Significance
Rising temperatures, drought, and growing human populations are increasing demand for reclaimed wastewater for agricultural use. However, wastewater often contains biologically active, pseudopersistent pharmaceuticals, even after passage through a water treatment facility. We determined that the biology, life histories, and microbial communities of an agricultural pest insect were altered when reared on artificial diets or plants irrigated by these chemicals. In this study, pharmaceuticals translocated through plants have been shown to negatively affect the biology of an agriculturally important insect. The responses to these pharmaceuticals could have implications for integrating pest management practices and for understanding the effects of reclaimed water on agricultural ecosystems that are critical for the sustainability of our food supply.
Keywords: CECs, microbial communities, pollution, hormones, wastewater
Abstract
Many countries are utilizing reclaimed wastewater for agriculture because drought, rising temperatures, and expanding human populations are increasing water demands. Unfortunately, wastewater often contains biologically active, pseudopersistent pharmaceuticals, even after treatment. Runoff from farms and output from wastewater treatment plants also contribute high concentrations of pharmaceuticals to the environment. This study assessed the effects of common pharmaceuticals on an agricultural pest, Trichoplusia ni (Lepidoptera: Noctuidae). Larvae were reared on artificial diets spiked with contaminants of emerging concern (CECs) at environmentally relevant concentrations. Trichoplusia ni showed increased developmental time and mortality when reared on artificial diets containing antibiotics, hormones, or a mixture of contaminants. Mortality was also increased when T. ni were reared on tomatoes grown hydroponically with the same concentrations of antibiotics. The antibiotic-treated plants translocated ciprofloxacin through their tissues to roots, shoots, and leaves. Microbial communities of T. ni changed substantially between developmental stages and when exposed to CECs in their diets. Our results suggest that use of reclaimed wastewater for irrigation of crops can affect the developmental biology and microbial communities of an insect of agricultural importance.
Pharmaceuticals have been increasingly prescribed for the past 30 y, and prescription rates have almost tripled in just the past 14 y (1, 2). In 2013, animals grown for human consumption were treated with 9.1 × 106 kg of antibiotics; of those, 6.6 × 106 kg were used for the purpose of increasing production (3). Many antibiotics and other common contaminants of emerging concern (CECs) (pharmaceuticals, mental stimulants, surfactants, etc.) can be excreted by both humans and animals with little change in their chemical structure (4–6). Not surprisingly, pharmaceuticals have been appearing in wastewater, surface waters, and in some cases tap water, over the past few years (7, 8).
Standard wastewater treatment facilities are not equipped to completely remove pharmaceuticals (9, 10), resulting in these compounds being found in effluent. In addition, even higher concentrations of many pharmaceuticals are released during heavy storms in the untreated wastewater overflow, which then directly contaminate the environment (11). These pharmaceuticals have been found at biologically active concentrations in surface waters around the world (12–16). There is also an increasing effort to use reclaimed wastewater in drought-affected areas, such as Southern California (17, 18). In agriculture/livestock operations, pharmaceuticals are found in manure that is used as fertilizer, effectively compounding the pharmaceutical concentrations (12, 19, 20). Current research shows these chemicals tend to be both pseudopersistent in soil and detrimental to soil and rhizosphere microbes (13, 21–24).
Our recent studies of the effects of pharmaceuticals on aquatic insects show that, at concentrations found in reclaimed water, these CECs can alter development of the mosquito Culex quinquefasciatus, its susceptibility to a common larvicide, and its larval microbial communities (25, 26). Watts et al. (27) showed 17α-ethinylestradiol, a common birth control agent, and Bisphenol-A, a common plasticizer, can cause deformities in the midge Chironomus riparius. However, because larval forms of aquatic insects develop directly in the contaminated water, their constant exposure is likely greater than most terrestrial insects. Interestingly, many CECs, which were not designed specifically to impact microbes, have been shown to affect microbial communities. For example, caffeine, a common mental stimulant, can alter biofilm respiration, and diphenhydramine, an antihistamine, has been shown to modify the microbial community of lake biofilms (28). Due to such unexpected effects, accurately predicting the consequences of specific CECs, even in model insects, is not yet possible. This problem is exacerbated by a lack of information regarding effects of pharmaceuticals and other CECs on the microbial communities of any terrestrial insects.
Arthropods, such as insects and crustaceans, rely on hormones to grow, develop, mate, and produce pigmentation (29–31). However, many pharmaceuticals, especially mammalian sex hormones, are structurally similar to chemicals that these organisms rely on for growth and development. These pharmaceuticals then bind to receptors and either overexpress or suppress their counterparts’ natural function. This has been seen in birds, reptiles, and arthropods where endocrine disruption occurs, primary and secondary sexual characteristics are modified, and courtship behaviors are changed (29, 32–36). Although most arthropod hormones do not closely match those of mammals, their molting hormone (ecdysone) is very similar in structure to the mammalian female sex hormone 17β-estradiol. In crustaceans, mammalian hormones have been known to cause both increased molting events and inhibition of chitobiase, the enzyme responsible for digestion of the cuticle during insect molting (37, 38). In insects, 17α-ethinylestradiol, a common synthetic birth control hormone, has been shown to alter molting and lead to deformities of C. riparius (27, 39). In addition to these effects, pharmaceuticals have been shown to have delayed cross-generational effects (39).
The cabbage looper (Trichoplusia ni; Hübner, Lepidoptera: Noctuidae) is a well-studied polyphagous insect native to North America and is found throughout much of the world (40, 41). T. ni are yellow-green to green in color and can complete their life cycle in as little as 21 d depending on temperature (42). This species is a pest on many agricultural crops including crucifers and a variety of other vegetables in both field and greenhouse settings (43). Potential agricultural losses are exacerbated by a history of pesticide resistance development (44–46).
Currently, there is little to no information regarding pharmaceutical effects at the concentrations found in reclaimed water on the growth or microbial community composition of any terrestrial herbivore. Many herbivores can be exposed to these contaminants after the CECs enter surface waters, soil, and plants from wastewater reuse and unintended discharge. To investigate the function of the gut microbes in insects, several studies have used antibiotics applied at high doses (47, 48). There is also no information regarding effects of CECs when translocated through plants to terrestrial insects. To test the hypothesis that common pharmaceuticals affect mortality, development, and microbial communities of T. ni, we conducted a series of bioassays in artificial diet and on a key host plant utilizing surface water concentrations of common important pharmaceuticals. We used a culture-independent approach by performing a 16S rRNA gene survey on both diet and whole-body insects. Any effects would have potentially important implications from agricultural perspectives. Also, as there is currently no information on effects of CECs on terrestrial insects acquired through a plant matrix, our findings would have possible interest for integrated pest management (IPM) research.
