TCA TCA TCA CAG CAG CGG and GAC ATC TTC GAA GCC CAG AAG ATC GAG TGG CAC GAG GAC GTT TCT TAC CTG CTC GAC AGC. The entire UvrD gene was sequenced to confirm no undesired mutations. BL21(DE3)ΔUvrD cells were transformed with the UvrD Avitag (pB773) and pB232 (BirA, biotin ligase that is in a pET21a that has the pBR322 origin replaced with the pSC101 origin to allow for compatibility of the two plasmids). UvrD proteins were purified as previously described (26, 35). The UvrD concentration was determined in 10 mM Tris, pH 8.1, 200 mM NaCl, 20% (vol/vol) glycerol using an extinction coefficient of ε280 = 1.06 × 105 M−1⋅cm−1. For UvrD with biotin/His6 tags, the Poros Q and Heparin columns were replaced by a Pierce Monomeric Avidin agarose (ThermoFisher Scientific) or a Ni-NTA agarose (Qiagen) column. The Rep protein was expressed and purified as previously described (49).
The biotin–UvrDΔCys(A100C, A473C) construct was randomly labeled using equal amounts of monoreactive maleimide Cy3 and Cy5 dyes on a Histrap HP column (GE Healthcare) (35). The extent of fluorophore labeling were determined spectrophotometrically using extinction coefficients of 150,000 M−1⋅cm−1 at 550 nm for Cy3 and 250,000 M−1⋅cm−1 at 650 nm for Cy5 (GE Healthcare). The absorbance at 280 nm was used to calculate the protein concentration after correcting for the absorbance of the dyes at 280 nm (5% of the absorbance at 650 nm for Cy5 and 8% of the absorbance at 550 nm for Cy3). Absorbance of Cy3 at 550 nm was corrected for 5% of absorbance at 650 nm of Cy5. The labeling efficiencies are 62% Cy3 and 60% Cy5 for biotin–UvrDΔCys(A100C, A473C).
Single-Molecule Total Internal Reflection Microscopy Experiments.
Single-molecule experiments were performed on an Olympus IX71 microscope (model IX2_MPITIRTL) as described (47). Typical movies were recorded at a 32-ms resolution (30.3-ms exposure) at 25 °C for 1–5 min (>95% of Cy5 has photobleached). The Cy3 fluorophore was excited with a 532-nm laser, and the fluorescence emissions of Cy3 and Cy5 were collected on separated channels. The FRET efficiency was calculated from the ratio of corrected Cy3 and Cy5 signals as described. FRET efficiencies were corrected for leakages of donor into the acceptor channels and instrumental detection efficiencies as described (47, 48).
The conformation of the 2B subdomain relative to the 1B subdomain was examined by monitoring Cy3 and Cy5 fluorescence. Under different solution conditions, UvrD can exist in number of oligomeric states (45). To study the monomeric form, fluorescently biotin–UvrDΔCys(Cy3/Cy5) was sparsely immobilized onto a Neutravidin (ThermoFisher Scientific)–PEG surface at picomolar concentrations (40 pM). Under this condition, UvrD is monomeric as confirmed from the fluorescence intensities and photobleaching steps. Data were collected in the imaging buffer with 0.1 mg/mL BSA and 20 to 600 mM NaCl. Due to the nature of random labeling of two dyes at two positions, a mixture of populations is present. The population of interest has a FRET signal between the 2B and 1B subdomains. Thus, only molecules that have one Cy3 and one Cy5 were included in the analyses and were used to construct population FRET histograms (n = number of molecules). These molecules constitute less than 10% of total observable molecules, and their FRET values were subjected to hidden Markov analyses (41) to find the number of FRET states and the rates between states. The FRET histograms were normalized to an area of 1 and fitted to Gaussian distributions to obtain the mean FRET value and the fraction of individual components. The average FRET of the entire population was calculated by summing the product of the species fraction and the FRET value for all species.
The 2B subdomain conformational distribution upon the binding of ATP-ɣ-S, DNA, and a second UvrD was studied in the imaging buffer with 2 mM MgCl2 and 20 mM NaCl. The 2B conformational distribution on the DNA binding was monitored by adding a 300-nM solution of ss-dsDNA oligomer 3′-(dT)20ds18 to a surface of immobilized labeled biotin–UvrDΔCys(Cy3/Cy5). The FRET distribution is the same in the presence of 300 nM DNA or absence of free DNA (by gently washed with two flow channel volumes). The 2B conformational distribution on the binding of DNA and a second molecule [UvrD, UvrD(K35I), or Rep] was examined by adding an excess amount of the second species (two volumes 0.5 or 1.0 µM).
