Significance
In eukaryotic cells, membranous organelles exchange trafficking vesicles that bud from the precursor and are transported to the target membrane where they fuse. Many of the required proteins are known. Moreover, some steps such as budding and fusion have been reconstituted using artificial membranes and a minimalistic set of proteins. However, it is not known whether these vesicles are functional in the complex environment of a cell. Here, we have introduced such artificial vesicles into living cells and find that only few components are needed for function in membrane traffic, with all other factors recruited from the cytoplasm. Thus, eukaryotic cells have a remarkable capacity for self-organization that can be exploited for targeting tags to specific regions in the cell.
Keywords: microinjection, liposome, SNARE proteins, membrane traffic, vesicle transport
Abstract
There is still a large gap in our understanding between the functional complexity of cells and the reconstruction of partial cellular functions in vitro from purified or engineered parts. Here we have introduced artificial vesicles of defined composition into living cells to probe the capacity of the cellular cytoplasm in dealing with foreign material and to develop tools for the directed manipulation of cellular functions. Our data show that protein-free liposomes, after variable delay times, are captured by the Golgi apparatus that is reached either by random diffusion or, in the case of large unilamellar vesicles, by microtubule-dependent transport via a dynactin/dynein motor complex. However, insertion of early endosomal SNARE proteins suffices to convert liposomes into trafficking vesicles that dock and fuse with early endosomes, thus overriding the default pathway to the Golgi. Moreover, such liposomes can be directed to mitochondria expressing simple artificial affinity tags, which can also be employed to divert endogenous trafficking vesicles. In addition, fusion or subsequent acidification of liposomes can be monitored by incorporation of appropriate chemical sensors. This approach provides an opportunity for probing and manipulating cellular functions that cannot be addressed by conventional genetic approaches. We conclude that the cellular cytoplasm has a remarkable capacity for self-organization and that introduction of such macromolecular complexes may advance nanoengineering of eukaryotic cells.
Intracellular delivery of macromolecular cargo into living cells constitutes an important approach for both fundamental research and therapeutic applications (1). For many years, efficient procedures for introducing nucleic acids into cells for gene expression or gene silencing have served as fundamental tools to advance molecular biology and cell biology, and some of them have shown promise in clinical trials for gene therapy of genetic diseases (1). In contrast, much less has been invested in the intracellular delivery of other macromolecular complexes such as multiprotein complexes, isolated organelles, liposomes, nanoparticles, or nanodevices due to the fact that the development of convenient and efficient delivery technologies has been lagging behind (1). So far, comparatively few studies are available in which intracellular delivery of macromolecular complexes is used for probing cellular functions. For instance, isolated mitochondria were microinjected to manipulate cellular mtDNA, which can be used to compensate metabolic defects due to mutations in mtDNA (2, 3). In another example, microinjection of beads in living cells was used to study particle movement, which was mediated by an interaction of the beads with the intracellular cytoskeleton (4, 5).
While these studies are promising, we believe that the enormous potential of such approaches for both basic research and therapeutic applications still remains to be exploited. For example, “bottom-up approaches” involving a combination of biochemistry and nanoengineering have recently been used for reconstructing model membranes in vitro that exhibit properties resembling their biological counterparts (6, 7). It remains to be established whether such devices retain their functionality in the environment of a living cell, which constitutes the ultimate test for the engineering quality of such devices. Indeed, testing such devices in the environment of a living cell may provide an approach toward their functional improvement. Thus, it may allow for bridging the still large gap between in vitro reconstitution and functional studies in intact cells, a major challenge presently facing the field of synthetic biology.
In this study, we have used intracellular delivery of engineered membranes with the goal to assess whether artificial vesicles can be created in the test tube, which are functional in the highly complex cellular process of membrane trafficking and which can be used to sense the intracellular environment and to modulate intracellular function. For delivering large vesicles and beads with a diameter of a few hundred nanometers, we have adapted a traditional microinjection system. While this method cannot easily be scaled up, it is reliable, of low toxicity, and it allows for obtaining proof of principle.
Our results reveal a surprising capacity of cells to integrate foreign artificial membranes into their endogenous transport pathways and probably also into their metabolism. For instance large unilamellar vesicles (LUVs) devoid of proteins and with a simple membrane lipid composition are slowly recognized by the dynein/dynactin complex and are transported to the Golgi apparatus as long as the membranes contain phosphatidylserine. In addition, incorporation of purified endosomal SNARE proteins into LUVs suffices to render them fully functional as trafficking vesicles that fuse with the correct endogenous compartment, with all other required components self-assembling from pools in the surrounding cytoplasm. Furthermore, by incorporating appropriate sensors or affinity tags into LUVs, it is possible to detect pH and enzymatic activity in endosomes and to change the dynamics of endosomal trafficking. Together, our results demonstrate that minimalistic nanodevices such as artificial vesicles allow for uncovering basic self-regulation mechanisms that appear to be fundamental properties of life cells and that cannot be accessed with more conventional approaches.
Results
Micoinjection of Nanoparticles Does Not Affect Cell Viability.
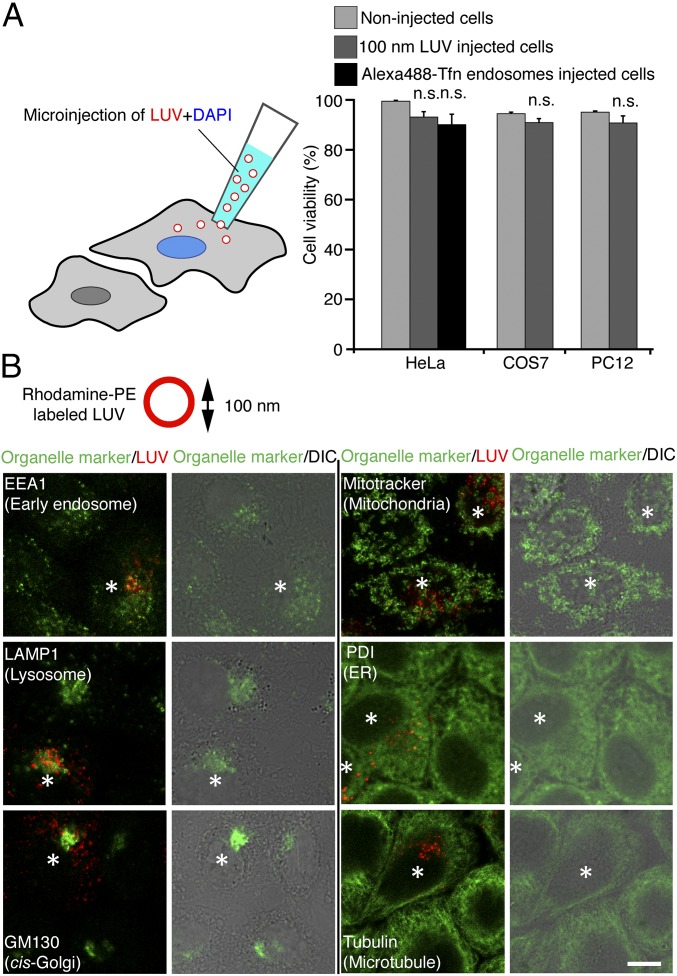
In the first set of experiments, we established a protocol for maximizing the efficiency of intracellular delivery while minimizing stress that is known to occur upon microinjection (8). To mark injected cells, injection solutions contained the DNA dye DAPI (4′,6-diamidino-2-phenylindole) (Fig. 1A). Cell viability was assessed by incubation with calcein-acetoxymethyl ester (calcein-AM), a cell-permeant dye precursor that is rendered fluorescent only in live cells due to the action of an intracellular esterase. HeLa cells injected with 100-nm LUVs or isolated Alexa 488-transferrin (Tfn) positive endosomes (Fig. S1A) using the optimized protocol did not show lower viability than noninjected cells 24 h after injection (Fig. 1A). Similar results were obtained with COS7 cells and PC12 cells (Fig. 1A). To exclude the possibility that the injection pressure changes the distribution of organelles, injected cells were immunolabeled for various organellar markers. Note that rhodamine-labeled phosphatidylethanolamine (PE) used for labeling LUVs was preserved during the fixation and immunostaining procedure (Fig. S1B). Again, no change in the staining pattern was observable when comparing cells injected with LUVs to noninjected (Fig. 1B). Moreover, injection did not activate autophagocytosis, since we were unable to detect any up-regulation of the autophagocytosis marker LC3 for up to 3 h after injection (Fig. S1C), in contrast to starved cells used as positive control (Fig. S1D). These results indicate that our microinjection protocol does not have any negative impact on cell viability or organelle distribution.