Results
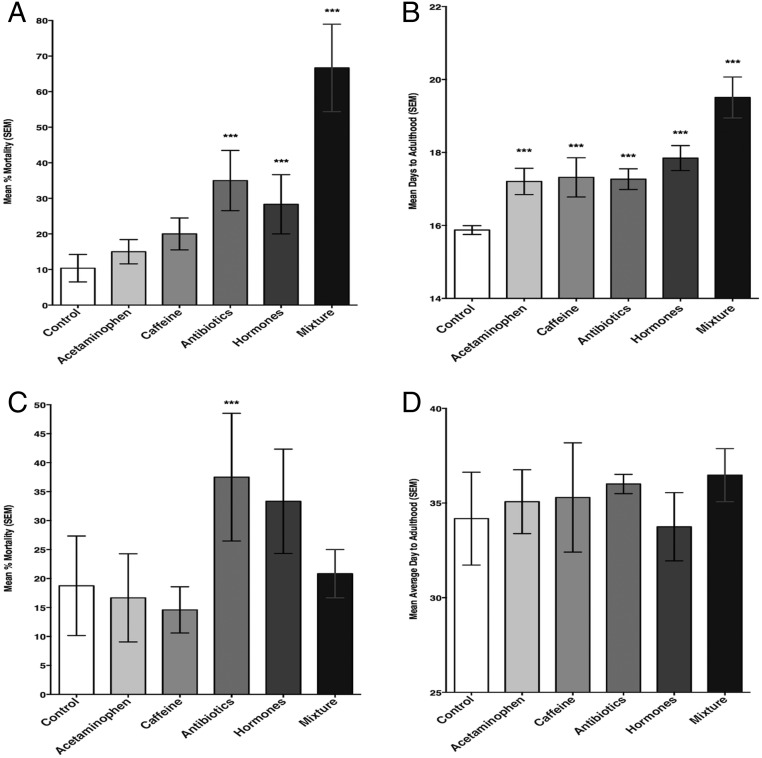
When T. ni were reared on artificial diet treated with pharmaceutical concentrations found in surface waters, mortality was increased (χ2 = 44.99; df = 5; P < 0.001) in the antibiotic (z value = 2.836; P = 0.0046), hormone (z value = 2.513; P = 0.0120), and mixture (z value = 5.387; P < 0.001) treatments compared with the controls (Fig. 1A). The time to adulthood (χ2 = 62.27; df = 5; P < 0.001) was increased in all treatments relative to the control (z values < −5.43; all P < 0.001) (Fig. 1B). When reared on plants, there was no significant difference for growth index (P = 0.99) between treatment groups; however, there was a significant effect on mortality (χ2 = 11.69; df = 5; P = 0.041), predominately when reared on plants treated with antibiotics (z value = 2.01; P = 0.044) (Fig. 1C). Chemical extraction data show increased concentrations of ciprofloxacin in leaves of the tomato plants relative to the control (Table 1). Because only the larvae on plants exposed to the antibiotic treatments showed any significant developmental responses, ultraperformance liquid chromatography (UPLC)/MS-MS analyses were limited to treatments containing antibiotics. There was no significant effect of treatment on time to adult for T. ni when reared on tomato plants (χ2 = 7.76; df = 5; P = 0.17) (Fig. 1D).
Fig. 1.
Effects of treatment on (A) mortality of Trichoplusia ni reared on artificial diets; (B) days to adulthood of T. ni on artificial diets; (C) mortality of T. ni reared on tomato plants treated with contaminated hydroponic growth solution; and (D) days to adulthood of T. ni reared on tomato plants grown in contaminated hydroponic solution. ***Significant difference (α = 0.05) relative to the control.
Table 1.
Concentration of antibiotics in treated plants measured by UPLC-MS/MS
| Treatment | Chemical | Roots | Stems | Old leaves | New leaves | Flowers | Fruit |
| Control | Lincomycin | ND | ND | ND | ND | ND | ND |
| Ciprofloxacin | NQ | NQ | NQ | NQ | NQ | NQ | |
| Oxytetracycline | NQ | NQ | NQ | NQ | NQ | NQ | |
| Antibiotics | Lincomycin | ND | ND | ND | ND | ND | ND |
| Ciprofloxacin | 23,524.58 (4,131.78) | NQ | 23.78 (3.21) | 161.70 (59.53) | NQ | NQ | |
| Oxytetracycline | 274.72 (64.32) | NQ | NQ | NQ | NQ | NQ | |
| Mixture | Lincomycin | ND | ND | ND | ND | ND | ND |
| Ciprofloxacin | 13,589.40 (5,654.30) | NQ | 124.36 (26.44) | 16.05 (15.57) | 17.31 (10.66) | NQ | |
| Oxytetracycline | NQ | NQ | NQ | NQ | NQ | NQ |
Chemical extraction data of tomatoes, variety “yellow pear,” per micrograms per kilogram are shown with standard error of the mean (SEM) in parentheses. ND denotes nondetectable, NQ denotes detectable but nonquantifiable, and parenthetical numbers designate SEM.
There were 865,142 quality-filtered 16S rRNA gene sequences, with an average of 9,721 (SD = 3,443) sequences per sample. Sample sequences were rarefied to a depth of 2,040 reads with a Good’s coverage of 99.56% (SD = 0.60%). Treatments [permutational multivariate ANOVA (PERMANOVA): F = 7.17; df = 5, 79; P < 0.001] and life stages (PERMANOVA: F = 25.26; df = 3, 81; P < 0.001) had significant effects on the T. ni microbial community β diversity, as determined by the weighted UniFrac distances. However, there was no overall difference between the microbial community of the insects compared with the diet (PERMANOVA: F = 0.42; df = 1, 83; P = 0.67). There were also significant interactions of treatment by life stage (PERMANOVA: F = 2.84; df = 15, 69; P < 0.001) and by type of community (PERMANOVA: F = 5.35; df = 5, 79; P < 0.001).
When analyzing the unweighted UniFrac distances, there was a significant effect of treatment (PERMANOVA: F = 1.62; df = 5, 79; P < 0.01) and stage (PERMANOVA: F = 3.26; df = 3, 81; P < 0.001), and an interaction of treatment and diet of the microbial β diversity (PERMANOVA: F = 1.44; df = 5, 79; P < 0.01). There was no significant effect of type (PERMANOVA: F = 0.54; df = 1, 83; P = 0.96), and there was no significant interaction of treatment and stage (PERMANOVA: F = 5.35; df = 15, 69; P = 0.14). As treatment was not significantly different in either weighted or unweighted UniFrac analyses, it was removed from further analyses.
When only third instar was considered, all treatments had significantly (Padj < 0.01) different microbial communities. However, sixth instars, pupae, and adult life stages (Padj ≥ 0.84, 0.56, 0.14, respectively) were not significantly different between treatments for both weighted and unweighted UniFrac. In all of the life stages, the treatments acetaminophen, caffeine, antibiotics, and hormones were all significantly different (P < 0.05) for weighted UniFrac. For unweighted UniFrac matrices, control and mixture treatment-fed insects were significantly different (Padj < 0.01).
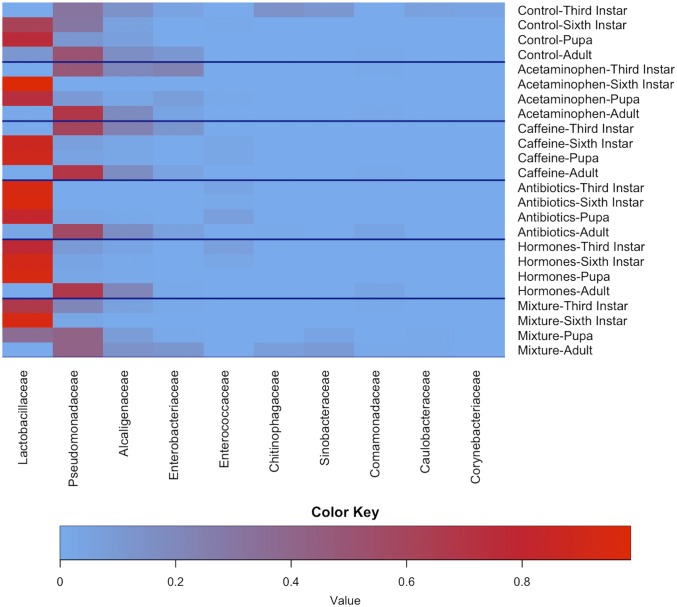
Fig. 2 and Tables 2 and 3 describe the overall communities of T. ni reared on treated artificial diets. The top 10 families, by average proportional abundance (Fig. 2), account for over 86% of the entire microbial community of T. ni (Tables 2 and 3). For the first three life stages (third instars, sixth instars, and pupae) in all treatments, the majority of microbes belong to the family Lactobacillaceae (Table 2). Lactobacillaceae’s proportional abundance increases in third and sixth instar; remains high, but decreases in the pupae; and then decreases further in adult insects where the majority of microbes were Pseudomonadaceae. Alcaligenaceae, Pseudomonadaceae, and Enterobacteriaceae, the next three families with the highest average percentages of rarefied operational taxonomic unit (OTU) counts, have similar patterns of high percentages in third instars followed by a decline in sixth instars and pupae, and then a spike in the adult life stage. This same pattern was seen in both Chitinophagaceae and Sinobacteraceae, as well. For the control, acetaminophen, caffeine, and mixture treatment groups containing all life stages (Table 3), the trends of average percentages follow the same patterns. The families with highest average percentage of rarefied OTU counts were Lactobacillaceae, Pseudomonadaceae, Alcaligenaceae, and Enterobacteriaceae, respectively. Interestingly, this pattern changes for antibiotic, hormone, and mixture treatment groups. For insects fed diets containing antibiotics and hormones, the average most proportionate families were Lactobacillaceae, Pseudomonadaceae, Alcaligenaceae, and Enterococcaceae.