Single-Molecule DNA Unwinding Experiments.
The biotin–UvrD (10 µL of 40 pM) was immobilized onto the NeutrAvidin surface for 5 min and then was washed to remove unbound biotin–UvrD. The Cy3/Cy5 labeled DNAs (10 µL of 50 nM) was added and incubated for 5–10 min and washed to remove free DNA. This process was repeated as needed so that the binding density reaches 100–400 molecules per 34 µm × 68 µm surface. In the absence of biotin–UvrD, no nonspecific binding of DNA on the surface was observed. To test for monomer unwinding, the imaging buffer with 2 mM MgCl2 and 20 mM NaCl containing up to 1 mM ATP was added onto the surface. Less than 1% of DNA molecules are unwound by UvrD monomers. A DNA unwinding event is distinguished from a photobleaching or a DNA dissociation event by its anticorrelated changes in Cy3/Cy5 intensities during unwinding. This unwinding process occurs immediately before the disappearance of Cy5 signal (releasing of the Cy5 strand). The surface was regenerated by washing with 1 M NaCl and repeating the above surface binding procedure. DNA unwinding experiments by dimers were performed by adding 100 nM UvrD at various ATP concentrations. To extract the stepping rates and the number of steps at every ATP concentration, the distribution of DNA unwinding times, t, was fitted to a gamma distribution describing an n-step unwinding model (43, 44):
where k is the stepping rate (steps per s) and n is the number of steps. Since k = ku + kd (where ku = unwinding rate and kd = dissociation rate), k ∼ ku when ku >> kd. Consequently, the kinetic step size (bp) d = L/n and the unwinding rate (bp/s) r = kd or Lk/n (L is the unwinding length in bp) can be calculated. Without fitting the distribution (i.e., independent of data binning), the DNA unwinding rates can also be computed from the mean and variance of unwinding times [k = mean/var, n = mean2/var, d = L(var/mean2), and r = L/mean] (44). This method is based on the FRET change upon DNA unwinding. In the absence of ATP, we did not observe any FRET increase from the baseline FRET value of ∼0.3.
Supplementary Material
Acknowledgments
We thank A. Soranno, K. Maluf, and E. Tomko for discussions and T. Ho for oligodeoxynucleotides. This work was supported by NIH Grant GM045948 (to T.M.L.) and American Cancer Society Award PF-15-040-01-DMC (to J.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.S.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712882114/-/DCSupplemental.
References
- 1.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: Mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 2.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epshtein V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 5.Bruand C, Ehrlich SD. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol. 2000;35:204–210. doi: 10.1046/j.1365-2958.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- 6.Florés MJ, Sanchez N, Michel B. A fork-clearing role for UvrD. Mol Microbiol. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- 7.Lestini R, Michel B. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 2007;26:3804–3814. doi: 10.1038/sj.emboj.7601804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGlynn P. Helicases that underpin replication of protein-bound DNA in Escherichia coli. Biochem Soc Trans. 2011;39:606–610. doi: 10.1042/BST0390606. [DOI] [PubMed] [Google Scholar]
- 9.Arthur HM, Lloyd RG. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet. 1980;180:185–191. doi: 10.1007/BF00267368. [DOI] [PubMed] [Google Scholar]
- 10.Veaute X, et al. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrova V, et al. Active displacement of RecA filaments by UvrD translocase activity. Nucleic Acids Res. 2015;43:4133–4149. doi: 10.1093/nar/gkv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomko EJ, Fischer CJ, Lohman TM. Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J Mol Biol. 2012;418:32–46. doi: 10.1016/j.jmb.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomko EJ, et al. 5′-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. EMBO J. 2010;29:3826–3839. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KS, Balci H, Jia H, Lohman TM, Ha T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat Commun. 2013;4:1878. doi: 10.1038/ncomms2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matson SW. Escherichia coli helicase II (urvD gene product) translocates unidirectionally in a 3′ to 5′ direction. J Biol Chem. 1986;261:10169–10175. [PubMed] [Google Scholar]
- 18.Runyon GT, Bear DG, Lohman TM. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc Natl Acad Sci USA. 1990;87:6383–6387. doi: 10.1073/pnas.87.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Runyon GT, Lohman TM. Escherichia coli helicase II (uvrD) protein can completely unwind fully duplex linear and nicked circular DNA. J Biol Chem. 1989;264:17502–17512. [PubMed] [Google Scholar]
