Significance
Meiotic recombination is a process in plants, animals, and fungi during which chromosomes exchange their parts. It generates new genetic variation in the progeny and is one of the reasons why progeny are both similar to and different from their parents. Recombination is initiated by formation of breaks in chromosomal DNA. We generated a high-resolution map of sites where these breaks are formed in the genome of maize. Surprisingly, we found that DNA breaks are abundant in all genome regions, including sites where recombination was thought to be limited, such as repetitive DNA. The map will allow understanding of how recombination patterns shape the genome and aid development of more efficient breeding methods.
Keywords: recombination, meiosis, double-strand breaks, chromosomes, maize
Abstract
Meiotic recombination is the most important source of genetic variation in higher eukaryotes. It is initiated by formation of double-strand breaks (DSBs) in chromosomal DNA in early meiotic prophase. The DSBs are subsequently repaired, resulting in crossovers (COs) and noncrossovers (NCOs). Recombination events are not distributed evenly along chromosomes but cluster at recombination hotspots. How specific sites become hotspots is poorly understood. Studies in yeast and mammals linked initiation of meiotic recombination to active chromatin features present upstream from genes, such as absence of nucleosomes and presence of trimethylation of lysine 4 in histone H3 (H3K4me3). Core recombination components are conserved among eukaryotes, but it is unclear whether this conservation results in universal characteristics of recombination landscapes shared by a wide range of species. To address this question, we mapped meiotic DSBs in maize, a higher eukaryote with a large genome that is rich in repetitive DNA. We found DSBs in maize to be frequent in all chromosome regions, including sites lacking COs, such as centromeres and pericentromeric regions. Furthermore, most DSBs are formed in repetitive DNA, predominantly Gypsy retrotransposons, and only one-quarter of DSB hotspots are near genes. Genic and nongenic hotspots differ in several characteristics, and only genic DSBs contribute to crossover formation. Maize hotspots overlap regions of low nucleosome occupancy but show only limited association with H3K4me3 sites. Overall, maize DSB hotspots exhibit distribution patterns and characteristics not reported previously in other species. Understanding recombination patterns in maize will shed light on mechanisms affecting dynamics of the plant genome.
Meiotic recombination is responsible for generating genetic variation and facilitates purging deleterious mutations from genomes. However, despite their importance, recombination events in most species are not distributed uniformly across the genome. Instead, they are predominantly clustered at recombination hotspots, which are interspersed with coldspots, regions of low recombination (1–3). Studies in yeast and mammals have linked initiation of meiotic recombination to the presence of open chromatin sites located upstream from genes and to trimethylation of lysine 4 in histone H3 (H3K4me3), a mark of active chromatin (2, 4–7). Core components of the meiotic recombination pathway are well conserved among species (8), and it is conceivable that this conservation would result in universal characteristics of recombination landscapes. However, as yeast and mammals possess unique characteristics, such as a very small genome size (yeast) or the presence of species-specific recombination mechanism features, such as PRDM9 (mammals) (9), analyzing a wider range of taxa will be helpful to distinguish between species-specific and universal aspects of recombination landscape. Here, we examined recombination initiation patterns in maize (Zea mays L.), a large-genome higher eukaryote with a set of recombination proteins that are widely shared by many species (8).
Meiotic recombination is universally initiated early in meiosis by formation of double-strand breaks (DSBs) in chromosomal DNA by a topoisomerase VI-like complex (10, 11). In plants, this complex contains three paralogous proteins, SPO11-1, SPO11-2, and MTOPVIB (11). The DSBs are subsequently resected to create single-stranded (ssDNA) ends, which become coated by two proteins, RAD51 and DMC1, to form a nucleoprotein filament, which then invades the homologous double-stranded DNA (dsDNA) region (12). Meiotic DSB repair results in the formation of two product types, crossovers (COs) and noncrossovers (NCOs), the latter including gene conversions (Fig. 1A). CO formation involves formation and resolution of double Holliday junctions (13). In contrast, most NCOs are produced through a separate pathway that does not use Holliday junction intermediates but utilizes a synthesis-dependent strand annealing mechanism (14, 15). Some NCOs are also formed through alternative resolution of double Holliday junctions (16, 17). Repair of the majority of meiotic DSBs leads to NCOs. In maize male meiocytes, there are ∼500 DSBs, yet their repair leads to fewer than ∼20 COs (18). The distribution of DSB hotspots may not necessarily mirror the CO hotspot distribution. In budding yeast and mice, the fraction of DSBs repaired in COs and NCOs varies in different genomic regions (19, 20). This, however, is not the case in humans (21).