Fig. 2.
Heatmap of the top 10 most proportionally abundant bacterial families by average OTUs of treatment life stage pairing. Increased red coloration is indicative of increased proportional abundance.
Table 2.
Average proportional abundance of bacterial families in Trichoplusia ni by life-stage
| Phylum | Family | Third instar | Sixth instar | Pupa | Adult |
| Acidobacteria | Corynebacteriaceae | 0.56 | 0.05 | 0.06 | 0.39 |
| Bacteriodetes | Chitinophagaceae | 1.75 | 0.00 | 0.1 | 1.40 |
| Firmicutes | Enterococcaceae | 2.11 | 1.50 | 1.86 | 0.28 |
| Lactobacillaceae | 43.07 | 88.70 | 75.45 | 2.56 | |
| Proteobacteria | Alcaligenaceae | 10.44 | 1.89 | 3.52 | 15.6 |
| Caulobacteraceae | 0.64 | 0.02 | 0.12 | 0.27 | |
| Comamonadaceae | 0.65 | 0.12 | 0.35 | 1.93 | |
| Enterobacteriaceae | 6.53 | 0.09 | 1.86 | 6.85 | |
| Pseudomonadaceae | 26.39 | 7.02 | 9.79 | 56.53 | |
| Sinobacteraceae | 1.36 | 0.00 | 0.39 | 1.61 | |
| Percentage total OTUs | 93.5 | 99.39 | 93.5 | 87.42 | |
| Average OTU count | 4,075.25 | 4,079.56 | 4,062.23 | 4,057.89 |
Table 3.
Average proportional abundance of bacterial families in Trichoplusia ni by treatment
| Phylum | Family | Control | Acetaminophen | Caffeine | Antibiotics | Hormones | Mixture |
| Acidobacteria | Corynebacteriaceae | 0.69 | 0.16 | 0.17 | 0.23 | 0.24 | 0.10 |
| Bacteriodetes | Chitinophagaceae | 2.37 | 0.16 | 0.00 | 0.27 | 0.01 | 2.36 |
| Firmicutes | Enterococcaceae | 0.87 | 0.61 | 1.41 | 2.58 | 2.39 | 0.32 |
| Lactobacillaceae | 41.88 | 36.95 | 47.31 | 65.37 | 63.71 | 50.04 | |
| Proteobacteria | Alcaligenaceae | 9.77 | 11.16 | 9.35 | 4.00 | 7.08 | 7.12 |
| Caulobacteraceae | 0.90 | 0.04 | 0.02 | 0.01 | 0.00 | 0.66 | |
| Comamonadaceae | 0.61 | 0.96 | 0.51 | 0.88 | 0.89 | 0.85 | |
| Enterobacteriaceae | 4.42 | 9.66 | 3.79 | 1.26 | 0.39 | 4.41 | |
| Pseudomonadaceae | 28.82 | 34.09 | 34.18 | 11.87 | 21.48 | 24.72 | |
| Sinobacteraceae | 1.92 | 0.00 | 0.04 | 0.00 | 0.01 | 3.49 | |
| Percentage total OTUs | 92.25 | 93.79 | 96.78 | 86.47 | 96.2 | 94.07 | |
| Average OTU count | 3,440.69 | 3,448.69 | 3,448.38 | 3,464.85 | 3,496.29 | 3,394.25 |
When examining the differential abundance of the individual OTUs by life stage in each treatment (Table S1), an interesting pattern appears. There were significant differences (Padj < 0.05) in controls between third instars and all other life stages, between acetaminophen-fed third instars and sixth instars, and between caffeine-fed third instars and sixth instars and pupae. However, in these three treatment groups, there were no significant differences (Padj > 0.05) between sixth instar and pupae versus their adult life stages. In the T. ni treated with antibiotics and the mixture treatment, microbes were significantly different (Padj < 0.05) between any early life stage and adults. For insects fed a hormone-contaminated diet, microbes were only different (Padj < 0.05) for third instars compared with adults.
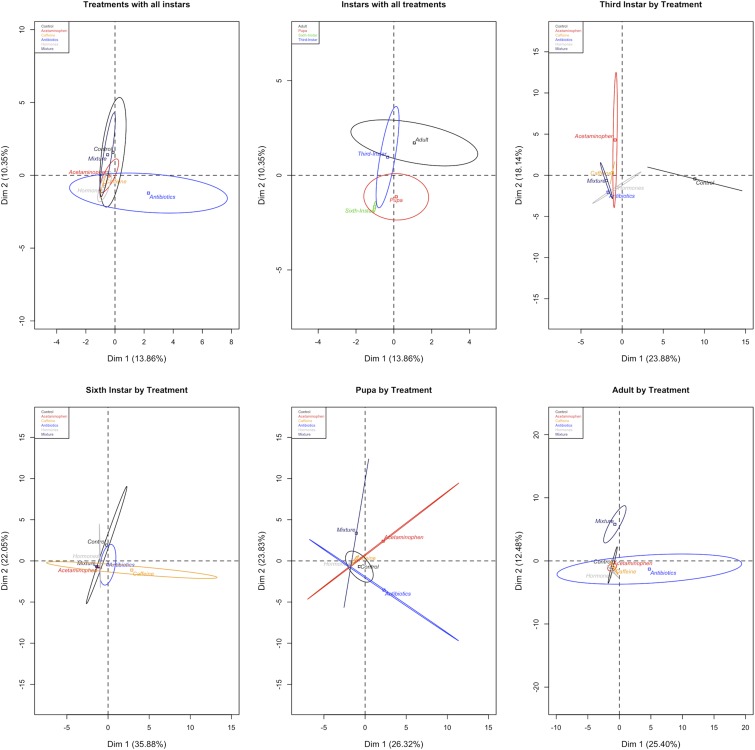
There were similar trends when individual OTUs were examined for each life stage comparing differences in treatments (Table S2). For third-instar insects, controls had significant differences (Padj < 0.05) in all other treatment groups; acetaminophen was significantly different (Padj < 0.05) compared with antibiotic, hormone, and mixture treatments, but not the caffeine treatment; caffeine-fed insect microbes followed a similar trend to acetaminophen. For third instar, there were no significant differences between antibiotic, hormone, and mixture treatments. Between all of the multiple treatment comparisons with OTUs, there were no significant differences in the sixth-instar insects. Microbe composition in the pupal and adult life stages followed similar trends. For pupae, acetaminophen and caffeine-fed insects’ microbes were different compared with antibiotic treatments. The microbial communities in the antibiotic treatment were significantly different from hormone and mixture treatments. Adults had significant differences (Padj < 0.05) in microbe composition in all treatments versus the mixture treatment. Principal-component analyses (PCAs) visualize these findings (Fig. 3).
Fig. 3.
Principal-component analyses of treatments with all instars, instars with all treatments, and by individual instars with all treatments. Ellipses denote the range of individuals around a centroid barycenter.
Discussion
In our study, CECs at concentrations found in reclaimed wastewater were shown to increase mortality of T. ni, especially on artificial diets contaminated with antibiotics, hormones, and a mixture of the chemicals. The mortality effect was also evident when T. ni were reared on plants grown in antibiotic-containing hydroponic growth media. Because plants grown in the hydroponic system contained quantifiable levels of ciprofloxacin in the leaf tissue (Table 1), and the antibiotic treatments significantly changed the microbial community of the insect (Fig. 2), we think this is possibly a cause of the mortality but we cannot exclude direct effects of the CECs on the insects or indirect effects through the plants. Ciprofloxacin is a quinolone topoisomerase IV and DNA gyrase inhibitor that acts by stabilizing the DNA-topoisomerase IV and DNA-girase so that it is no longer reversible (49). This blocks DNA replication and eventually causes cell death of bacteria. However, unlike bacteria, when higher-level organisms evolved, the A and B subunits of the topoisomerases fused, creating homodimers that cannot be targets of ciprofloxacin (50), and thus damage to the ribosomes of insects is not a possible mechanism of toxicity.