Fig. 1.
Efficient introduction system of macromolecular complexes. (A) Large unilamellar vesicles (LUVs) labeled with rhodamine-phosphatidylethanolamine (PE) or isolated Alexa 488-transferrin (Tfn) positive endosomes were microinjected into HeLa cells with DAPI (injection marker). Cell viability of injected HeLa cells was measured by calcein-AM, which is converted to green fluorescent calcein in living cells. Cell viability was not affected by injection into HeLa, COS7, and PC12 cells 24 h after injection. n.s., no statistical difference. (B) HeLa cells were injected with LUVs [100-nm average diameter, labeled with rhodamine-PE (red)], fixed at 15 min after injection, and immunostained for organelle markers (green). Asterisks mark injected cells. (Scale bar, 10 µm.) DIC, differential interference contrast.
Next, we extended our experiments to other nanoparticles. Liposomes of various sizes (average diameter 30 nm, 100 nm, and 200 nm), 100 nm latex beads, and isolated Alexa 488-Tfn positive endosomes (∼300 nm) could be smoothly injected into HeLa cells (Fig. S1E). However, microinjection of isolated labeled mitochondria or larger liposomes (∼500 nm liposomes) was less efficient because of easy clogging of the needle (Fig. S1E), suggesting that our injection system is suitable for nanoparticles with a diameter not exceeding a few hundred nanometers.
Injected Liposomes Are Targeted to the Golgi Apparatus Depending on Size and Composition.
It was shown previously that incubation of perforated cells with small unilamellar liposomes (SUVs) resulted in fusion with Golgi membranes 5–30 min after their addition (9, 10). Golgi targeting was found to be more effective with SUVs than with larger liposomes and did not require microtubule-based transport (9, 10). Furthermore, it was shown that the amphipathic lipid packing sensor (ALPS) motif of GMAP-210 (a golgin on the cis-Golgi membrane) contributes to capturing of SUVs (10).
To further explore the fate of liposomes introduced into intact mammalian cells, we injected protein-free LUVs (average diameter 100 nm) composed of phosphatidylcholine (PC, 79.7%), phosphatidylserine (PS, 20%), and rhodamine-PE (0.3%) into HeLa cells and monitored their fate over time using fluorescence microscopy. Five minutes after injection, the LUVs were distributed randomly across the cytoplasm and did not colocalize with any of the examined organelle markers (Fig. 2 A and B and Fig. S2A). One hour after injection, and even more conspicuously 3 h after injection, the rhodamine signal was dimmer and concentrated mainly around the perinuclear region, exhibiting significant colocalization with Golgi markers [Golgi matrix protein of 130 kDa (GM130) and Golgin97] but also, albeit to a lesser extent, with other organellar markers such as LAMP1 (lysosomes) and PDI (endoplasmic reticulum) (Fig. 2 A and B and Fig. S2A). Moreover, encapsulation of 50 mM calcein to monitor fusion by dequenching indicated that injected LUVs fused with the Golgi labeled with Alexa 594-cholera toxin B (Fig. S2B). In contrast, injection of SUVs (average diameter of 30 nm) with the same lipid composition resulted in much faster sequestration at the Golgi apparatus, in agreement with the previous studies (Fig. 2C). As control, we injected fluorescent latex beads (100-nm diameter) and a fraction enriched in early/recycling endosomes that were isolated from HeLa cells preincubated with Alexa 488-Tfn (Fig. S1A). Neither beads nor the endosomes showed any colocalization with GM130 for up to 180 min after injection (Fig. 2C), suggesting that Golgi targeting is specific for liposomes. We next tested whether Golgi targeting of LUVs is dependent on the presence of acidic lipids (PS). Indeed, targeting was enhanced by ∼20% when the PS concentration was increased to 50%, and was significantly reduced when PS was omitted (Fig. 2D). This result was confirmed by the simultaneous injection of LUVs containing either 20% PS or no PS that were labeled with different dyes, respectively (Fig. S2C).
Fig. 2.
Fate of injected macromolecular complexes in HeLa cells. (A) HeLa cells were fixed at the indicated time after injection of 20% PS/PC LUVs (100-nm average diameter, labeled with rhodamine-PE) and immunostained for organelle markers (green) (see Fig. S2A for views of larger fields). Note that 5 min after injection, no significant colocalization with any of the tested organellar markers is observable. However, 1 h after injection, a high degree of colocalization with GM130, and to a lesser extent with some other of the tested markers including LAMP1 and PDI, became evident. (Scale bar, 3.5 µm.) (B) Quantification of colocalization between injected 20% PS/PC LUVs and organelle markers. The value of Pearson’s correlation coefficient was calculated from at least 12 injected cells for each point. (C) Quantification of colocalization between injected 20% PS/PC LUVs, 20% PS/PC SUVs, isolated Alexa 488-transferrin (Tfn) endosomes, and latex beads with GM130. The value of Pearson’s correlation coefficient was calculated from at least 12 injected cells for each point. Because fluorescent signals of Alexa 488-Tfn in injected endosomes were too weak to measure colocalization at 180 min after injection, the value could not be calculated at the point (n.a., not applicable). (D) Effect of phosphatidylserine (PS) concentration on targeting of LUVs toward the Golgi (monitored by determining the degree of colocalization with GM130). The value of Pearson‘s correlation coefficient was calculated from at least 14 injected cells for each point. (E) Targeting LUVs to mitochondria using the HaloTag system. To bind LUVs to the surface of mitochondria, labeled LUVs containing HaloTag ligand were injected into cells expressing HaloTag-GFP-MAO. Immunostaining of HaloTag-GFP-MAO–expressing cells for HaloTag or for mitofilin show that the fusion construct is properly localized to mitochondria (images at Right). (Scale bar, 10 µm.) (F) HaloTag ligand labeled 20% PS/PC LUVs were injected into HeLa cells expressing HaLoTag-GFP-MAO. The LUVs signal (red) colocalized with mitochondria expressing HaLoTag-GFP-MAO (blue) as well as with the Golgi marker GM130 (green) 60 min after injection. (Scale bar, 5 µm.) (G) Percentage of the fluorescent signal of injected HaloTag ligand LUVs colocalizing with GFP expressing mitochondria or with GM130-positive Golgi membranes 60 min after injection. Colocalization of HaloTag-GFP-MAO with GFP increased, whereas the percentage of colocalization with GM130 decreased. *P < 0.05, ***P < 0.001, all determined by unpaired t test. (H) Time course of the degree of colocalization between HaloTag ligand LUVs with GM130. The value of Pearson’s correlation coefficient was calculated from 9 to 14 injected cells for each point. *P < 0.05, **P < 0.01, all determined by unpaired t test.