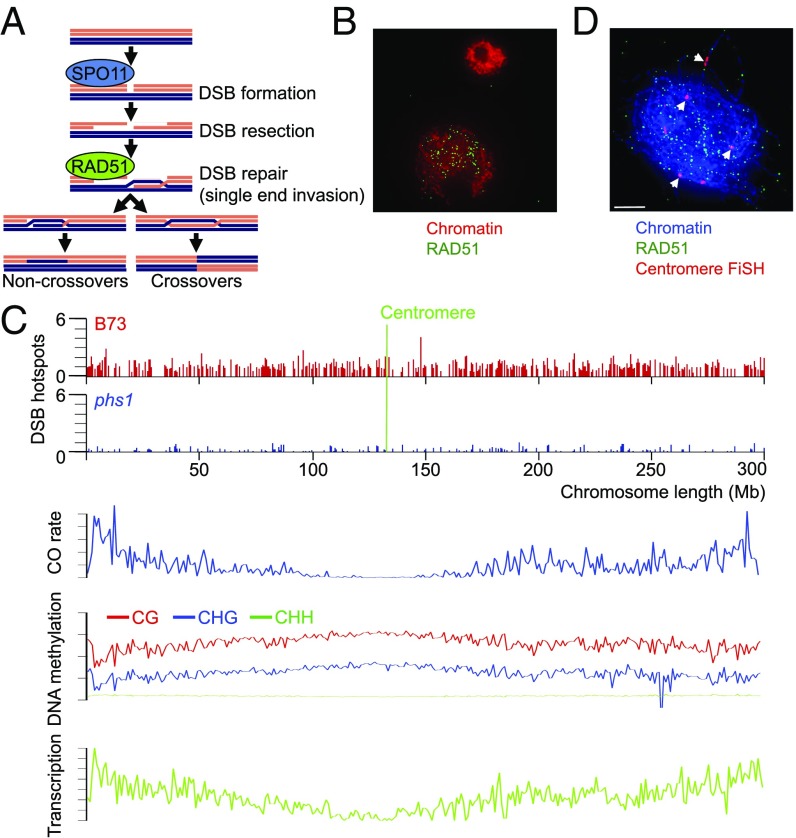
Fig. 1.
Mapping meiotic DSB hotspots in maize. (A) Diagram of the main steps of the meiotic recombination pathway. (B) Comparison of RAD51 antibody staining in a meiocyte (large cell) and a somatic cell (small cell) of the maize anther showing that RAD51 foci are absent from somatic cells. (C) Map of DSB hotspots on chromosome 1 of the B73 inbred of maize (Top) and the phs1 mutant (Bottom). The y axis represents hotspot strength compared to input control. Position of the centromere is marked with a green line. See SI Appendix, Fig. S4, for the other chromosomes. Shown below are patterns of CO distribution, DNA methylation, and location of genes expressed during meiosis, calculated using a 1-Mb sliding window. (D) Colocalization of RAD51 foci with maize centromeres in a zygotene meiocyte of B73 using immuno-FiSH. RAD51 foci overlapping centromeres identified in 3D images are marked with arrows. (Scale bar: 5 µm.) Images in B and D are flat projections of 5–10 consecutive optical sections of 3D image stacks.
In mice and humans, the key determinant of the recombination initiation landscape is PRDM9, a histone methyl transferase that also contains a series of DNA-binding zinc finger domains (9). Through the zinc finger domains, PRDM9 targets H3K4 trimethylation to genome sites containing a guanine-cytosine (GC)-rich degenerate DNA sequence motif, which become DSB hotspot locations (9). PRDM9 homologs have been identified in many vertebrates but not in plants or fungi (22, 23). In contrast to mammals, yeast does not have DNA sequence motifs associated with meiotic DSB formation, and its DSB hotspots tend to be located in promoters of genes (2, 24). However, yeast has a small and very compact genome characterized by short intergenic spaces and small amounts of repetitive DNA. These features make it distinct from many higher eukaryotes, which possess large genomes rich in repetitive DNA, both known to affect recombination event distribution (3, 25).
To elucidate recombination landscape features that are common to a broad spectrum of eukaryotes, we mapped sites of recombination initiation in maize. The genome of maize is typical for many higher eukaryotes in terms of size (2.4 Gbp) and the content of repetitive DNA (∼85%) (26). Most COs in maize are formed near chromosome ends (27) and close to genes (3). CO formation is suppressed at centromeres. Extensive pericentromeric regions also exhibit few COs, even though they contain a significant fraction, ∼20%, of maize genes (27). Mapping DSB hotspots in maize revealed that recombination is initiated in all regions of chromosomes, including centromeric and pericentromeric regions. Furthermore, the majority of DSBs are in repetitive DNA, mainly retrotransposons, whereas only one-quarter of DSBs are in genes. Our data suggest that DSBs in repetitive DNA and those in genic regions exhibit distinct features and that only DSBs formed in genic regions contribute to CO formation.
Results
DSB Hotspots Are Ubiquitous in All Chromosome Regions.
To generate a map of DSB sites, we used a chromatin immunoprecipitation (ChIP) approach in which chromatin from flowers containing zygotene meiocytes was enriched in fragments associated with the RAD51 protein. RAD51 forms distinct foci on chromosomes at the sites of meiotic DSBs, facilitating their repair (12, 28). In maize, the foci are visible from early zygotene to late pachytene, with a peak number of roughly 500 per meiocyte at midzygotene (SI Appendix, Fig. S1). RAD51 foci have been reported absent from somatic cells in maize plants grown under normal conditions (28), which we confirmed by examining over 200 somatic cells from zygotene anthers (Fig. 1B). Thus, the RAD51-associated chromatin originated exclusively from meiocytes.
DNA fragments recovered from ChIP were sequenced on the Illumina platform, and the sequence reads were mapped to the reference genome (26). To identify sites in the genome occupied by RAD51, we compared the distribution of RAD51 ChIP-seq reads to the following: (i) meiotic chromatin that was not subjected to ChIP (input chromatin); (ii) ChIP products generated from meiotic chromatin using preimmune IgG, instead of the anti-RAD51 antibody; and (iii) ChIP products generated from leaf tissue chromatin using the anti-RAD51 antibody. As expected, the two latter treatments yielded very low amounts of DNA (∼2 and ∼5% of those recovered in RAD51 ChIP on meiotic flowers, respectively). Using a conservative false discovery rate of <0.01, we identified 3,126 meiotic RAD51 peaks across the maize genome (SI Appendix, Fig. S2).