Interestingly, we did not see the increased time to adulthood in T. ni reared on plants compared with those reared on contaminated artificial diet. We postulate the discrepancy is possibly due to a number of factors such as dilution of CECs, as they were acquired from the water by the plants or there was biodegradation of the chemicals occurring in the plant (51) or by photodegradation. However, recent studies have shown pharmaceutical concentrations in surface waters, which appear to remain constant over the course of several years (15, 52, 53). More studies would be needed to determine how CECs at concentrations found in reclaimed water for agriculture would interact with current IPM strategies (particularly pesticide application and use of beneficial insects), and how soil matrices would affect the chemical acquisition and translocation by plants.
Many insects rely on microbial communities and endosymbionts to grow and develop; however, it has been shown that Lepidoptera species do not have a vertically transmitted microbial community (54, 55). In addition, because the effects of microbial communities on T. ni survival and development have not been documented, we present these data only to show that microbial communities change when exposed to CECs, and not as a proven factor influencing survival. We found significant shifts in the microbial community in the various life stages examined within the control treatments notably from third instar to subsequent life stages. A similar result has been reported for mosquitoes (26) and other insects (56–58). However, there is one family, Lactobacillaceae, which appears in all treatments and life stages in high proportions, except for adults. They are fairly common in insects (59–61) and can be responsible for at least 70% of the bacterial community (62). Lactobacillaceae is responsible for ∼42% of the bacteria in all life stages, followed by Pseudomonadaceae, Alcaligenaceae, and Enterobacteriaceae. Lactobacillaceae have been shown to act as beneficial bacteria in Drosophila (63); however, its function in T. ni is still unknown. Alcaligenaceae has been shown to be present in other moths (60), but Lepidopterans are not thought to have a functional microbiome (55).
There are clear patterns regarding the changes in microbial community proportionality according to the heat map (Fig. 2). In controls, third-instar microbial communities are relatively evenly spaced by family. The microbial community becomes predominately Lactobacillaceae for sixth instars and pupae. Once the insects reach the adult stage, their most predominant family is Pseudomonadaceae. This pattern holds in the acetaminophen and caffeine treatment groups as well. Interestingly, the other treatment groups do not share this pattern. For antibiotic- and hormone-treated T. ni, Lactobacillaceae is the predominant microbial family in the immature stages, but at the adult stage microbial community reverts to predominantly Pseudomonadaceae. We suspect that this is because, once the larvae undergo metamorphosis and shed their gut contents in preparation for pupation, they are no longer exposed to the pressures exerted by the CECs on the microbial community.
Fig. 3 provides a visual indication of the changes in the bacterial communities over time. The increase in β diversity after eclosion could be due to the larvae no longer being exposed to CECs or diet-borne bacteria after being moved to sterile containers. Also, when bacteria are lost as larvae digest their gut contents during pupation, the microbial β diversity could change. Interestingly, the hormone-treated T. ni follow a similar pattern to those exposed to antibiotics, but their ellipses are always much smaller, suggesting the entire insect population is showing a uniform response within their microbial communities. However, in the mixture-treated insects, larvae displayed a greater average diversity in their microbial community structure than either pupae or adults. This finding has not been shown in any single category of treatment, and we suspect the microbes exposed to mixtures could be experiencing potential interactive effects among chemicals (for example, synergistic, additive, or antagonistic effects). Such interactions should be the focus of future studies along with investigations of plant rhizosphere bacteria, particularly since we found a difference in the Bradyrhizobiaceae family for all treatments.
These results show that a terrestrial insect pest of commercial crops can be affected by CECs found in reclaimed wastewater for agricultural use. Our results suggest that CECs found in wastewater can impact T. ni growth and development, survivorship, and alter their microbial communities. Because T. ni is a common agricultural pest found around the world, feeds on a wide variety of plants, and has a history of developing pesticide resistance, its ability to deal with toxins is likely higher than many other insects. In addition, the responses we observed to CECs could have interesting implications for IPM practices on plants such as lowering the amount of pesticides needed or increasing susceptibility to insect pathogens, as has been shown in mosquitoes (25). These potential effects may be understated because some insects cannot detect the presence of the pharmaceuticals (64). However, we do not recommend purposefully exposing crops to CECs specifically for the control of insects because our study documented that these pharmaceuticals are translocated into crops and we do not yet know their possible effects on humans if consumed (65). We specifically want to note that ingestion of these compounds through uptake and translocation by a plant is not the only way T. ni or any other insect would be exposed to these compounds. Overhead sprinkler irrigation could cause contact absorption by the plants or insects, and simply drinking water on leaves at contaminated sites could expose insects to higher concentrations than were found in plant tissues. In fact, the ciprofloxacin concentration used was less than one-third of the highest rate (16). We urge caution in extrapolating to plants growing in soil, because variation in soil type and potential soil bacterial degradation could affect persistence [although soil bacteria are often negatively impacted by CECs (66)]. However, CEC exposures are considered pseudopersistent because they are reapplied with each irrigation. Thus, the effects reported here are likely to be conservative. Additional studies with other insects, particularly those with other feeding strategies, will be necessary before any patterns can be discerned.
Materials and Methods
Insect Rearing.
Insects were acquired from Benzon Research. Eggs clusters were cut on wax paper sheets and taped to the lids of 237-mL Styrofoam cups containing artificial diet (Southland Products) to emerge. For experiments that required larvae to be moved, loopers were allowed to develop to second instars before initiating trials to minimize handling damage.
Artificial Diet.
Dry T. ni artificial diet mix and raw linseed oil were purchased from Southland Products and mixed following the manufacturer’s directions. For artificial diet treatments, 100 mL of deionized (DI) water was spiked with one of five CECs treatment stocks dissolved in methanol or an untreated control of 0.5 mL of methanol. Environmentally relevant CEC treatments were used as in Pennington et al. (26), and based on surface water concentrations described by Kolpin et al. (15) and Mutiyar and Mittal (16) (Table 4). Briefly, treatment groups consisted of a control with only artificial diet for T. ni; an acetaminophen treatment (MP Biomedicals; purity ≥ 90%); a caffeine treatment (Fisher Scientific; laboratory grade purity); an antibiotic treatment of lincomycin, oxytetracycline, and ciprofloxacin (Alfa Aesar; purity ≥ 98%); a hormone treatment of estrone, 19-norethindrone, 17β-estradiol, and 17α-ethynylestradiol (Sigma-Aldrich; purity ≥ 98%); and a mixture of all pharmaceuticals. Hydrochloric acid, NaOH (prepared to a 1 M stock solution) (Fisher Scientific; 12.1 M, anhydrous pellets, respectively), and a pH adjuster (JR Peters Laboratory) were used to adjust the final pH of all treatments and experimental solutions to 7 ± 0.5. Distilled water and treatments were heated and mixed by magnetic stirrer with hot diet. Approximately 158 mL of liquid artificial diet was added to each cup, allowed to solidify at room temperature, and stored at 4 ± 2 °C until used. The amount of artificial diet provided was more than enough for all insects to fully develop. As the boiling temperature of water is below all of the contaminants’ melting point, concentrations in the final media are assumed to be those in Table 4. Excess water from the sides of the containers was removed with a KimWipe before approximately 20 T. ni eggs were taped on the lids of the artificial diet cups. After emergence, larvae were removed until only 10 were left for each diet cup with six replicates for each of the six treatments (n = 10 per replicate, total T. ni = 360). Data including day to adulthood and mortality were taken daily until all larvae reached the adult stage or died. The experiments ended once all insects either emerged as adults or perished. Adults were then frozen in a −62 ± 2 °C in an ultracold freezer (Forma Scientific), and adult mass was recorded.
Table 4.