Finally, we investigated whether microinjected LUVs can be directed to specific intracellular sites using affinity capturing. To do so, we used the HaloTag system (11). We first synthesized PE-conjugated HaloTag ligand (Fig. S2D), which covalently binds to the HaloTag protein fusion tag. Then, 20% PS/PC LUVs containing 2% HaloTag ligand-PE were injected into cells expressing HaloTag fused to the mitochondria-targeting signal of monoamine oxidase (MAO), a resident of the outer mitochondrial membrane (HaloTag-GFP-MAO) (Fig. 2E). As expected, the HaloTag protein was selectively localized to mitochondria (Fig. 2E). Sixty minutes after injection, HaloTag ligand-LUVs were found preferentially associated with mitochondria, whereas the association with the Golgi complex was reduced in comparison with untagged liposomes (Fig. 2 F–H). To confirm the specificity of the interaction, we prepared cell-free extracts (postnuclear supernatants) of cells expressing HaloTag-GFP-MAO and incubated them with HaloTag ligand-LUVs, resulting in a specific binding that was dependent on the presence of the HaloTag system (Fig. S2E).
LUVs Are Transported Along Microtubules via Dynein/Dynactin Complexes, but only with Considerable Delay After Microinjection.
The slow kinetics by which LUVs are targeted to the Golgi apparatus prompted us to investigate whether targeting involves microtubule-based transport. Indeed, Golgi targeting was completely inhibited when cells were incubated with nocodazole (a tubulin polymerization inhibitor) and significantly retarded upon incubation with ciliobrevin D (an inhibitor of dynein) (Fig. 3A). We therefore investigated the movement of injected LUVs in more detail using confocal live cell imaging. As reference, we monitored the movement of endogenous endosomes, labeled by preincubating the cells with Alexa 488-Tfn for 5 min. Vesicles showing unidirectional displacement over 1 µm at least once during a time period of 40 s were classified as undergoing directional movement. Moreover, we distinguished vesicles that remained stationary (less than 0.2-µm displacement over 40 s) from vesicles that exhibited random movement, i.e., nondirected, diffusion-like motility. Five minutes after injecting 20% PS/PC LUVs, most liposomes (64.3% of 235 LUVs) exhibited random movement, whereas the rest remained stationary, with directed movement not being observable at this time (Fig. 3B and Movie S1). Mean-square displacement (MSD) analysis confirmed that randomly moving LUVs are mainly driven by Brownian motion (Fig. S3A). In contrast, most endosomes remained stationary, whereas 15.6% of 742 endosomes showed directional movement (Fig. 3 B and C and Fig. S3A and Movie S2). Fifteen minutes after injection, the population of stationary LUVs increased to 89.2% of 185 LUVs but still no directed movement was observed (Fig. 3B and Movie S3), while most of PC liposomes still moved randomly (Fig. 3B). However, 60 min after injection, 18.1% (668 LUVs were measured) of injected LUVs showed directional movement, approximately two-thirds (66.1%) toward the perinuclear region, i.e., in the minus-end direction (Fig. 3 B and C and Movie S4). This resembled the directional movement of endosomes of which 69.2% were traveling toward the minus end of the microtubules (Fig. 3C). Directional movement of both liposomes and endosomes was inhibited by 1 µM nocodazole (Fig. 3C and Fig. S3 B and C and Movies S5 and S6), confirming that the movement is mediated by microtubule-dependent transport. Furthermore, almost no directional movement was observed in the absence of PS over 60 min after injection (Fig. 3 B and C), suggesting that PS is essential for microtubule-dependent directional movement of LUVs. As outlined above, the movement of PS-containing LUVs 1 h after injection resembled that of endogenous endosomes. However, we noted that both speed and traveling distance (displacement) of individual events were lower than those of endogenous early endosomes (Fig. 3 D and E). The average speed of LUVs containing 20% PS was 0.32 ± 0.033 μm/s, that of Tfn-positive endosomes was 0.77 ± 0.05 μm/s [in agreement with a previous study (12)]. The average displacement was: LUVs, 2.44 ± 0.18 μm; endosomes, 3.45 ± 0.16 μm (Fig. 3 D and E), suggesting that the link between LUVs and the microtubule-based transport system is not as effective as that of the endogenous organelles. The data described so far suggest that dynein is recruited to PS-containing LUVs, allowing directional movement toward the perinuclear region, but with a considerable delay after injection. Immunoblotting confirmed the selective binding of several components of the presumed transport complex [dynein intermediate chain, p150Glued (a component of dynactin), and tubulin] to PS-containing LUVs, whereas no binding was observable to LUVs devoid of PS (Fig. 3F). Taken together, injected SUVs undergo random diffusion and are quickly captured by the Golgi complex with no involvement of cytoskeleton-based transport. In contrast, injected LUVs show reduced diffusion, giving them time to recruit the dynein/dynactin complex that then transports them toward the cell interior. The latter process is slow, and while most of the membrane appears to end up in the Golgi area of the cell, the lipids are also recovered in other membranes.
Fig. 3.
Injected LUVs display directional movement after injection. (A) Targeting of 20% PS/PC LUVs to the Golgi is inhibited by 1 μM nocodazole (disassembling microtubules) or 20 μM ciliobrevin D (blocking dynein). The drug concentration was kept constant during the experiment. Colocalization was measured after fixation and staining for the Golgi marker GM130. The value of Pearson’s correlation coefficient was calculated from at least 12 injected cells for each time point. **P < 0.01, ***P < 0.001, all determined by unpaired t test. (B) Representative trajectories of vesicle movement (see also Movies S1–S4). Time-lapse imaging was performed with one acquisition every 200 ms in HeLa cells. The first frame is shown as a background, with the trajectories plotted in different colors to better distinguish individual trajectories. Trajectories represent vesicles that could be tracked over 10 s during 40-s acquisition. Alexa 488-Tfn–positive endosomes were imaged at 5 min after internalization of Tfn. LUVs were imaged at the indicated time after injection. White arrows show typical examples of directional movement. White dotted lines show the cell boundary. N, nucleus. (Scale bar, 3 µm.) (C) The distribution of directional movements toward the cell periphery (probably plus-end directed) and toward the cell center (probably minus-end directed) (percentage of all vesicles). For the liposomes, the measurements were carried out 60 min after injection, and for endogenous endosomes 5 min after internalization of Alexa 488-Tfn. Gray bars indicate the percentage of vesicles showing directional movement in cells pretreated with 1 μM nocodazole. Numbers on Top of the bar indicate the number of analyzed vesicles. Data are from a representative experiment. (D) Histogram of the mean velocities of directional movements. See C for details. For liposomes, the data represent 53 tracks from 13 cells, for the endosomes, the data represent 57 tracks obtained from 16 cells. (E) Histogram showing the traveling distance of each directional movement. See D for details. (F) LUVs containing PS bind dynein intermediate chain (IC), dynactin (p150Glued), and α-tubulin when incubated with a cytosolic fraction isolated from HeLa cells. LUVs containing either 20% PS and 80% PC or only PC were incubated for the indicated time at 37 °C, with binding being measured by flotation gradients. The top fractions of each gradient containing the LUVs were used for analysis by immunoblotting.
SNARE Proteins Suffice for Specific Targeting of Liposomes to Endosomes.
The data described so far suggest that cells possess a “default” pathway that recognizes large vesicles devoid of any targeting information and moves them toward the Golgi area of the cell as long as they contain acidic lipids. In contrast, all endogenous vesicles possess an identity that is determined by their precursor history and that defines their intracellular destination with high precision (13). In particular, the endosomal system contains multiple trafficking routes connecting the trans-Golgi network with the plasma membrane and the degradative limb of the pathway, with the identity and targeting of shuttle vesicles being governed by “zip code” proteins and lipids such as Rabs, SNAREs, and phosphoinositides (14–16). In the following experiments we therefore asked whether cells can be tricked into accepting an injected synthetic liposome as a specific trafficking vesicle if it is labeled with a minimal set of such zip code molecules. To address this issue, we focused on the early endosomal system since it constitutes a dynamic and flexible sorting platform and since many of the molecules responsible for targeting specificity are known (17, 18).