We utilized a recombination-deficient meiotic mutant to validate that the 3,126 peaks are indeed sites of RAD51 present at meiotic DSBs. As a spo11 mutant in maize has not been described yet, we employed the phs1 mutant, which is defective in early steps of meiotic recombination. The phs1 mutant lacks RAD51 foci; instead of the 500 foci observed in the wild type, phs1 meiocytes show only about three RAD51 foci per cell (29). Very little DNA was recovered in the phs1 ChIP experiments, ∼2% of the amount obtained in the wild type. Furthermore, we found only about 60 RAD51 peaks in the ChIP-seq data in phs1, with none of them at the same location as in the wild type. Thus, we concluded that the 3,126 wild-type peaks indeed represent hotspots of meiotic DSB formation.
We found that DSB hotspots were, on average, 1.2 kb long (SI Appendix, Fig. S3A) and exhibited relatively random spacing (SI Appendix, Fig. S3B). The number of hotspots per chromosome was strongly correlated with chromosome length (r = 0.95; P < 2.2e−16) (SI Appendix, Fig. S3C). Hotspots were present in all chromosome regions, including centromeric and pericentromeric locations (Fig. 1C and SI Appendix, Fig. S4). Despite constituting a large fraction of the genome, most repetitive elements in maize are unique sequence-wise due to mutations as well as transposon insertions into repetitive DNA, which create unique sequence borders (30). Due to the substantial sequence diversity, maize centromeres are well assembled (31), even though they consist predominantly of tandemly arranged CentC repeats interspersed with retrotransposons (31).
Finding DSBs in centromeres/pericentromeres was interesting, as these regions lack COs (3, 27). To confirm the DSB presence at centromeres, we conducted experiments combining the anti-RAD51 antibody with a fluorescence in situ hybridization (FISH) probe detecting the maize centromere repeat (29). These experiments demonstrated the presence of RAD51 foci on centromeres in midzygotene meiocytes (Fig. 1D).
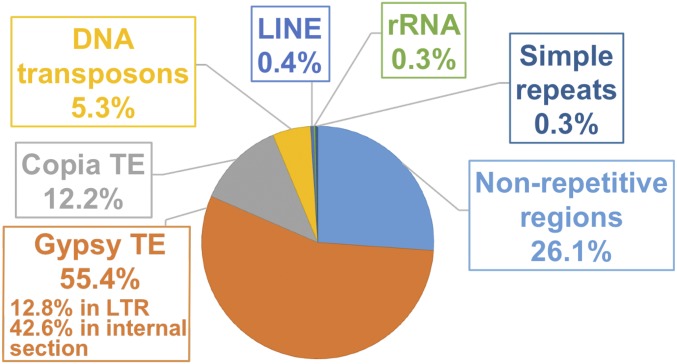
Another type of highly repetitive region in which we found evidence of meiosis-specific DSBs was rRNA loci (Fig. 2 and SI Appendix, Figs. S5 and S6). This finding was also not anticipated as rRNA loci have been reported to be excluded from DSB formation in yeast (32).
Fig. 2.
Percentage of DSB hotspots present in various genome components.
Most DSB Hotspots Are in Repetitive DNA.
To further dissect the distribution of DSB hotspots, we examined their genomic context. We found that the majority of DSB hotspots were located in repetitive DNA, primarily Gypsy retrotransposons (Fig. 2 and SI Appendix, Fig. S5). To determine if there were any specific regions within Gypsy in which DSB hotspots were more prevalent, we aligned the hotspots in Gypsy elements to sequences of the ∼220 Gypsy retrotransposon types identified in maize (33). About 77% of Gypsy-located hotspots primarily matched internal retrotransposon regions, while the remaining 23% primarily matched the long terminal repeats (LTRs). However, when the relative length of LTRs versus the internal region was taken into account, DSB hotspots appeared to be 2.4-fold more frequent in LTRs than in the internal region.
DSB Hotspot and Chromatin State.
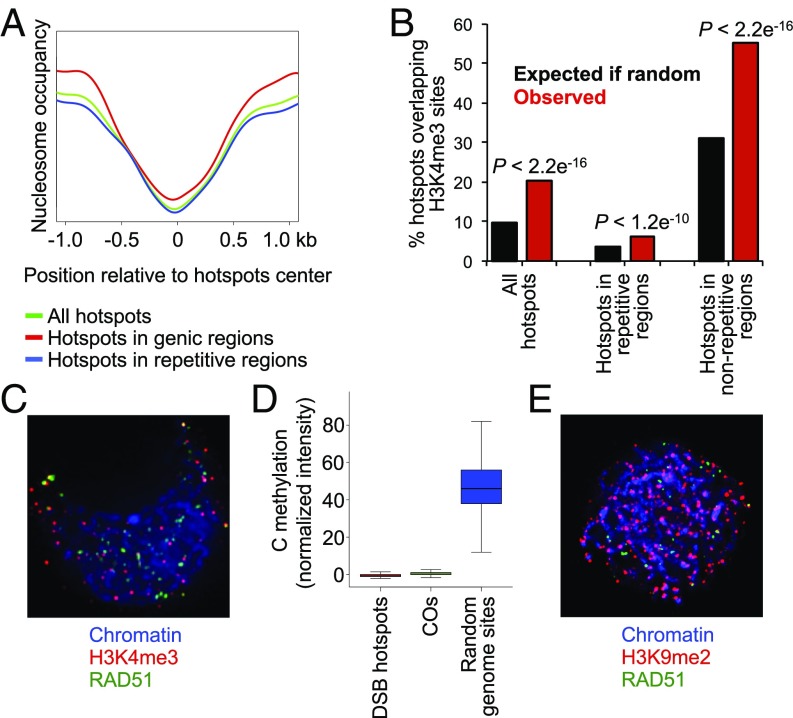
To search for factors controlling DSB location, we examined chromatin features of DSB hotspot regions. To do this analysis, we first investigated the positions of maize hotspots relative to open chromatin sites marked by low nucleosome occupancy. This experiment was conducted using micrococcal nuclease digestion of chromatin extracted from meiotic anthers. We found a strong association between hotspot presence and nucleosome-free chromatin for both hotspots located in genic regions and those in repetitive DNA (Fig. 3A).