CEC treatment group components and nominal concentrations
| Contaminant | Concentration, μg/L | Ref. |
| Antibiotics | ||
| Oxytetracycline | 72.90 | 6 |
| Lincomycin | 0.730 | 6 |
| Ciprofloxacin | 6,500 | 7 |
| Hormones | ||
| 17α-Ethynylestradiol | 0.831 | 6 |
| 17β-Estradiol | 0.200 | 6 |
| 19-Norethindrone | 0.872 | 6 |
| Estrone | 0.112 | 6 |
| Mixture | ||
| Acetaminophen | 10.00 | 6 |
| Caffeine | 6.000 | 6 |
| Antibiotics | Concentration as above | |
| Hormones | Concentration as above |
Three separate replicates (n = 20 per replicate, total T. ni = 360) of artificial diet for each of the six treatment groups were prepared, as previously stated, and used for microbial DNA extractions. At least three individuals were removed at third instar, sixth instar, the pupal stage, and the adult life stages. A subset of those that pupated were washed in 2.5% bleach solution and transferred to sterile cups so that upon adult emergence they would not be exposed to the bacteria in diets. Following removal of pupae from the artificial diet, diet samples were taken and stored at −62 ± 2 °C for microbial DNA extractions. Each whole-body insect was washed three times in clean 200-proof EtOH and stored in clean 200-proof EtOH at −62 ± 2 °C until DNA extraction (67).
Host Plant.
Tomatoes (Solanum lycopersicum L., variety “yellow pear”) were grown from seeds in 10.16-cm pots in UC soil mix no. 3 (68) and fertilized with Miracle Gro nutrient solution (Scotts Company) at labeled rate and watered as needed. At ∼10 cm, tomato plants’ roots were washed with water and transplanted to sand culture as in Hladun et al. (64). Transplants were treated with CECs in hydroponic growth media (Oasis Hydroponic Fertilizer 16-4-17; Oasis Grower Solutions) with concentrations described in Table 4 with average pH of 7.0 ± 0.5. Treatment media were prepared utilizing similar methanol stock solutions of treatment compounds as described previously; however, for this experiment, stock solutions were four times more concentrated to reduce the amount of methanol required. Plants were watered by an air-powered, in-house, automatic-watering system every 2 h from 6:00 AM to 6:00 PM and every 4 h thereafter every day for 5 min and fully drained upon conclusion. Hydroponic solutions were kept in 120-L containers below the plants and received no sunlight exposure except during the watering times. The hydroponic solutions in all containers were drained and refilled every 2 wk to maintain CEC concentrations. Each container included one of five CEC treatments or an untreated control hydroponic solution and was used to water four plants. Plants grew 4–6 wk before cabbage loopers were bagged onto whole leaves with white mesh organza bags (20.32 cm × 30.48 cm). There were four replicate hydroponic containers for each of the six treatments with four plants per 120-L container and three T. ni per plant (n = 12 individuals per container; n = 288 total T. ni). The organza bags were checked daily to assure food availability, and food was not allowed to be entirely consumed. If the available food became limited, the larvae were immediately moved to a leaf of similar location on the plant. Data regarding growth and development and mortality were collected daily, and the experiment was ended when all individuals had either eclosed as adults or perished. Growth index data were calculated as in Zhang et al. (65). The index is the sum of stages attained by individuals under experimental conditions divided by the sum of the most mature stage that would be reached in the control population. Data were then analyzed in R. Plants were separated into parts (roots, stems, old leaves, etc.) and immediately frozen at −62 ± 2 °C. Plant material was freeze-dried within 1 mo of collection and stored in light-proof boxes at room temperature until they underwent solid-phase extraction.
DNA Extractions and Illumina Sequencing of Whole-Body Trichoplusia ni Bacteria.
All DNA extractions and Illumina preparations were performed as in McFrederick and Rehan (62) within 1 mo of −62 ± 2 °C storage. Briefly, DNA extractions were performed using a DNeasy Blood and Tissue kit (Qiagen). An individual from each life stage (n = 4), each treatment group (n = 6), and replicate group (n = 3), along with triplicates of a pooled blank for each treatment group (n = 24; total n = 96), were placed in individual wells of a 96-well plate provided in the kit. We then added 180 μL of the supplied buffer ATL, a sterile 3.2-mm chrome-steel bead and 100 μL of 0.1-mm glass beads (Biospec) to each well. A Qiagen TissueLyzer was then used to bead-beat each sample for 6 min at 30 Hz. After addition of 20 μL of Proteinase K to each sample, samples were incubated samples at 57 °C overnight. The standard DNeasy extraction protocol was then followed.
Dual-index inline barcoding was used to prepare libraries for sequencing on the MiSeq sequencer (Illumina). We used primers that included either the forward or reverse Illumina sequencing primer, a unique 8-nt-long barcode, and the forward or reverse genomic oligonucleotide as in Kembel et al. (69). The bacterial 16S rDNA sequence primers used were 799F-mod3 CMGGATTAGATACCCKGG (70) and 1115R AGGGTTGCGCTCGTTG (69), which have been shown to minimize contamination from plastids.
Following the generation of 16S rDNA amplicons by these primers, PCR was performed to generate Illumina amplicons. PCRs were performed using 10 μL of ultrapure water, 10 μL of 2× Pfusion High-Fidelity DNA polymerase (New England Biolabs), 0.5 μL of each 10 μM primer stock, and 4 μL of DNA. We used a 52 °C annealing temperature, 35 cycles, and negative controls for each reaction. We used the Ultraclean PCR cleanup kit (MoBio) to remove unincorporated primers and dNTPs. Then, 1 μL of the clean PCR product was used as a template for another PCR, using HPLC-purified primers to complete the Illumina sequencing construct as in Kembel et al. (69): CAAGCAGAAGACGGCATAC GAGATCGGTCTCGGCATTCCTGC and AATGATACG GCGACCACCGAGATCTACACTCTTTCCCTACACGACG. For these reactions, a 58 °C annealing temperature for 15 cycles and negative controls were used. Once the PCR cycles were finished, 18 μL of the PCR product and SequalPrep Normalization plates (Thermo Fisher Scientific) were used to normalize the amount of DNA in each sample. Five microliters of each normalized sample were pooled together and used to performed another cleanup. Next, a 2100 Bioanalyzer (Agilent) was used to assess library quality. After quality control, the libraries were sequenced using a MiSeq sequencer (Illumina) and MiSeq Reagent kit, version 3 (Illumina), with 2 × 300 cycles. Raw data are available on the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession no. SRP099237).
Bioinformatics.
All genomic data were processed in macQIIME, version 1.9.1-20150604 (71, 72). USEARCH, version 6.1 (73), was used to identify and remove chimeric sequences, and SUMACLUST (74) was used to cluster OTUs with at least 97% sequence identity, and remove any with less than two reads per sample (75). We then used macQIIME to perform standard α and β diversity analyses. To assign taxonomy to OTUs, Greengenes taxonomy (76) and the RDP naive Bayesian classifier (77) were utilized. Because training set can influence these taxonomic assignments (78), BLASTN searches against NCBI’s online Nucleotide Collection (nr/nt) and 16S ribosomal RNA sequences (Bacteria and Archea) databases (accessed 03/02/2017) were performed. Any mitochondria or chloroplast OTUs and other obvious contaminants (as determined by the blank controls) were removed from the dataset as in McFrederick and Rehan (62). We aligned the quality-filtered dataset using the pynast aligner (79) and the Greengenes database (76). The phylogeny of the bacterial OTUs was reconstructed using FASTTREE, version 2.1.3 (80), and generated weighted and unweighted UniFrac distance matrices (81) using the phylogeny and OTU tables. Adonis analyses (82) and PCA (83) graphs were performed or created in R, version 3.3.1 (84), utilizing the UniFrac matrices. For α diversity, rarefaction analysis was conducted for to obtain the maximum depth with the least amount of sample loss. The R package “gplots” (85) was used to create a heatmap of the most abundant bacterial families; a 2.5% abundance in at least one sample was used as the cutoff.
Solid-Phase Extraction.