To establish proof of principle, we first examined whether endosomes isolated by subcellular fractionation retain their capacity of selective targeting when reinjected. Early/recycling endosomes were prepared from cells that were prelabeled with Alexa 488-Tfn (Fig. S1A). These endosomes were functional since they underwent homotypic fusion in vitro (19). We have already shown above that in contrast to liposomes, injected endosomes are not targeted to the Golgi (Fig. 2C). Rather, already 5 min after injection they conspicuously colocalized with endogenous endosomes that were pulse labeled with Alexa 568 (Fig. 4A). To quantify the degree of colocalization, the center of all spots in both channels was calculated and the distance of every green endosome to the closest red particle was measured (19). As shown in Fig. 4B, a peak with a maximum at 100 nm was observed which, as shown previously (19), is indicative of fusion between the injected and endogenous endosomes (see also below). To control for accidental overlap, we carried out experiments in which cells were kept at 4 °C. No peak was discernible (Fig. 4B, gray curve). Moreover, no significant overlap was seen with other organelles such as lysosomes (Fig. 4C, and not shown). Similarly, targeting specificity was reduced when the endosomes were treated with trypsin before injection (Fig. S4 A–C).
Fig. 4.
SNARE proteins determine targeting specificity. (A) Representative 3D images of HeLa cells fixed 5 min after injection of early/recycling endosomes containing internalized Alexa 488-Tfn and DAPI (injection marker). Endogenous early/recycling endosomes were labeled by internalization of Alexa 568-Tfn during microinjection for 5 min. Arrows indicate colocalization between injected (green) and endogenous endosomes (red). (Scale bar, 5 µm.) (B) Histogram of the minimum distance between injected endosomes and the closest Alexa 568-Tfn–positive endosomes (black, gray shows the histogram for a control experiment at 4 °C). The blue shaded area is defined as colocalized in this study and taken as indicative for fusion (see text for details). (C) The degree of colocalization between injected isolated endosomes (labeled with Alexa 488-Tfn) and endogenous Alexa 568-Tfn–positive endosomes or LAMP1-positive lysosomes (in percentage of injected endosomes). ***P < 0.001, all determined by unpaired t test. (D) Twenty percent PS/PC LUVs reconstituted early endosomal SNAREs (4-EE-SNARE) preferentially colocalized with endogenous early/recycling endosomes after injection into HeLa cells. (Left) Analysis of 4-EE-SNARE LUVs by flotation density gradient reveals efficient incorporation of the SNARE proteins. Proteoliposomes were loaded at the bottom and overlaid with a discontinuous Nycodenz gradient, followed by analysis of the fractions by SDS/PAGE and immunoblotting for the SNARE proteins. All proteins are recovered in the low-density top fractions containing liposomes. The Right figure shows representative confocal microscopy images of HeLa cells, prelabeled with Alexa 488-Tfn and fixed 5 min after injection of 4-EE-SNARE LUVs. Protein-free LUVs were used as control. White arrows indicate colocalization between injected LUVs and Alexa 488-Tfn–positive endosomes. (Scale bar, 2.5 µm.) (E) Histogram of the minimum distance between endogenous Alexa 488-Tfn endosomes and protein-free LUVs or 4-EE-SNARE LUVs. The blue shaded area is defined as colocalized in this study and taken as indicative of fusion. (F) Colocalization between injected 4-EE-SNARE LUVs (black) and LUVs (gray), respectively, and endogenous Alexa 488-Tfn endosomes or LAMP1-positive lysosomes (in percentage of injected liposomes). **P < 0.01, all determined by unpaired t test.
Together, these observations establish that isolated endosomes retain all components that are necessary for directed movement, targeting, and fusion with their endogenous counterparts when reintroduced into intact cells. The question then arises as to which of these components is necessary and sufficient to endow a vesicle with the targeting and fusion selectivity of an early endosome. The only integral membrane constituents that are known to be essential for all trafficking organelles of the secretory pathway include the SNARE proteins that not only catalyze fusion but, at least in some instances, also participate in the recruitment of specific tethering factors (15, 20). To confirm that the fusion between injected and endogenous endosomes is indeed dependent on SNAREs, we inhibited SNARE disassembly required for SNARE activation. Disassembly is carried out by N-ethylmaleimide sensitive factor (NSF) that uses soluble NSF attachment protein (SNAP, with αSNAP being the most abundant isoform) as cofactor (21). Preincubation of the cells with N-ethylmaleimide (NEM) or coinjection of a dominant-negative αSNAP mutant (22) strongly reduced colocalization (Fig. S4 D and E).
We then asked whether SNAREs alone are sufficient to convey correct targeting information to an otherwise naive artificial vesicle. To this end, we prepared proteoliposomes containing the set of SNAREs shown previously to be responsible for the homotypic fusion of early endosomes (EE), including syntaxin 13, vti1a, syntaxin 6, and VAMP4, referred to as EE-SNAREs (18, 23). Proteoliposomes were generated by detergent removal and adjusted to an average diameter of 100 nm and a SNARE:lipid molar ratio corresponding closely to the estimates for isolated endosomes (refs. 18 and 24 and see Methods for details). LUVs included 20% PS/0.3% rhodamin-PE/PC lipids as described above, because PS is necessary for reconstitution of SNARE proteins into liposomal membrane. Efficient membrane incorporation of the proteins was confirmed by immunocytochemistry (Fig. S4F) and coflotation gradients (Fig. 4D). For control, protein-free LUVs were used. Note that the presence of SNARE proteins did not change the mobility of LUVs after injection (Fig. S4G).
LUVs were microinjected into HeLa cells whose endosomes were labeled with Alexa 488-Tfn. After 5 min (a time point at which LUVs are not yet targeted to the Golgi complex, see Fig. 2B), the cells were fixed and analyzed for colocalization between LUVs and internalized Alexa 488-Tfn. Indeed, LUVs containing the four EE-SNAREs displayed colocalization with Alexa 488-Tfn (Fig. 4D, white arrows). The histogram of the minimum distance between 4-EE-SNARE–containing LUVs and Alexa 488-Tfn exhibited a broad peak between 100 nm and 350 nm (Fig. 4E) that was clearly absent when the 4-EE-SNARE–containing LUVs were replaced with protein-free LUVs. Accordingly, 4-EE-SNARE LUVs significantly colocalized with Tfn-positive endosomes, but with lower efficiency than isolated Tfn-positive endosomes (Fig. 4F). In contrast, colocalization with LAMP1 was low (Fig. 4F). These data indicate that SNARE proteins suffice for targeting liposomes to the correct domain of the secretory pathway.
Injected EE-SNARE–Containing LUVs Fuse with Endogenous Endosomes.
One important question is whether these EE-SNARE–containing LUVs only associate with endogenous endosomes (docking) or whether they also fuse with them. Using colocalization by fluorescence microscopy we have shown earlier that appropriate thresholds can be applied to the minimum distance histograms shown above that allow for discriminating between docking and fusion with reasonable accuracy (19). According to these criteria, the majority of the liposomes associated with endosomes had undergone fusion. For independent confirmation of our threshold settings we carried out a control experiment in which LUVs containing syntaxin 6 were injected, followed by colocalization analysis between the lipid dye and immunostaining for syntaxin 6 (“fusion control”). A peak at about 75 nm was observed (Fig. S4H), which shows that our threshold was accurately set.