Fig. 3.
Relationship between DSB hotspot location and chromatin state. (A) Nucleosome occupancy at the sites of DSB hotspots measured using micrococcal nuclease digestion. (B) Overlap of DSB hotspots with H3K4me3 sites. (C) Immunocolocalization of H3K4me3 marks and RAD51 foci in a maize meiocyte at zygotene. Image is a flat projection of five consecutive optical sections of a 3D nucleus. (D) Comparison of cytosine methylation levels (CG, CHG, and CHH combined) detected using a ChIP-microarray analysis in a 2-kb window around DSB hotspots, CO sites, and random genome sites. (E) Immunocolocalization of H3K9me2 and RAD51 in a maize meiocyte at zygotene showing limited overlap of DSBs with H3K9me2 sites. Image is a flat projection of five consecutive optical sections through a 3D nucleus.
We also examined H3K4me3 as another mark of active chromatin. However, we discovered that only about 20% of hotspots overlapped with H3K4me3 sites detected using ChIP with an anti-H3K4me3 antibody on chromatin from isolated zygotene meiocytes (Fig. 3B). We confirmed this conclusion using cytological immunolocalization, which showed that only about 5% of RAD51 foci at midzygotene overlapped with H3K4me3 foci (Fig. 3C; n = 20). As immunolocalization detects a much smaller number of H3K4me3 sites than ChIP, these two results are essentially in agreement in showing limited colocalization of RAD51 and H3K4me3 sites. We then examined the association of ChIP H3K4me3 sites with DSB hotpots located in different genome contexts. Overlap with H3K4me3 was low (∼5%) for DSB hotspots located in repetitive DNA but higher for those in genic regions (∼55%) (Fig. 3B).
Since cytosine methylation has been shown to affect the CO landscape in plants (3, 23, 34–38), we examined the DNA methylation status of DSB hotspot sites. To do this, we analyzed total levels of C methylation in a 2-kb window around DSB hotspot centers. We found that DNA methylation was dramatically reduced at DSB hotspots (P = 0) (Fig. 3D). We also examined a set of 104 maize CO events mapped with a resolution of less than 2 kb (SI Appendix, SI Methods) and found a similarly significant reduction (P < 2.2e−16).
Finally, to examine the association of DSBs with heterochromatin, as suggested by the large fraction of DSB hotspots located in repetitive DNA, we conducted immunocolocalization experiments with RAD51 and a heterochromatin marker, H3 histone lysine 9 dimethylation (H3K9me2) (39). We found that, at zygotene, about 5% of RAD51 foci colocalized with H3K9me2-marked heterochromatin (Fig. 3E).
Overall, our data indicate that DSBs in maize are formed at nucleosome-free and DNA-hypomethylated sites. However, the larger context of DSBs varies and only a minority of DSBs overlap with sites marked by H3K4me3. Even though a large fraction of DSB hotspots are located within transposons, they are present in nucleosome-free regions of transposon DNA rather than in condensed heterochromatin.
Hotspot Presence in Genes Is Not Correlated with Transcription.
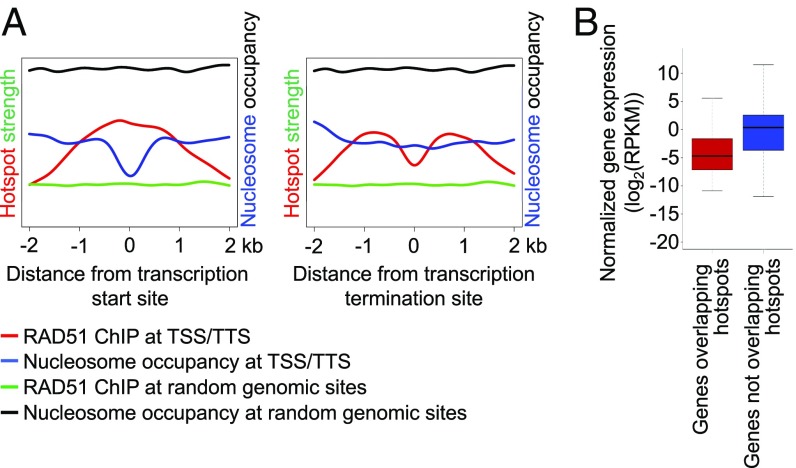
About 26% of maize DSB hotspots were in single-copy genic DNA. However, taking into account that genic DNA amounted to only about 14% of the Illumina reads that could be aligned to the maize genome, the relative frequency of hotspots was still higher in genic regions than in repetitive DNA (SI Appendix, Fig. S5). Within genes, there were two strong DSB formation peaks, located around the transcription start and transcription termination sites (Fig. 4A).
Fig. 4.