Dried plant material (0.25 ± 0.05 g) was ground by mortar and pestle with liquid nitrogen to a fine powder and extracted by Environmental Protection Agency guidelines 1694 (86) within 6 mo of storage. Briefly, samples spiked with deuterated standards were added to powdered plant material to determine recovery of chemicals. Samples were extracted with two rounds of sonication (30 min) in 20 mL of HPLC-grade acetonitrile (Fisher Scientific), and then centrifuged in a Beckman model J2-21M Induction Drive Centrifuge with a JA-17 tube holder for 15 min at 13,800 × g. Supernatant was decanted into a glass 40-mL vial and dried under N2 gas in a hot water bath at 32.5 ± 2.5 °C. In between both rounds of organic solvent extraction, plant material was sonicated for 30 min with 20 mL of phosphate buffer at pH 2.0 ± 0.2 and centrifuged as before, and the supernatant decanted into a separate 40-mL glass vial. Extracts from dried acetonitrile were resuspended in 1 mL of HPLC-grade methanol (Fisher Scientific) and added to the phosphate buffer extracts. Phosphate buffer was then loaded an Oasis Waters solid-phase extraction (SPE) cartridge HLB 6 cc vacuum cartridge, 200 mg sorbent per cartridge, 30 µm particle size, preconditioned with 6 mL of HPLC methanol, and then 12 mL of DI water, and eluted under gravity. After all of the phosphate buffer had passed, the SPE column was dried under vacuum for 45 min. Once dried, 20 mL of HPLC methanol was passed through the SPE column by gravity into a glass 20-mL vial. The methanol was then dried under N2 gas in a hot-water bath (32.5 ± 2.5 °C). Dried extracts were resuspended in 1.5 mL of Ultima-grade H2O (18 Ω)/methanol (95:5 by vol/vol) and transferred to a 2-mL centrifuge tube. Extracts were centrifuged at 15,100 × g for 15 min in a tabletop SciLogex d2012. Supernatants were passed through a 13-mm polytetrafluoroethylene syringe filter with a 0.22-μm pore size (Restek) and stored at −4.0 ± 2.0 °C until UPLC LC-MS/MS analysis.
LC-MS/MS Analysis.
Instrument analysis was performed on a Waters ACQUITY UPLC combined with a Waters micromass triple-quadrupole mass spectrometer (qQq) equipped with an electrospray ionization (ESI) interface (Waters). Separation was achieved using an ACQUITY UPLC C18 column (2.1 mm × 100 mm, 1.7 μm; Waters) at 40 °C and a binary gradient system of mobile phase A, DI water (18 Ω) acidified using 0.2% formic acid (FA), and mobile phase B composed of MeOH/acetonitrile (50:50 vol/vol) and 0.2% FA was used to separate analytes. The solvent gradient program, in terms of mobile phase A, was as follows: initial condition began with 80% until 1 min when it was decreased linearly to 60% for 0.5 min, it was further decreased to 15% where it was held for 3 min, then decreased linearly to 10% for 1 min, then increased linearly to 90% for 1.5 min and held for 1 min after which time it equilibrated for 1 min for a total run time of 7:00 min. The flow rate was 0.3 mL⋅min−1, and the injection volume was set to 5 μL. Mass data were acquired using Intellistart (Waters) in the multiple reactions monitoring mode and product ion scan in the positive ESI mode. Calibration curves were used for determining concentrations, and blanks were included in all runs and between each treatment. Limit of detection and limit of quantification were determined using signal/noise ratios of 3 and 8, respectively. Control plant extractions with standards were used to determine matrix effect corrections. These corrections when applied to everything could result in a nonquantifiable (detected but unable to quantify) reading when it should have been nondetectable. The specific instrument settings were as follows: capillary source voltage, 1.34 kV; dwell time, 0.008 s; source temperature, 150 °C; desolvation temperature, 500 °C; desolvation gas, 900 L⋅h−1; and cone gas, 50 L⋅h−1; the collision gas was argon 99.9% pure. Cone voltage and collision voltage for each compound are described in Table S3.
Statistics.
All statistical analyses were performed using R (version 3.3.1). Normality was determined using Shapiro–Wilk normality tests, quantile–quantile plots, and histograms. Effects of treatments on mortality were determined using a generalized linear model with a binomial distribution and comparisons between treatments were performed using R’s “summary” function for each model as a post hoc test. ANOVA was used to determine differences of growth index by treatment. Days to adulthood overall differences were determined with Kruskal–Wallis tests with differences by treatment determined using the “survival” and “OIsurv” packages (87, 88). In all cases, when data were not considered normal, either a Poisson distribution or a negative binomial generalized linear model was used and best fit was determined from Akaike’s “An Information Criterion” and followed with R’s “summary” function for pairwise comparisons of treatment. Adonis within the R package “vegan” (82) was used for all PERMANOVA analyses. As there is no post hoc (89) test for Adonis, adjusted P values were obtained from metagenomeHIT_zig in R through macQIIME (71, 90) to determine differentially abundant OTUs between treatments and between life stages. All Adonis analyses were conducted on weighted and unweighted UniFrac distance matrices. Weighted UniFrac places more emphasis on more abundant microbiota, whereas unweighted UniFrac is based solely on presence or absence; therefore, rare microbiota have similar weight to common microbiota. The Adonis groups that were significantly different were broken into their component differences, which were compared by adjusted P values using p.adjust function and Benjamini–Hochberg adjustment in R. For UPLC work, individual compound peaks were detected and integrated using TargetLynx XS software (Waters).
Supplementary Material
Acknowledgments
We thank G. Kund for his knowledge and guidance in experimental design and methodology and S. Chin, T. Dang, and S. Truong for edits on this manuscript. We also thank the University of California, Riverside, Genomics Core facility staff for sequencing expertise. This research was supported by Environmental Protection Agency Award R835829 (to J.G. and J.T.T.) and through a fellowship awarded by the National Aeronautics and Space Administration Minority University Research and Education Project Institutional Research Opportunity Fellowships in Extremely Large Data Sets (Award NNX15AP99A to J.A.R.). This work was also supported in part by the US Department of Agriculture National Institute of Food and Agriculture, Hatch Project (Project 1011669). S.L.D. was funded by National Science Foundation Integrative Graduate Education and Research Traineeship Grant DGE-1144635.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP099237).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713385114/-/DCSupplemental.
References
- 1.National Center for Health Statistics . Health, United States, 2013: With Special Feature on Prescription Drugs. National Center for Health Statistics; Hyattsville, MD: 2014. [PubMed] [Google Scholar]
- 2.Schumock GT, et al. National trends in prescription drug expenditures and projections for 2014. Am J Health Syst Pharm. 2014;71:482–499. doi: 10.2146/ajhp130767. [DOI] [PubMed] [Google Scholar]
- 3.Department of Health and Human Services . Antimicrobials Sold or Distributed for Use in Food-Producing Animals. Department of Health and Human Services; Washington, DC: 2013. [Google Scholar]
- 4.Ternes TA, Joss A, Siegrist H. Scrutinizing pharmaceuticals and personal care products in wastewater treatment. Environ Sci Technol. 2004;38:392A–399A. doi: 10.1021/es040639t. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch R, Ternes T, Haberer K, Kratz K-L. Occurrence of antibiotics in the aquatic environment. Sci Total Environ. 1999;225:109–118. doi: 10.1016/s0048-9697(98)00337-4. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto H, et al. Persistence and partitioning of eight selected pharmaceuticals in the aquatic environment: Laboratory photolysis, biodegradation, and sorption experiments. Water Res. 2009;43:351–362. doi: 10.1016/j.watres.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 7.Monteiro S, Boxall ABA. Occurrence and fate of human pharmaceuticals in the environment. Rev Environ Contam Toxicol. 2010;202:53–154. doi: 10.1007/978-1-4419-1157-5_2. [DOI] [PubMed] [Google Scholar]
- 8.Sklerov F, Saucier M. August 19, 2011 Follow-Up Study Confirms No Risk From Pharmaceuticals and Personal Care Products in NYC Drinking Water (NYC Department of Environmental Protection Public Affairs, Flushing, NY). Available at www.nyc.gov/html/dep/html/press_releases/11-77pr.shtml#.WJaFNXc-JR4. Accessed November 24, 2016.