To prove beyond doubt that the injected liposomes are indeed fusing with endogenous endosomes, we used several independent approaches. First, we blocked SNARE function using an approach similar to that described above for early endosomes. Indeed, preincubation of the cells with NEM or coinjection of a dominant-negative αSNAP mutant strongly reduced colocalization (Fig. 5A), confirming that SNARE function is required.
Fig. 5.
Injected EE-SNARE containing LUVs fuse with endogenous endosomes. (A) Effect of NEM (10 µM) and of a dominant negative αSNAP mutant (L294A, coinjected at 10 mg/mL) on colocalization between injected 4-EE-SNARE LUVs and endogenous endosomes, labeled with Alexa 488-Tfn. (B) Snapshots from a movie (Movie S7) showing approach between an Alexa 488-Tfn–labeled endosome (white arrow) and a 4-EE-SNARE LUV (white arrowhead). After contact (time: 0 s), the red and green spot merged and moved together. (Scale bar, 2.5 µm.) Movement trajectories of the endosome (green) and the LUV (red) are shown in the Upper Right plot. After contact the movements are identical. (Scale bar, 1 µm.) Lower Right shows the fluorescence intensities of the tracked endosome (green) and the LUV (red). Shortly after contact there is a sharp drop in red fluorescence, probably caused by the dilution of the labeled lipid after fusion and thus indicative of fusion. Only a slight change was observed in the intensity of the green dye, possibly due to the fact that the dilution factors between the membrane and the content markers are different. (C) Snapshots from a movie (Movie S8) showing increase of calcein fluorescence due to dilution-induced fluorescence dequenching upon fusion of a liposome with an endogenous vesicle. An endosome (blue) docked to a calcein-containing LUV reconstituted with 4-EE-SNAREs (red) exhibited a sudden increase in calcein fluorescence (green) signal (0 s time point) in the endosome. (Scale bar, 2 µm.) The graph shows the fluorescence intensities of the tracked endosome, labeled with Alexa 633-Tfn (blue) and calcein (green). There is a sharp increase in green fluorescence, caused by the dilution of calcein by fusion (0 s time point). (D) C12FDG, a fluorescein-based β-galactosidase substrate, was loaded into 4-EE-SNARE LUVs, followed by injection into HeLa cells that were preincubated with a β-galactosidase–Tfn fusion protein and with Alexa 633-Tfn. The cells were incubated for 15 min and fixed. A fluorescein (green) signal, generated by cleavage of C12FDG, was detectable in Alexa633-Tfn–positive endosomes (blue), whereas no such signal was observed when the injected and preloaded liposomes did not contain the four EE-SNARE proteins. (Scale bar, 5 µm.) (E) The fluorescent pH sensor pHrodo was incorporated into 4-EE-SNARE LUVs that were also labeled with 0.3% nitrobenzoxadiazole (NBD)-PE. These liposomes were injected into HeLa cells preincubated with Alexa 633-Tfn to label endogenous endosomes. Fifteen minutes after injection, an increase in pHrodo fluorescence was observable that overlapped with Alexa 633 and that was absent when the SNARE proteins were omitted. (Scale bar, 5 µm.) (F) The increase in pHrodo fluorescence is sensitive to the V-ATPase inhibitor bafilomycin. Cells injected with 4-EE-SNARE LUVs containing pHrodo were incubated for 15 min and then the capturing of time-lapse imaging was started. After addition of bafilomycin (200 nM) a decrease in fluorescence intensity was observable. The plot shows fluorescence intensity changes of pHrodo in individual cells before and 30 s after addition of bafilomycin. For all plots in this figure, error bars indicate SEM, *P < 0.05, **P < 0.01, ***P < 0.001, determined by unpaired t test.
Second, we monitored encounters between LUVs and endosomes by live cell imaging. Fig. 5B and Movie S7 show a green-labeled endosome approaching an 4-EE-SNARE LUV from a different direction, followed by contact (time: 0 s). Thereafter, the red and green spot moved together. Furthermore, the fluorescence intensity of the red vesicle decreased just after the collision, which is probably due to the dilution of the labeled lipid into the unlabeled membrane (dequenching) and thus indicative of fusion (Fig. 5B). Only a slight change was observed in the intensity of the green dye, possibly due to the fact that the dilution factors between the membrane and the content markers are different. Note that before fusion the fluorescence intensity of the red particle showed stronger fluctuations, probably due to Brownian movement along the z axis.
Third, we encapsulated calcein (50 mM) into the SNARE-containing LUVs before injection and observed encounters with Alexa 488-Tfn–labeled endogenous endosomes using live cell imaging. In several cases, we observed a sudden increase of calcein fluorescence in Alexa 633-Tfn–positive endosomes due to dilution-induced dequenching at the contact site between 4-EE-SNARE LUVs and endosomes. No such dequenching was observable when protein-free LUVs were used (Fig. 5C and Movie S8).
As a fourth approach, we loaded liposomes and endogenous endosomes with an enzyme–substrate pair that generates a fluorescent signal only when both components are in the same compartment. The 4-EE-SNARE LUVs were loaded with a quenched fluorescein-tagged β-galactosidase substrate (C12FDG) and then injected into HeLa cells that had internalized Tfn conjugated with β-galactosidase and labeled with Alexa 633, simultaneously. If fusion between LUVs and endosomes occurs, the substrate is cleaved by the enzyme, liberating fluorescein in the lumen. Indeed, in Alexa 633-Tfn–positive endosomes of cells injected with 4-EE-SNARE LUVs contained a significantly higher fluorescein signal than endosomes in cells injected with control LUVs (4-EE-SNARE LUVs without C12FDG, protein-free C12FDG LUVs) (Fig. 5D).
In the final approach, we took advantage of the fact that the interior of endosomes is acidified by the action of a V-type proton pump (25). The 4-EE-SNARE LUVs were prepared that contain pHrodo conjugated to PE (26). pHrodo is a fluorescent pH sensor which increases fluorescence in acidic environments (27). When these vesicles were injected into cells, we detected an increase in fluorescence intensity exclusively in Alexa 633-Tfn–positive endosomes (Fig. 5E), which was not observed when protein-free LUVs were injected (Fig. 5E). Moreover, the signal was reduced if cells were treated with the V-ATPase inhibitor bafilomycin (Fig. 5F).
Taken together, we conclude that it suffices to incorporate purified EE-SNAREs into artificial vesicles to induce targeting and fusion with endogenous endosomes.
Interference with Endosomal Trafficking by Incorporation of an Affinity Tag into Endosomes via Fusion with Liposomes.
Above we have shown that liposomes can be directed to mitochondria if both membranes are equipped with complementary affinity tags. Moreover, the data above show that liposomes containing 4-EE-SNAREs fuse with early endosomes, thus providing a tool for introducing external molecules into endosomes (scheme in Fig. 6A). Therefore, we asked whether a combination of both approaches can be used to divert early endosomes to mitochondria, thus interfering with the recycling of transferrin. First, we tested in vitro whether liposomes containing both HaloTag ligand and 4-EE-SNAREs can recruit early endosomes to mitochondria derived from cells expressing HaloTag-GFP-MAO. Indeed, when HaloTag ligand + 4-EE-SNARE LUVs were mixed with a postnuclear supernatant obtained from cells expressing HaloTag-GFP-MAO, a marker of early/recycling endosomes (Tfn receptor) robustly associated with mitochondria immunoisolated with anti-GFP antibodies (Fig. 6B). Next, we microinjected HaloTag ligand + 4-EE-SNARE LUVs into HaloTag-GFP-MAO–expressing cells. No drastic changes in the distribution of EEA1-positive endosomes, lysosomes, and the Golgi were observable at low magnification (Fig. S5). However, at high magnification, Tfn-positive endosomes were found to overlap with liposomes recruited to the surface of HaloTag-expressing mitochondria (Fig. 6C). No such association was observable if EE-SNAREs were omitted or if HaloTag ligand LUVs were injected in GFP-MAO–transfected cells, regardless of whether they contained SNAREs or not (Fig. 6C).