DSB hotspot location within genes. (A) Patterns of DSB formation around gene transcription start and transcription termination sites. Random RAD51 ChIP and nucleosome occupancies were calculated based on 1,000 randomly selected genes. (B) Comparison of expression levels of genes overlapping and not overlapping DSB hotspots.
Based on the association between hotspots and active chromatin, we hypothesized that DSB formation in genic DNA may take place in or near highly expressed genes, as gene expression is generally associated with active chromatin. To test this hypothesis, we examined the transcriptome of isolated male meiocytes. We discovered that genes overlapping DSB hotspots were expressed at lower levels than genes not containing hotspots (P = 1.13e−17; Fig. 4B). We also conducted a similar analysis for hotspots located in Gypsy elements and found that no more than 19 of the 1,732 Gypsy-associated hotspots were in expressed elements. We found a similar pattern when considering all transposable elements expressed in isolated meiocytes. Of 22,924 transposable elements expressed at a level higher than five counts-per-million, 23 overlapped 14 DSB hotspots. Overall, these data indicate that DSB formation in maize is not directed to sites of highly transcriptionally active chromatin.
Maize Hotspot Sequence.
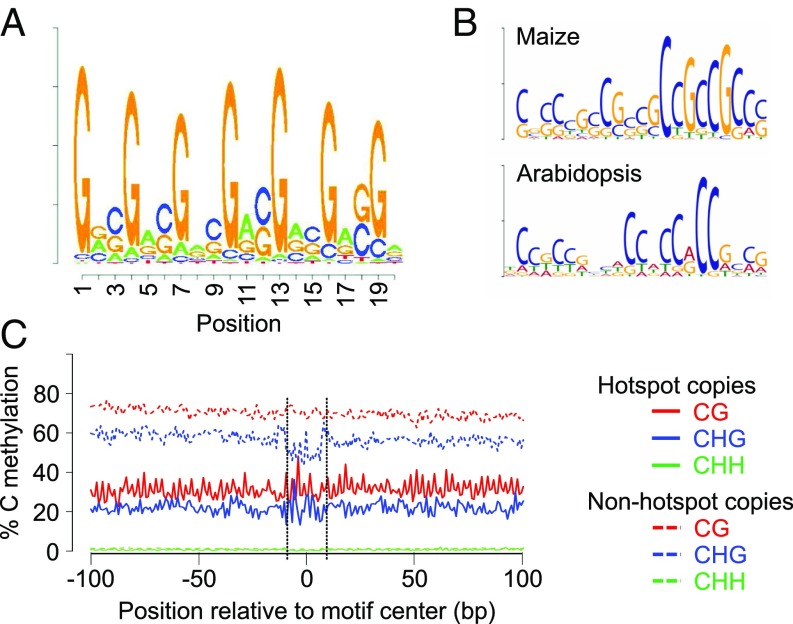
Maize DSB hotspot regions showed a higher GC content than the average for the Illumina reads that could be aligned to the maize genome (SI Appendix, Fig. S7). Further investigations of DNA sequence of DSB hotspots identified a 20-bp-long GC-rich degenerate DNA sequence motif (named MHS, Maize Hotspot Sequence) present in about 72% of genic hotspots (Fig. 5A). In contrast, we have not been able to find a statistically significant motif in repetitive DNA hotspots. By examining the dataset of 104 maize high-resolution-mapped CO events (SI Appendix, SI Methods), we detected a sequence motif associated with CO sites with similarity to MHS (Fig. 5B). The maize CO motif was also significantly similar (P = 2.25e−5) to a previously identified CO motif in Arabidopsis (Fig. 5B) (38).
Fig. 5.
DNA sequence context of DSB hotspots. (A) MHS, a DNA sequence motif associated with DSB hotspots located in maize genic regions. (B) CO motifs in maize (this study) and Arabidopsis (38). (C) Methylation status of MHS copies associated with DSB hotspots (hotspot copies) and copies that do not become hotspot sites (nonhotspot copies).
MHS copies are distributed along the entire chromosome length but tend to be more frequent near telomeres (SI Appendix, Fig. S8). The RAD51 signal was enriched around MHS (SI Appendix, Fig. S9). However, presence of MHS alone is insufficient for DSB hotspot formation, as the B73 genome contains 27,371 sequences that match the MHS sequence consensus at 95% identity or more. To examine if there are differences in MHS copy numbers among maize inbreds, we generated whole-genome Illumina sequences of two other inbreds, Mo17 and CML228. The Mo17 genome contained 28,947 MHS copies, while the CML228 had 6,614 copies. Interestingly, CML228 exhibits much lower DSB and CO numbers than B73 or Mo17 (18), suggesting that the MHS copy number may be a determinant of recombination frequencies.
An intriguing question is what distinguishes MHS copies associated with hotspots (hotspot copies) from those that are not (nonhotspot copies). As MHS is cytosine-rich, we examined the DNA methylation status of the motif. We analyzed the three types of DNA methylation present in plants, CG, CHG, and CHH (where H is any nucleotide other than G), which are controlled by distinct genetic pathways (40). CG and CHG methylation levels were lower at hotspot copies compared with nonhotspot copies (Fig. 5C). The high-GC methylation levels of nonhotspot copies suggested that they were located in heterochromatin. Some DSB formation was present at MHS copies in methylated DNA, although it was not sufficient to result in formation of detectable hotspots (SI Appendix, Fig. S10). However, this observation suggests that the DNA methylation level of MHS might be a regulator of hotspot strength. The presence of DSB formation in regions of reduced DNA methylation implies sensitivity of the DSB machinery to methylation, perhaps involving SPO11, as it was recently found in mice (41). Altering DNA methylation levels could activate new DSB hotspots, which may provide a mechanistic explanation for the observations that reduced DNA methylation alters CO patterns in plants (34–37).