- 20.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 21.Maluf NK, Fischer CJ, Lohman TM. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 22.Comstock MJ, et al. Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015;348:352–354. doi: 10.1126/science.aaa0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokoloski JE, Kozlov AG, Galletto R, Lohman TM. Chemo-mechanical pushing of proteins along single-stranded DNA. Proc Natl Acad Sci USA. 2016;113:6194–6199. doi: 10.1073/pnas.1602878113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali JA, Maluf NK, Lohman TM. An oligomeric form of E. coli UvrD is required for optimal helicase activity. J Mol Biol. 1999;293:815–834. doi: 10.1006/jmbi.1999.3185. [DOI] [PubMed] [Google Scholar]
- 25.Yokota H, Chujo YA, Harada Y. Single-molecule imaging of the oligomer formation of the nonhexameric Escherichia coli UvrD helicase. Biophys J. 2013;104:924–933. doi: 10.1016/j.bpj.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runyon GT, Wong I, Lohman TM. Overexpression, purification, DNA binding, and dimerization of the Escherichia coli uvrD gene product (helicase II) Biochemistry. 1993;32:602–612. doi: 10.1021/bi00053a028. [DOI] [PubMed] [Google Scholar]
- 27.Maluf NK, Ali JA, Lohman TM. Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J Biol Chem. 2003;278:31930–31940. doi: 10.1074/jbc.M304223200. [DOI] [PubMed] [Google Scholar]
- 28.Cheng W, Hsieh J, Brendza KM, Lohman TM. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J Mol Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 29.Ha T, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 30.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 31.Chisty LT, et al. Monomeric PcrA helicase processively unwinds plasmid lengths of DNA in the presence of the initiator protein RepD. Nucleic Acids Res. 2013;41:5010–5023. doi: 10.1093/nar/gkt194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 33.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 34.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 35.Jia H, et al. Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. J Mol Biol. 2011;411:633–648. doi: 10.1016/j.jmb.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 37.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng W, et al. The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc Natl Acad Sci USA. 2002;99:16006–16011. doi: 10.1073/pnas.242479399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brendza KM, et al. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci USA. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arslan S, Khafizov R, Thomas CD, Chemla YR, Ha T. Protein structure. Engineering of a superhelicase through conformational control. Science. 2015;348:344–347. doi: 10.1126/science.aaa0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabanayagam CR, Eid JS, Meller A. Using fluorescence resonance energy transfer to measure distances along individual DNA molecules: Corrections due to nonideal transfer. J Chem Phys. 2005;122:061103. doi: 10.1063/1.1854120. [DOI] [PubMed] [Google Scholar]
- 43.Lucius AL, Maluf NK, Fischer CJ, Lohman TM. General methods for analysis of sequential “n-step” kinetic mechanisms: Application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys J. 2003;85:2224–2239. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neuman KC, et al. Statistical determination of the step size of molecular motors. J Phys Condens Matter. 2005;17(47):S3811–S3820. doi: 10.1088/0953-8984/17/47/012. [DOI] [PubMed] [Google Scholar]
- 45.Maluf NK, Lohman TM. Self-association equilibria of Escherichia coli UvrD helicase studied by analytical ultracentrifugation. J Mol Biol. 2003;325:889–912. doi: 10.1016/s0022-2836(02)01276-7. [DOI] [PubMed] [Google Scholar]
- 46.Liu B, Baskin RJ, Kowalczykowski SC. DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate. Nature. 2013;500:482–485. doi: 10.1038/nature12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen B, et al. Diffusion of human replication protein A along single-stranded DNA. J Mol Biol. 2014;426:3246–3261. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joo C, Ha T. Single molecule FRET with total internal reflection microscopy. In: Selvin PR, Ha T, editors. Single Molecule Techniques: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. pp. 3–36. [Google Scholar]
- 49.Lohman TM, Chao K, Green JM, Sage S, Runyon GT. Large-scale purification and characterization of the Escherichia coli rep gene product. J Biol Chem. 1989;264:10139–10147. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.