Fig. 6.
Interference with endosomal trafficking by diverting endosomes to mitochondria via fusion with targeted liposomes. (A) Overview over the experimental design. LUVs were prepared that contain both 4-EE-SNAREs and HaloTag ligand. These liposomes are then injected into HeLa cells expressing HaloTag-GFP targeted to the outer mitochondrial membrane as shown in Fig. 2. (B) In vitro recruitment of endosomes incubated with 4-EE-SNARE LUVs containing HaloTag ligand to mitochondria. Postnuclear supernatants (PNSs) were used as source for endosomes and mitochondria that were obtained from HeLa cells expressing either GFP-MAO or GFP-MAO fused to HaloTag, and incubation was carried out for 30 min with the liposomes. Mitochondria were then immunoisolated using an anti-GFP antibody. Note that transferrin receptor (TfnR) showed significant coprecipitation with mitochondria when 4-EE-SNARE + HaloTag ligand proteoliposomes (PL), but not when HaloTag ligand liposomes (Lipo) were used for microinjection. (C) HaloTag-GFP-MAO–expressing HeLa cells were injected with HaloTag ligand LUVs containing 4-EE-SNARE proteins (Top) or with HaloTag ligand LUVs devoid of 4-EE-SNAREs (Bottom). Fifteen minutes after injection of the LUVs, Alexa 633-Tfn was internalized for 5 min and the cells were chased for 30 min. Note that Alexa 633-positive endosomes were associated with mitochondria and showed colocalization with liposomes only when the liposomes contained both 4-EE-SNARE and HaloTag ligand. (Scale bar, 2.5 µm.) The panels on the Right show intensity plots of the line scans (a purple line) in the pictures on the Left. (D) Alexa 633-Tfn (green) trafficking in 4-EE-SNARE + HaloTag ligand LUV (red)-injected cells expressing HaloTag-GFP-MAO (blue). Fifteen minutes after injection of the LUVs, Alexa 633-Tfn was internalized for 5 min and the cells were chased for 3 or 15 min. Asterisks indicate the HaloTag-GFP-MAO–expressing cells injected with 4-EE-SNARE LUVs containing HaloTag ligand. (Scale bar, 20 µm.) (E) Fluorescence intensity ratio of internalized Alexa 633-Tfn between HaloTag-GFP-MAO–expressing HeLa cells injected with 4-EE-SNARE + HaloTag ligand LUVs and noninjected cells. Microinjection of 4-EE-SNARE + HaloTag ligand LUVs resulted in retention of the fluorescence signal that was significantly higher than in control cells. (F) Quantification of the distribution of Alexa 633-Tfn–positive endosomes between the peripheral region and the perinuclear region after 15-min chasing. The perinuclear index (see Methods for details) decreased in HaloTag-GFP-MAO–expressing cells after microinjection of 4-EE-SNARE + HaloTag ligand LUVs, suggesting that Alexa 633-Tfn–positive endosomes were scattered. For all plots in this figure, error bars indicate SEM, *P < 0.05, **P < 0.01, determined by unpaired t test.
Together, these data show that liposomes containing EE-SNAREs and an artificial mitochondrial targeting signal can be used to deliver at least some endosomes to mitochondria. We then asked whether mitochondrial trapping interferes with endosomal recycling. To this end, we loaded cells with Alexa 633-Tfn and then monitored fluorescence for different chase times. After 3 min, no change was observable between injected and noninjected cells (Fig. 6 D and E). However, upon increasing chase time, selective retention by injected cells of the labeled Tfn became obvious (Fig. 6 D and E) and the distribution of Alexa 633-Tfn–positive endosomes were scattered (Fig. 6F), suggesting that the early endosomes are effectively trapped, thus interfering with the recycling of Tfn.
Discussion
A major goal of modern synthetic biology is to reconstruct features of life cells from scratch using purified components that are capable of forming dynamic molecular networks copying those of intact cells. To achieve this, it is necessary to better understand the self-organizing capacity of the cytoplasm. In this study, we have probed this capacity by introducing synthetic nanoparticles with defined compositions, focusing on artificial vesicles. Our results show that cells are remarkably adept in dealing with such foreign material.
Injected protein-free liposomes accumulate in the Golgi region of the cell, but both kinetics and mechanism of targeting depended on the properties of the liposomes. SUVs undergo random diffusion and are rapidly captured by the Golgi apparatus, in agreement with previous observations (9). As shown recently (10), capture is mediated by the ALPS motif of GMAP-210 located on the cis-Golgi membrane. The ALPS motif has a high affinity for small vesicles enriched in unsaturated lipids (10). In contrast, LUVs reach the Golgi more slowly and require transport along microtubules as random diffusion of these larger particles is hindered (28). Transport is mediated by a dynein/dynactin complex that is recruited from the cytoplasm by phosphatidylserine in the liposome membrane. This complex pulls LUVs toward the perinuclear region that contains resident early/late endosomes, lysosomes, and the Golgi. As expected, removal of PS from liposomal membrane, or functional inhibition of dynein, decreased LUV targeting to this region.
Why does the Golgi serve as default target for protein-free liposomes? The Golgi is a hub for intracellular trafficking, collecting, packaging, and sorting of numerous lipids and proteins (29). Additionally, the Golgi remodels membrane lipids by lipid-synthesizing enzymes, lipid flippases, and lipid-transport proteins (LTPs). In particular, LTPs are thought to be lipid sensors important for maintaining lipid homeostasis in cells (29, 30). Therefore, we think that the Golgi collects vesicles that are not transported elsewhere and thus integrates them into cellular pathways by remodeling of their lipid constituents. Moreover, our data suggest that injected LUVs fuse with Golgi cisternae. Again, this agrees with earlier work showing fusion of SUVs with Golgi membranes when added to perforated cells (9), and also in vitro when liposomes were incubated with isolated Golgi membranes (31). The mechanism of fusion is not known. Both studies showed that protease treatment decreased the fusion efficiency. However, it is not clear which proteins are involved and particularly whether SNAREs participate. Most SNAREs are tail-anchored (TA) proteins that are posttranslationally inserted into the ER membrane by the GET pathway (32). This pathway requires a specific membrane receptor, and thus SNAREs are not expected to be inserted into membranes devoid of the receptor, although some insertion may also occur in the absence of the GET system (see ref. 33 for a more comprehensive discussion). Alternatively, it is possible that both lipid mixing and content dequenching used for monitoring fusion are due to disintegration of the liposomes and redistribution of the lipids rather than to fusion. For instance, it has recently been reported that autophagosome-like structures can be generated under certain conditions at the Golgi, followed by degradation of engulfed molecules by fusion with lysosomes (34). This may explain why some of the labeled lipids from injected LUVs end up in LAMP1-positive structures (Fig. 2B). Whichever of the scenarios is correct, it is evident that cells have the capacity to remodel zip code-free membranes by various mechanisms that probably are too slow to affect functional trafficking vesicles but become apparent after extended residence time of a stranded vesicle in the cytoplasm.
Intriguingly, the slow default targeting to the Golgi is completely overridden if the injected vesicles contain targeting information. Endosomes that are isolated from cells and then reinjected largely maintain their targeting specificity and fuse with their endogenous counterparts, showing that both targeting and fusion information is imprinted on the trafficking vesicle and survives disruption of cells and purification of organelles. Thus, SNARE proteins are both necessary and sufficient to convert liposomes into trafficking vesicles endowed with both correct targeting information and the capacity for fusion. Note, however, that dysfunction of SNARE proteins does not interfere with docking of early endosomes in vitro (19). Apparently, vesicle targeting results from cooperative interaction of several regulators (including Rab proteins, polyphosphoinositides, and their effectors) that may partially compensate for each other, whereas in the simplified system used here, the role of single regulators can be investigated. Indeed, targeting can be partially “overridden” using an artificial system such as HaloTag, which allows targeting of injected liposomes to predefined sites.