Only DSBs in Genic Regions Are Likely to Contribute to CO Formation.
As the number of meiotic DSBs in maize is over 25-fold higher than the number of COs (28), most DSBs do not become CO sites. To examine how DSB distribution corresponds to CO distribution, we compared the DSB hotspot map to a CO map based on recombination data from the maize Nested Association Mapping (42). This comparison showed that CO distribution in maize does not follow the distribution of DSBs (r = 0.045; SI Appendix, Fig. S11). However, the correlation was much stronger (r = 0.41; SI Appendix, Fig. S11) and similar to the one reported in mouse (r = 0.46–0.64) (1) when only genic DSB hotspots were considered. CO site and MHS distributions were also strongly correlated (r = 0.72; SI Appendix, Fig. S11). These analyses imply that COs in maize are predominantly drawn from DSBs formed in genic regions while DSBs formed in repetitive DNA do not result in COs. Previous studies showing that COs in maize are generally formed in gene-rich regions (3, 27, 43) are consistent with this conclusion.
Of note, we found a strong DSB hotspot upstream from the bronze1 (bz1) locus on chromosome 9 (SI Appendix, Fig. S12). bz1 is the location of one of the few known maize CO hotspots, which exhibits CO frequencies over 100-fold higher than the genome average (44). Presence of this hotspot may be responsible for the high CO and gene conversion activities at this locus.
Discussion
DSB Hotspots in Maize Are Frequent in Repetitive DNA.
We found a ubiquitous presence of DSB hotspots in centromeric and pericentromeric regions of maize chromosomes. As these regions are devoid of COs (3, 27), the DSBs must be repaired as NCOs. Indeed, presence of gene conversions has been reported in maize centromeric DNA (45, 46). Nevertheless, finding DSB hotspots at centromeres was surprising, as DSB formation in centromeric regions has not been reported in either yeast or mammals (1, 2, 21, 47). It is highly unlikely that these hotspots could be ChIP-seq mapping artifacts because (i) maize centromeres are well assembled (31), which enables accurate mapping of ChIP-seq reads; and (ii) the ChIP-seq mapping data are corroborated by immuno-FISH experiments showing colocalization of RAD51 foci with centromeric repeats. Our data are consistent with previous reports of the presence of early recombination nodules in pericentromeric regions of maize chromosomes (48). Early nodules are cytological structures visible in transmission electron microscopy that correspond to RAD51/DMC1 protein complexes (48). Furthermore, signatures of recombination during centromere evolution have been found in rice, which is another species with well-assembled centromere region sequences (49).
We also discovered that the majority of DSBs were formed in repetitive DNA, which was unexpected as well. DSB formation in retrotransposons has been reported in budding yeast (50). Thus, the presence of a large fraction of DSBs in retrotransposons could be a reflection of the abundance of these elements in the maize genome (26). However, other maize transposons, such as Copia, LINE, and DNA transposons, exhibited fewer DSB hotspots relative to their ubiquity in the genome (SI Appendix, Fig. S5). Furthermore, yeast lack the highly condensed heterochromatin ubiquitous in maize (51), suggesting that DSB formation in retrotransposons in maize may differ at the mechanistic level from that in yeast.
Two DSB Classes in Maize.
The differing characteristics of DSB hotspots located in genic vs. repetitive genome regions suggest that they represent two distinct classes of DSBs. DSB hotspots in genes were associated with the presence of MHS, while hotspots in repetitive DNA were not. Hotspots in genic regions were also more likely to overlap active chromatin sites marked with H3K4me3 than hotspots in repetitive DNA.
Differences in characteristics between genic and nongenic hotspots may reflect differences in how the two DSB classes are formed. They also likely affect the way in which DSBs are repaired, as mainly genic DSBs contribute to CO formation. It has been proposed that DSB fate may depend on the timing of DSB formation; DSBs made earlier are more likely to be repaired as COs than those made later (52, 53). DSBs in euchromatin regions may form earlier than those in heterochromatin and thus be more prone to result in COs.
Mechanistic Underpinnings of DSB Hotspot Recognition in Maize.
Comparing recombination patterns in maize with those of yeast and mammals will be helpful for identifying universal recombination landscape features that are shared by a broad spectrum of species. Several general characteristics of maize DSB hotspot distribution patterns, such as hotspot width and spacing, are similar to those reported in mammals and yeast (1, 2, 21, 47). Association of DSB hotspots with nucleosome-free chromatin is also shared among species (2, 7), including maize hotspots located in genic regions and repetitive DNA. These commonalities suggest that there are conserved aspects of the DSB-targeting mechanism, which may have implications for genome evolution.
A striking difference in DSB distribution patterns between maize vs. yeast and mammals is the relatively low overlap of DSB hotspots with H3K4me3 sites in maize. In mice, the 93.9% of DSB-hotspot overlap with H3K4me3 (1) is a result of the action of PRDM9 (7). In PRDM9-lacking mouse mutants, DSBs are still produced but show altered distribution (54). These data suggest that PRDM9 functions primarily to sequester DSB formation away from 5′ regulatory regions of genes. In budding yeast, colocalization between DSB hotspots and H3K4me3 marks is also strong, but it may be a result of the compactness of the yeast genome rather than a causal relationship (24). Our data in maize suggest that the presence of H3K4me3 is not an absolute requirement for DSB formation.