- 9.Hedgespeth ML, et al. Pharmaceuticals and personal care products (PPCPs) in treated wastewater discharges into Charleston Harbor, South Carolina. Sci Total Environ. 2012;437:1–9. doi: 10.1016/j.scitotenv.2012.07.076. [DOI] [PubMed] [Google Scholar]
- 10.Gros M, Petrović M, Ginebreda A, Barceló D. Removal of pharmaceuticals during wastewater treatment and environmental risk assessment using hazard indexes. Environ Int. 2010;36:15–26. doi: 10.1016/j.envint.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Phillips PJ, et al. Combined sewer overflows: An environmental source of hormones and wastewater micropollutants. Environ Sci Technol. 2012;46:5336–5343. doi: 10.1021/es3001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei R, Ge F, Huang S, Chen M, Wang R. Occurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere. 2011;82:1408–1414. doi: 10.1016/j.chemosphere.2010.11.067. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez DA, et al. Bioassay of estrogenicity and chemical analyses of estrogens in streams across the United States associated with livestock operations. Water Res. 2013;47:3347–3363. doi: 10.1016/j.watres.2013.03.028. [DOI] [PubMed] [Google Scholar]
- 14.Huang B, et al. Occurrence, removal and bioaccumulation of steroid estrogens in Dianchi Lake catchment, China. Environ Int. 2013;59:262–273. doi: 10.1016/j.envint.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Kolpin DW, et al. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: A national reconnaissance. Environ Sci Technol. 2002;36:1202–1211. doi: 10.1021/es011055j. [DOI] [PubMed] [Google Scholar]
- 16.Mutiyar PK, Mittal AK. Risk assessment of antibiotic residues in different water matrices in India: Key issues and challenges. Environ Sci Pollut Res Int. 2014;21:7723–7736. doi: 10.1007/s11356-014-2702-5. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, Conkle JL, Gan J. Multi-residue determination of pharmaceutical and personal care products in vegetables. J Chromatogr A. 2012;1254:78–86. doi: 10.1016/j.chroma.2012.07.041. [DOI] [PubMed] [Google Scholar]
- 18.Brown E, et al. Policy for Water Quality Control for Recycled Water (Recycled Water Policy) California Water Boards; Palm Desert, CA: 2013. [Google Scholar]
- 19.Shappell NW, et al. Estrogenic activity and steroid hormones in swine wastewater through a lagoon constructed-wetland system. Environ Sci Technol. 2007;41:444–450. doi: 10.1021/es061268e. [DOI] [PubMed] [Google Scholar]
- 20.Kumar K, Gupta SC, Baidoo SK, Chander Y, Rosen CJ. Antibiotic uptake by plants from soil fertilized with animal manure. J Environ Qual. 2005;34:2082–2085. doi: 10.2134/jeq2005.0026. [DOI] [PubMed] [Google Scholar]
- 21.Gan J, Bondarenko S, Oki L, Haver D, Li JX. Occurrence of fipronil and its biologically active derivatives in urban residential runoff. Environ Sci Technol. 2012;46:1489–1495. doi: 10.1021/es202904x. [DOI] [PubMed] [Google Scholar]
- 22.Kinney CA, Furlong ET, Werner SL, Cahill JD. Presence and distribution of wastewater-derived pharmaceuticals in soil irrigated with reclaimed water. Environ Toxicol Chem. 2006;25:317–326. doi: 10.1897/05-187r.1. [DOI] [PubMed] [Google Scholar]
- 23.Thiele-Bruhn S. Pharmaceutical antibiotic compounds in soils—a review. J Plant Nutr Soil Sci. 2003;166:145–167. [Google Scholar]
- 24.Chefetz B, Mualem T, Ben-Ari J. Sorption and mobility of pharmaceutical compounds in soil irrigated with reclaimed wastewater. Chemosphere. 2008;73:1335–1343. doi: 10.1016/j.chemosphere.2008.06.070. [DOI] [PubMed] [Google Scholar]
- 25.Pennington MJ, Rivas NG, Prager SM, Walton WE, Trumble JT. Pharmaceuticals and personal care products alter the holobiome and development of a medically important mosquito. Environ Pollut. 2015;203:199–207. doi: 10.1016/j.envpol.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Pennington MJ, Prager SM, Walton WE, Trumble JT. Culex quinquefasciatus larval microbiomes vary with instar and exposure to common wastewater contaminants. Sci Rep. 2016;6:21969. doi: 10.1038/srep21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watts MM, Pascoe D, Carroll K. Exposure to 17 alpha-ethinylestradiol and bisphenol A—effects on larval moulting and mouthpart structure of Chironomus riparius. Ecotoxicol Environ Saf. 2003;54:207–215. doi: 10.1016/s0147-6513(02)00029-5. [DOI] [PubMed] [Google Scholar]
- 28.Rosi-Marshall EJ, et al. Pharmaceuticals suppress algal growth and microbial respiration and alter bacterial communities in stream biofilms. Ecol Appl. 2013;23:583–593. doi: 10.1890/12-0491.1. [DOI] [PubMed] [Google Scholar]
- 29.Jindra M, Palli SR, Riddiford LM. The juvenile hormone signaling pathway in insect development. Annu Rev Entomol. 2013;58:181–204. doi: 10.1146/annurev-ento-120811-153700. [DOI] [PubMed] [Google Scholar]
- 30.Knowles SFGW, Carlisle DB. Endocrine control in the Crustacea. Biol Rev. 1956;31:396–467. [Google Scholar]
- 31.Martín D, Wang SF, Raikhel AS. The vitellogenin gene of the mosquito Aedes aegypti is a direct target of ecdysteroid receptor. Mol Cell Endocrinol. 2001;173:75–86. doi: 10.1016/s0303-7207(00)00413-5. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez G, Sorci G, Smith LC. Testosterone and sexual signalling in male house sparrows (Passer domesticus) Behav Ecol Sociobiol. 2001;50:557–562. [Google Scholar]
- 33.Hoffmann F, Kloas W. Estrogens can disrupt amphibian mating behavior. PLoS One. 2012;7:e32097. doi: 10.1371/journal.pone.0032097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tompsett AR, Wiseman S, Higley E, Giesy JP, Hecker M. Effects of exposure to 17α-ethynylestradiol during larval development on growth, sexual differentiation, and abundances of transcripts in the liver of the wood frog (Lithobates sylvaticus) Aquat Toxicol. 2013;126:42–51. doi: 10.1016/j.aquatox.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Tompsett AR, et al. Effects of 17α-ethynylestradiol on sexual differentiation and development of the African clawed frog (Xenopus laevis) Comp Biochem Physiol C Toxicol Pharmacol. 2012;156:202–210. doi: 10.1016/j.cbpc.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Segner H, et al. Identification of endocrine-disrupting effects in aquatic vertebrates and invertebrates: Report from the European IDEA project. Ecotoxicol Environ Saf. 2003;54:302–314. doi: 10.1016/s0147-6513(02)00039-8. [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez EM, Medesani DA, Fingerman M. Endocrine disruption in crustaceans due to pollutants: A review. Comp Biochem Physiol A Mol Integr Physiol. 2007;146:661–671. doi: 10.1016/j.cbpa.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 38.Zou E, Fingerman M. Effects of estrogenic xenobiotics on molting of the water flea, Daphnia magna. Ecotoxicol Environ Saf. 1997;38:281–285. doi: 10.1006/eesa.1997.1589. [DOI] [PubMed] [Google Scholar]
- 39.Watts MM, Pascoe D, Carroll K. Chronic exposure to 17 alpha-ethinylestradiol and bisphenol A-effects on development and reproduction in the freshwater invertebrate Chironomus riparius (Diptera: Chironomidae) Aquat Toxicol. 2001;55:113–124. doi: 10.1016/s0166-445x(01)00148-5. [DOI] [PubMed] [Google Scholar]
- 40.Chippendale GM, Beck SD, Strong FM. Nutrition of the cabbage looper, Trichoplusia ni (Hübn)—I. Some requirements for larval growth and wing development. J Insect Physiol. 1965;11:211–223. [Google Scholar]
- 41.University of Alberta Museums 2017 Trichoplusia ni. Encyclopedia of Life. Available at www.eol.org. Accessed November 28, 2016.