Intriguingly, the reconstitution of SNARE proteins did not change the movement of LUVs after injection (Fig. S4G), suggesting that targeting specificity appears to be largely defined by target recognition and capture rather than by target-directed transport. It appears that traveling on microtubule tracks allows vesicles to explore the cytoplasmic space, i.e., to “look” for targets with which they interact. This “capture” probably involves tethering factors as documented in a recent study showing that ectopic expression of tethering proteins on mitochondria redirects targeting of Golgi-destined vesicles (35, 36). Our work shows that this feature can be exploited by steering artificial vesicles toward a predefined site using simple affinity tagging (as shown here by LUV targeting to mitochondria). Vesicles that are not captured eventually end up in the Golgi area via the default mechanism described above.
In summary, our results show that introduction of (proteo)liposomes as biomimetics into life cells may serve as a powerful tool to answer questions that cannot be accessed with more conventional approaches. The fact that only SNAREs are needed to convert a liposome devoid of any other trafficking component (such as Rab proteins, polyphosphoinositides, tethering factors, and coat proteins) into a functional trafficking vesicle impressively documents the power of self-organization of the cytoplasmic protein “soup”—evidently all other required factors are recruited from the cytoplasm. Moreover, as shown here in an exemplary fashion, the combination of basic biological information (e.g., vesicle targeting by SNARE proteins) with tools or sensors (e.g., lipids, fluorescent reporters, tags, and polymers) that cannot be expressed by conventional genetic approaches provides an opportunity for probing and manipulating cellular functions.
Materials and Methods
Materials.
All phospholipids were obtained from Avanti Polar Lipids. Primary antibodies were obtained from the following companies: anti-GM130, anti-EEA1, and anti-p150Glued from BD Biosciences; anti-LAMP1, anti–α-tubulin, and anti-mitofilin from Abcam; anti-FLAG from Sigma-Aldrich; anti-dynein intermediate chain from Millipore; anti-PDI from ABfinity; anti-GFP, anti-syntaxin 6, anti-syntaxin 13, anti-vti1a, anti-VAMP4, and anti-Rab5 from Synaptic Systems; anti-LC3 from MBL International. Cy2- or Cy3-conjugated goat anti-mouse and goat anti-rabbit IgG and HRP-conjugated goat anti-mouse and goat anti-rabbit IgG and rabbit anti-sheep IgG were obtained from Jackson ImmunoResarch Laboratories. Alexa Fluor-conjugated transferrins, streptavidin-conjugated β-galactosidase, and 5-dodecanoylaminofluorescein di-β-d-galactopyranoside (C12FDG) were from Molecular Probes. The 100-µm fluorescent polystyrene beads, biotin-conjugated transferrin, calcein, N-ethylmaleimide, and DAPI were from Sigma-Aldrich. Ciliobrevin D was from Millipore.
Chemical Synthesis.
HaloTag succinimidyl ester (O4) ligand (Promega) was used as a stock solution in chloroform. It was conjugated with DOPE lipid (Avanti Lipids) according to the following procedure: DOPE in chloroform was mixed with the HaloTag ligand stock solution, and triethylamine was added. The reaction mixture was stirred for 1 h and left for 3 h at room temperature. Afterward, an additional amount of HaloTag succinimidyl ester was added, and, after 4 h, the reaction mixture was concentrated by evaporation at 30 °C in the rotary evaporator. The residue was dissolved in a minimal amount of chloroform and applied onto a 10 × 10 cm high-performance TLC plate with silica gel 60 (Merck KGaA); layer thickness was 0.2 mm. The plate was developed with an eluent consisting of chloroform, methanol, and water (65/35/5, vol/vol/v). The required product zone was visualized with cerium-ammonium nitrate developer (applied onto a narrow side stripe of the plate; blue spot appeared upon heating). The product lane was scratched from the plate, and the product was extracted from silica gel with the eluent (see above), followed by sonification, centrifugation, and repeated extraction. The liquid extracts were combined, and volatile materials were evaporated in vacuum. The successful labeling of DOPE was confirmed by electrospray ionization MS (ESI-MS). The experiment showed peaks containing the proton adduct (m/z 1,137.9) with the expected molecular mass (C59H110ClN2O14P, M = 1,136.78). Synthesis and purification of pHrodo-PE is described in ref. 26.
Preparation of Proteoliposomes.
LUV preparation was performed as described (37). Lipids were mixed in chloroform:methanol (2:1) and the solvents were evaporated. The dried lipid film was redissolved in diethyl ether followed by addition of HB150 buffer [150 mM KCl, 20 mM Hepes (pH 7.5)] containing 1 mM DTT. After dispersal by sonication, the diethyl ether was removed by evaporation. Liposomes were extruded using polycarbonate membranes with a pore size of 100 nm (Avanti Polar Lipids) to give uniformly distributed large unilamellar vesicles. Incorporation of proteins into LUVs was achieved by n-octyl-β-d-glucoside (OG)-mediated reconstitution. Proteins in OG were mixed with LUVs and then detergent was removed by overnight dialysis at 4 °C in HB150 buffer containing SM-2 biobeads (BioRad). To prepare calcein encapsulated liposomes, the dried lipid film was resuspended with 50 mM calcein in 20 mM Hepes (pH 7.5) buffer (adjusted to an osmolarity of 330 mOsm with KCl). After five freeze–thaw cycles, liposomes were extruded using polycarbonate membranes of a pore size of 100 nm. To reconstitute proteins, proteins and OG were mixed with LUVs and then the detergent was removed by overnight dialysis at 4 °C in 50 mM calcein buffer. Just before microinjection, nonencapsulated calcein was removed through a Sephadex G-50 desalting column. SUV preparation was performed according to the comicellization method as described previously (38). The mixture of lipids and detergent (2% sodium cholate) was passed through a Sephadex G-50 column using the HP150 buffer to form unilamellar liposomes by detergent removal. The 20% PS/PC liposomes consist of 79.7% PC (l-α-phosphatidylcholine), 20% PS (l-α-phosphatidylserine) and, 0.3% rhodamine-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-lissamine rhodamine B sulfonyl ammonium salt) (molar ratios). Ten percent of C12FDG, 0.3% pHrodo-PE, and 2% HaloTag ligand-PE was added and PC contents were adjusted.
The SNARE protein-to-phospholipid molar ratio was adjusted to that of early endosomes (18, 24). Accordingly, for EE-SNARE LUVs, the protein-to-lipid ratio of syntaxin 13, vti1a, syntaxin 6, and Vamp4 was 1:2,000, 1:10,000, 1:1,200, and 1:15,400, respectively.
Microinjection.
A total of 2 mM (proteo)liposomes and 10 µg/mL DAPI (injection marker) in HB150 were filled in Femtotips (Eppendorf). The 1 × 104 HeLa cells were plated on poly-l-lysine (Sigma-Aldrich)-coated 12-mm coverslips (Marienfeld GmbH) and then the coverslip was placed into a 35-mm Petri dish (Becton Dickinson) filled with prewarmed injection medium [F12 medium (Invitrogen), supplemented with 10% FCS, 10 mM Hepes (pH 7.5) and 100 units/mL each of penicillin and streptomycin]. Microinjection was performed using a Femtojet (Eppendorf) and Injectman micromanipulator (Eppendorf) under a Leica DMIL inverted microscope for 5 min per coverslip. After microinjection, the cells were incubated at 37 °C in the cell culture medium and then fixed with 4% paraformaldehyde (PFA) (Sigma-Aldrich) in PBS for 10 min followed by immunocytochemistry using antibodies specific for organelles as indicated. To label transferrin-positive endosomes, microinjection was performed in 5 µg/mL Alexa Fluor 488-, Alexa Fluor 568-, or Alexa Fluor 633-transferrin–containing injection medium for 5 min. After microinjection, the cells were incubated at 37 °C in the cell culture medium, then fixed with 4% PFA in PBS for 10 min.