Similar to the studies of COs in maize and Arabidopsis (3, 23, 38, 55), our data point to a key role of DNA methylation in determining the location of recombination events. However, the relationship between DNA methylation and recombination in maize may be more complex than in Arabidopsis. Whereas recombination hotspots are hypomethylated in both species (3, 23, 38, 55), chromosome-wide methylation patterns differ relative to the recombination landscape. In wild-type Arabidopsis, CO suppression around the centromere closely reflects the increase in DNA methylation (23, 38, 55). In contrast, DNA methylation in maize (56) does not follow the same strongly U-shaped distribution pattern as COs (Fig. 1C). Furthermore, maize has much higher overall DNA methylation levels than Arabidopsis, and the presence of DNA methylation in maize is more critical to genome functioning. Disrupting the DNA methylation pathway in maize generally results in embryo lethality (57).
Overall, our analyses of DSB hotspots, along with studies of CO hotspots in maize and other plant species (3, 23, 38, 43, 55), do not unequivocally point to a single determinant of hotspot location in plants. It is possible that in plants, hotspots form at sites that harbor a combination of features, including open chromatin and reduced levels of DNA methylation. However, the presence of open chromatin may not be by itself sufficient for a site to become a DSB hotspot. We base this conclusion on the fact that highly transcribed genes, which harbor highly open chromatin, rarely become sites of DSB hotspots in maize.
Finding a hotspot motif in maize is intriguing also because the motif is present only at genic DSB hotspots. Although presence of a motif resembles the situation in mammals, it does not imply that the recognition mode in plants and mammals is the same due to the lack of PRDM9-equivalents in maize. It is, however, possible that the overall function of MHS is similar to that of the mammalian motif, despite a different hotspot recognition mechanism. MHS is also similar to the sequences identified as associated with CO hotspots in Arabidopsis, particularly in terms of the 3-bp G/C periodicity (23, 38, 55). The latter similarity is an indication of a conserved hotspot recognition mechanism in plants.
We anticipate that the high-resolution map of DSB hotspots in maize will be valuable for both basic plant biology and plant breeding. Understanding distribution of meiotic recombination events, and elucidating factors that control it, will aid studies of how recombination affects population dynamics and species evolution. This knowledge should also allow engineering recombination hotspots in genomic regions with limited COs.
Methods
DSB Mapping Using RAD51 ChIP.
Polyclonal antibodies were raised in rabbits against a recombinant protein produced by expressing a full-length coding sequence of the ZmRAD51A1 gene in Escherichia coli. Two rounds of immunoprecipitation were performed on chromatin extracted from male flowers at the zygotene stage of meiotic prophase I. Details of the experimental procedures are described in SI Appendix, SI Methods.
H3K4me3 ChIP-Seq.
Male meiocytes in leptotene and zygotene were collected using the capillary collection of meiocytes method as previously described (58). Ten microliters of rabbit polyclonal anti–trimethyl-histone (Lys4) antibody (EMD Millipore) was used on chromatin extracted from ∼30,000 meiocytes, following instructions of the MAGnify ChIP Kit (Invitrogen). Standard Illumina protocols were used for library construction and sequencing.
Nucleosome Occupancy Mapping.
Nuclei were prepared from zygotene-stage anthers as described in the ChIP protocol except that EDTA was omitted from buffers. Details of the procedure are described in SI Appendix, SI Methods.
Cytological Analyses.
Preparation of immunolocalization and immuno-FiSH microscopic slides is described in detail in SI Appendix, SI Methods. The slides were examined using a DeltaVision imaging station (Applied Precision). Three-dimensional stacks of images were collected across the entire thickness of the specimen with optical sections 150 nm apart. The image stacks were subjected to constrained iterative deconvolution and analyzed with the softWoRx software (Applied Precision). Colocalization analyses were conducted in 3D space to distinguish between actual colocalization and accidental overlap. For counting, protein foci were located automatically in the datasets by identifying local peak intensities in three dimensions.
Computational Analyses.
A computational pipeline was developed to process ChIP-seq datasets. Details of the pipeline procedure, as well as protocols for peak calling, generating, and analysis of the DSB hotspot map, are described in SI Appendix, SI Methods. Standard procedures were used to identify DNA sequence motifs associated with the presence of DSB hotspots and COs.
Supplementary Material
Acknowledgments
We thank R. Kelly Dawe for advice on maize centromeres and Teresa Pawlowska and Wayne Crismani for comments on the manuscript. This research was supported by National Science Foundation Grants IOS-1025881 and IOS-1546792 (to W.P.P.) and by United States–Israel Binational Agricultural Research and Development Fund Grant US-4828-15 (to W.P.P. and A.A.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RAD51 ChIP sequence data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=khqfyssmzzinrcj&acc=GSE55701 (accession no. GSE55701); H3K4me3 ChIP and RNA sequence data have been deposited in the NCBI database, https://www.ncbi.nlm.nih.gov/biosample?LinkName=bioproject_biosample_all&from_uid=185817 [accession nos. SAMN01884257–SAMN01884260 (H3K4me3 ChIP), and SAMN01889176 and SAMN01889179 (RNA)]; nucleosome occupancy sequence data have been deposited in the NCBI GEO database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84368 (accession no. GSE84368).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713225114/-/DCSupplemental.