- 42.Toba HH, Kishaba AN, Pangaldan R, Vail PV. Temperature and the development of the cabbage looper. Ann Entomol Soc Am. 1973;66:965–974. [Google Scholar]
- 43.Franklin MT, Ritland CE, Myers JH. Spatial and temporal changes in genetic structure of greenhouse and field populations of cabbage looper, Trichoplusia ni. Mol Ecol. 2010;19:1122–1133. doi: 10.1111/j.1365-294X.2010.04548.x. [DOI] [PubMed] [Google Scholar]
- 44.Janmaat AF, Myers J. Rapid evolution and the cost of resistance to Bacillus thuringiensis in greenhouse populations of cabbage loopers, Trichoplusia ni. Proc Biol Sci. 2003;270:2263–2270. doi: 10.1098/rspb.2003.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shikano I, Cory JS. Dietary mechanism behind the costs associated with resistance to Bacillus thuringiensis in the cabbage looper, Trichoplusia ni. PLoS One. 2014;9:e105864. doi: 10.1371/journal.pone.0105864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shikano I, Cory JS. Genetic resistance to Bacillus thuringiensis alters feeding behaviour in the cabbage looper, Trichoplusia ni. PLoS One. 2014;9:e85709. doi: 10.1371/journal.pone.0085709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chouaia B, et al. Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 2012;12(Suppl 1):S2. doi: 10.1186/1471-2180-12-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kafil M, Bandani AR, Kaltenpoth M, Goldansaz SH, Alavi SM. Role of symbiotic bacteria in the growth and development of the Sunn pest, Eurygaster integriceps. J Insect Sci. 2013;13:99. doi: 10.1673/031.013.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31(Suppl 2):S24–S28. doi: 10.1086/314056. [DOI] [PubMed] [Google Scholar]
- 50.Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huber C, Bartha B, Harpaintner R, Schröder P. Metabolism of acetaminophen (paracetamol) in plants–two independent pathways result in the formation of a glutathione and a glucose conjugate. Environ Sci Pollut Res Int. 2009;16:206–213. doi: 10.1007/s11356-008-0095-z. [DOI] [PubMed] [Google Scholar]
- 52.McEachran AD, Shea D, Bodnar W, Nichols EG. Pharmaceutical occurrence in groundwater and surface waters in forests land-applied with municipal wastewater. Environ Toxicol Chem. 2016;35:898–905. doi: 10.1002/etc.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batt AL, Kincaid TM, Kostich MS, Lazorchak JM, Olsen AR. Evaluating the extent of pharmaceuticals in surface waters of the United States using a national-scale rivers and streams assessment survey. Environ Toxicol Chem. 2016;35:874–881. doi: 10.1002/etc.3161. [DOI] [PubMed] [Google Scholar]
- 54.Dillon RJ, Dillon VM. The gut bacteria of insects: Nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 55.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci USA. 2017;114:9641–9646. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hail D, Dowd SE, Bextine B. Identification and location of symbionts associated with potato psyllid (Bactericera cockerelli) lifestages. Environ Entomol. 2012;41:98–107. doi: 10.1603/EN11198. [DOI] [PubMed] [Google Scholar]
- 57.Vasanthakumar A, Handelsman J, Schloss PD, Bauer LS, Raffa KF. Gut microbiota of an invasive subcortical beetle, Agrilus planipennis Fairmaire, across various life stages. Environ Entomol. 2008;37:1344–1353. doi: 10.1603/0046-225x(2008)37[1344:gmoais]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 58.Pennington MJ, et al. Effects of contaminants of emerging concern on Megaselia scalaris (Lowe, Diptera: Phoridae) and its microbial community. Sci Rep. 2017;7:8165. doi: 10.1038/s41598-017-08683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McFrederick QS, et al. Environment or kin: Whence do bees obtain acidophilic bacteria? Mol Ecol. 2012;21:1754–1768. doi: 10.1111/j.1365-294X.2012.05496.x. [DOI] [PubMed] [Google Scholar]
- 60.Xia X, et al. DNA sequencing reveals the midgut microbiota of diamondback moth, Plutella xylostella (L.) and a possible relationship with insecticide resistance. PLoS One. 2013;8:e68852. doi: 10.1371/journal.pone.0068852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Engel P, Moran NA. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 62.McFrederick QS, Rehan SM. Characterization of pollen and bacterial community composition in brood provisions of a small carpenter bee. Mol Ecol. 2016;25:2302–2311. doi: 10.1111/mec.13608. [DOI] [PubMed] [Google Scholar]
- 63.Storelli G, et al. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 64.Hladun KR, Parker DR, Trumble JT. Cadmium, copper, and lead accumulation and bioconcentration in the vegetative and reproductive organs of Raphanus sativus: Implications for plant performance and pollination. J Chem Ecol. 2015;41:386–395. doi: 10.1007/s10886-015-0569-7. [DOI] [PubMed] [Google Scholar]
- 65.Zhang M, Chaudhuri SK, Kubo I. Quantification of insect growth and its use in screening of naturally occurring insect control agents. J Chem Ecol. 1993;19:1109–1118. doi: 10.1007/BF00987372. [DOI] [PubMed] [Google Scholar]
- 66.Kong WD, Zhu YG, Fu BJ, Marschner P, He JZ. The veterinary antibiotic oxytetracycline and Cu influence functional diversity of the soil microbial community. Environ Pollut. 2006;143:129–137. doi: 10.1016/j.envpol.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 67.Hammer TJ, Dickerson JC, Fierer N. Evidence-based recommendations on storing and handling specimens for analyses of insect microbiota. PeerJ. 2015;3:e1190. doi: 10.7717/peerj.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matkin OA, Chandler PA. 1957. The U.C.-type mixes. The U.C. System for Producing Healthy Container-Grown Plants, Manual 23, ed Baker KF (University of Califonia Division of Agricultural Sciences, Agricultural Experiment Station, Extension Service, Berkeley, CA), pp 68–85.
- 69.Kembel SW, et al. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci USA. 2014;111:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanshew AS, Mason CJ, Raffa KF, Currie CR. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J Microbiol Methods. 2013;95:149–155. doi: 10.1016/j.mimet.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuczynski J, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol. 2012;Chapter 1:Unit 1E.5. doi: 10.1002/9780471729259.mc01e05s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 74.Mercier C, Boyer F, Bonin A, Coissac E. 2013 SUMATRA and SUMACLUST: Fast and exact comparison and clustering of sequences. SeqBio 2013 Workshop. Available at https://git.metabarcoding.org/obitools/sumatra/wikis/home. Accessed October 15, 2015.
- 75.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newton ILG, Roeselers G. The effect of training set on the classification of honey bee gut microbiota using the naïve Bayesian classifier. BMC Microbiol. 2012;12:221. doi: 10.1186/1471-2180-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caporaso JG, et al. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hamady M, Lozupone C, Knight R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oksanen J, et al. 2008 The Vegan Package. Available at cran.r-project.org/. Accessed October 15, 2015.
- 83.Husson F, Josse J, Le S, Mazet J. 2010 FactoMineR: Multivariate exploratory data analysis and data mining with R. R Package, Version 1.38. Available at www.cran.r-project.org/package=FactoMineR.fr. Accessed October 15, 2015.
- 84.R Core Team 2016 R: A language and environment for statistical computing. Available at www.r-project.org/. Accessed October 15, 2015.
- 85.Warnes GR, et al. 2016 gplots: Various R programming tools for plotting data. Available at https://cran.r-project.org/package=gplots. Accessed October 15, 2015.
- 86.US Environmental Protection Agency 2007. Method 1694: Pharmaceuticals and personal care products in water, soil, sediment, and biosolids by HPLC/MS/MS (US Environmental Protection Agency, Washington, DC) [PubMed]
- 87.Fox J. 2002 Cox proportional-hazards regression for survival data. Available at citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.110.2264&rep=rep1&type=pdf. Accessed October 15, 2015.
- 88.Therneau TM, Lumley T. 2014 Package “survival.” CRAN, pp 1–136. Available at https://cran.r-project.org/web/packages/survival/index.html. Accessed October 15, 2015.
- 89.Oksanen J. r-sig-ecology 2012 Post-hoc test for adonis PERMANOVA. r-sig-ecology. Available at r-sig-ecology.471788.n2.nabble.com/post-hoc-test-for-adonis-PERMANOVA-td7577679.html. Accessed January 1, 2017.
- 90.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.