Preparation of Early Endosomes.
Early endosomes were prepared as in ref. 39 with modifications. HeLa cells were harvested by trypsin/EDTA treatment (Lonza GmbH), followed by washing once with fresh culture medium and once with internalization medium (OptiMEM, containing 10 mM glucose; Invitrogen). To label early endosomes, the prewarmed HeLa cells were then incubated at 37 °C with 50 µg/mL Alexa Fluor 488- or Alexa Fluor 568-transferrin (Invitrogen) for 5 min, dissolved in internalization medium. The reaction was stopped by chilling on ice and the cells were washed three times with ice-cold PBS containing 5 mg/mL BSA and once with homogenization buffer (250 mM sucrose, 3 mM imidazole-HCl, pH 7.4). The cellular pellets were resuspended in homogenization buffer with protease inhibitor (Complete EDTA-free, Roche) and cracked using a ball homogenizer with a clearance of 0.02 mm. The homogenate was centrifuged at 2,000 × g for 15 min, and the postnuclear supernatant (PNS) fraction was layered on top of a Nycodenz gradient consisting of 3 mL each of ice-cold Nycodenz solutions of 28%, 19%, 7.3%, respectively, followed by centrifugation at 200,000 × g for 90 min at 4 °C in a Beckman SW41 rotor. The 7.3%/19% boundary (early endosome-rich fraction) was concentrated while changing the buffer HB150 using a VIVASPIN 2 (30,000 molecular weight cutoff) (Sartorius). Concentrated endosomes were snap frozen in liquid nitrogen.
Supplementary Material
Acknowledgments
We thank U. Ries for technical support, Dr. V. Belov (MPI for Biophysical Chemistry Göttingen) for chemical synthesis, Dr. D. Stamou (University of Copenhagen) for providing pHrodo-PE, Dr. S. Rizzoli (University Medical Center Göttingen) for kindly providing the Matlab algorithm for examining the colocalization, and Dr. M. Ratz (MPI for Biophysical Chemistry Göttingen) for providing the human MAO cDNA plasmid. S.K. was supported by the Uehara Memorial Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713524114/-/DCSupplemental.
References
- 1.Stewart MP, et al. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 2.King MP, Attardi G. Injection of mitochondria into human cells leads to a rapid replacement of the endogenous mitochondrial DNA. Cell. 1988;52:811–819. doi: 10.1016/0092-8674(88)90423-0. [DOI] [PubMed] [Google Scholar]
- 3.Wu TH, et al. Mitochondrial transfer by photothermal nanoblade restores metabolite profile in mammalian cells. Cell Metab. 2016;23:921–929. doi: 10.1016/j.cmet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams RJ, Bray D. Rapid transport of foreign particles microinjected into crab axons. Nature. 1983;303:718–720. doi: 10.1038/303718a0. [DOI] [PubMed] [Google Scholar]
- 5.Beckerle MC. Microinjected fluorescent polystyrene beads exhibit saltatory motion in tissue culture cells. J Cell Biol. 1984;98:2126–2132. doi: 10.1083/jcb.98.6.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buddingh’ BC, van Hest JCM. Artificial cells: Synthetic compartments with life-like functionality and adaptivity. Acc Chem Res. 2017;50:769–777. doi: 10.1021/acs.accounts.6b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagny TJ, Bassereau P. Bioinspired membrane-based systems for a physical approach of cell organization and dynamics: Usefulness and limitations. Interface Focus. 2015;5:20150038. doi: 10.1098/rsfs.2015.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Yu LC. Single-cell microinjection technology in cell biology. Bioessays. 2008;30:606–610. doi: 10.1002/bies.20759. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T, Pagano RE. ATP-dependent fusion of liposomes with the Golgi apparatus of perforated cells. Cell. 1988;55:797–805. doi: 10.1016/0092-8674(88)90135-3. [DOI] [PubMed] [Google Scholar]
- 10.Magdeleine M, et al. A filter at the entrance of the Golgi that selects vesicles according to size and bulk lipid composition. eLife. 2016;5:e16988. doi: 10.7554/eLife.16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Los GV, Wood K. The HaloTag: A novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 12.Das A, Nag S, Mason AB, Barroso MM. Endosome-mitochondria interactions are modulated by iron release from transferrin. J Cell Biol. 2016;214:831–845. doi: 10.1083/jcb.201602069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 14.Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 15.Jahn R, Scheller RH. SNAREs: Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 16.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 17.Jovic M, Sharma M, Rahajeng J, Caplan S. The early endosome: A busy sorting station for proteins at the crossroads. Histol Histopathol. 2010;25:99–112. doi: 10.14670/hh-25.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohya T, et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 19.Geumann U, Barysch SV, Hoopmann P, Jahn R, Rizzoli SO. SNARE function is not involved in early endosome docking. Mol Biol Cell. 2008;19:5327–5337. doi: 10.1091/mbc.E08-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24:35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 22.Barnard RJO, Morgan A, Burgoyne RD. Stimulation of NSF ATPase activity by alpha-SNAP is required for SNARE complex disassembly and exocytosis. J Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwilling D, et al. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 2007;26:9–18. doi: 10.1038/sj.emboj.7601467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bethani I, et al. The specificity of SNARE pairing in biological membranes is mediated by both proof-reading and spatial segregation. EMBO J. 2007;26:3981–3992. doi: 10.1038/sj.emboj.7601820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forgac M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 26.Kemmer GC, et al. Lipid-conjugated fluorescent pH sensors for monitoring pH changes in reconstituted membrane systems. Analyst (Lond) 2015;140:6313–6320. doi: 10.1039/c5an01180a. [DOI] [PubMed] [Google Scholar]
- 27.Veshaguri S, et al. Direct observation of proton pumping by a eukaryotic P-type ATPase. Science. 2016;351:1469–1473. doi: 10.1126/science.aad6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: Volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 29.Bankaitis VA, Garcia-Mata R, Mousley CJ. Golgi membrane dynamics and lipid metabolism. Curr Biol. 2012;22:R414–R424. doi: 10.1016/j.cub.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drin G. Topological regulation of lipid balance in cells. Annu Rev Biochem. 2014;83:51–77. doi: 10.1146/annurev-biochem-060713-035307. [DOI] [PubMed] [Google Scholar]
- 31.Kagiwada S, et al. In vitro fusion of rabbit liver Golgi membranes with liposomes. J Biol Chem. 1993;268:1430–1435. [PubMed] [Google Scholar]
- 32.Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borgese N, Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta. 2011;1808:937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi H, et al. Golgi membrane-associated degradation pathway in yeast and mammals. EMBO J. 2016;35:1991–2007. doi: 10.15252/embj.201593191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willett R, et al. COG complexes form spatial landmarks for distinct SNARE complexes. Nat Commun. 2013;4:1553. doi: 10.1038/ncomms2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong M, Munro S. Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science. 2014;346:1256898. doi: 10.1126/science.1256898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez JM, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336:1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 39.Barysch SV, Jahn R, Rizzoli SO. A fluorescence-based in vitro assay for investigating early endosome dynamics. Nat Protoc. 2010;5:1127–1137. doi: 10.1038/nprot.2010.84. [DOI] [PubMed] [Google Scholar]
- 40.Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 41.Li X, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol. 2016;18:404–417. doi: 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.