References
- 1.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan J, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodgers-Melnick E, et al. Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc Natl Acad Sci USA. 2015;112:3823–3828. doi: 10.1073/pnas.1413864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grey C, et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP. A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae. Genome Res. 2009;19:2245–2257. doi: 10.1101/gr.096297.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange J, et al. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell. 2016;167:695–708 e616. doi: 10.1016/j.cell.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- 9.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert T, et al. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science. 2016;351:943–949. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- 11.Vrielynck N, et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science. 2016;351:939–943. doi: 10.1126/science.aad5196. [DOI] [PubMed] [Google Scholar]
- 12.Kurzbauer MT, Uanschou C, Chen D, Schlögelhofer P. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell. 2012;24:2058–2070. doi: 10.1105/tpc.112.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitby MC. Making crossovers during meiosis. Biochem Soc Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 14.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 15.Hunter N, Kleckner N. The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 16.Chelysheva L, Vezon D, Belcram K, Gendrot G, Grelon M. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet. 2008;4:e1000309. doi: 10.1371/journal.pgen.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000285. doi: 10.1371/journal.pgen.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidhu GK, et al. Recombination patterns in maize reveal limits to crossover homeostasis. Proc Natl Acad Sci USA. 2015;112:15982–15987. doi: 10.1073/pnas.1514265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serrentino ME, Borde V. The spatial regulation of meiotic recombination hotspots: Are all DSB hotspots crossover hotspots? Exp Cell Res. 2012;318:1347–1352. doi: 10.1016/j.yexcr.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 20.de Boer E, Jasin M, Keeney S. Local and sex-specific biases in crossover vs. noncrossover outcomes at meiotic recombination hot spots in mice. Genes Dev. 2015;29:1721–1733. doi: 10.1101/gad.265561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker Z, et al. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. Elife. 2017;6:e24133. doi: 10.7554/eLife.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi K, et al. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet. 2013;45:1327–1336. doi: 10.1038/ng.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tischfield SE, Keeney S. Scale matters: The spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle. 2012;11:1496–1503. doi: 10.4161/cc.19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, et al. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 2009;5:e1000733. doi: 10.1371/journal.pgen.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 27.Gore MA, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 28.Franklin AE, et al. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pawlowski WP, et al. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science. 2004;303:89–92. doi: 10.1126/science.1091110. [DOI] [PubMed] [Google Scholar]
- 30.Liu R, et al. A GeneTrek analysis of the maize genome. Proc Natl Acad Sci USA. 2007;104:11844–11849. doi: 10.1073/pnas.0704258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider KL, Xie Z, Wolfgruber TK, Presting GG. Inbreeding drives maize centromere evolution. Proc Natl Acad Sci USA. 2016;113:E987–E996. doi: 10.1073/pnas.1522008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vader G, et al. Protection of repetitive DNA borders from self-induced meiotic instability. Nature. 2011;477:115–119. doi: 10.1038/nature10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 34.Melamed-Bessudo C, Levy AA. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:E981–E988. doi: 10.1073/pnas.1120742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirouze M, et al. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:5880–5885. doi: 10.1073/pnas.1120841109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colomé-Tatché M, et al. Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci USA. 2012;109:16240–16245. doi: 10.1073/pnas.1212955109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yelina NE, et al. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 2012;8:e1002844. doi: 10.1371/journal.pgen.1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shilo S, Melamed-Bessudo C, Dorone Y, Barkai N, Levy AA. DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis. Plant Cell. 2015;27:2427–2436. doi: 10.1105/tpc.15.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eichten SR, et al. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 2012;8:e1003127. doi: 10.1371/journal.pgen.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamudio N, et al. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 2015;29:1256–1270. doi: 10.1101/gad.257840.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Li L, Yan J. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat Commun. 2015;6:6648. doi: 10.1038/ncomms7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dooner HK, He L. Polarized gene conversion at the bz locus of maize. Proc Natl Acad Sci USA. 2014;111:13918–13923. doi: 10.1073/pnas.1415482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J, et al. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8:e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talbert PB, Henikoff S. Centromeres convert but don’t cross. PLoS Biol. 2010;8:e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014;24:1650–1664. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stack SM, Anderson LK. Crossing over as assessed by late recombination nodules is related to the pattern of synapsis and the distribution of early recombination nodules in maize. Chromosome Res. 2002;10:329–345. doi: 10.1023/a:1016575925934. [DOI] [PubMed] [Google Scholar]
- 49.Ma J, Bennetzen JL. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc Natl Acad Sci USA. 2006;103:383–388. doi: 10.1073/pnas.0509810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki M, Tischfield SE, van Overbeek M, Keeney S. Meiotic recombination initiation in and around retrotransposable elements in Saccharomyces cerevisiae. PLoS Genet. 2013;9:e1003732. doi: 10.1371/journal.pgen.1003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer NM, Lisch D, Li Q. Creating order from chaos: Epigenome dynamics in plants with complex genomes. Plant Cell. 2016;28:314–325. doi: 10.1105/tpc.15.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higgins JD, et al. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell. 2012;24:4096–4109. doi: 10.1105/tpc.112.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauppi L, et al. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013;27:873–886. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wijnker E, et al. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. Elife. 2013;2:e01426. doi: 10.7554/eLife.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He S, et al. Comparative analysis of genome-wide chromosomal histone modification patterns in maize cultivars and their wild relatives. PLoS One. 2014;9:e97364. doi: 10.1371/journal.pone.0097364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, et al. Genetic perturbation of the maize methylome. Plant Cell. 2014;26:4602–4616. doi: 10.1105/tpc.114.133140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dukowic-Schulze S, Sundararajan A, Ramaraj T, Mudge J, Chen C. Sequencing-based large-scale genomics approaches with small numbers of isolated maize meiocytes. Front Plant Sci. 2014;5:57. doi: 10.3389/fpls.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.