Significance
The essential roles of cytoplasmic E3 ligases in the protein quality control (PQC) pathways have been increasingly highlighted in yeast and animal studies. However, in plants, only CHIP E3 ligase has been characterized, while the knowledge of cytoplasmic PQC E3 ligases remains rudimentary. Misfolded Protein Sensing RING E3 ligase 1 (MPSR1), a self-regulatory sensor system that functions only in the occurrence of misfolded proteins, is an identified cytoplasmic PQC E3 ligase in plants that directly recognizes emergent misfolded proteins independently of chaperones. In addition, MPSR1 sustains the integrity and activity of the 26S proteasome under proteotoxic stress. Given that MPSR1 RING E3 ligase is well conserved in eukaryotes, this study sheds light on a PQC pathway that is present particularly in plants and beyond.
Keywords: PQC, E3 ligase, proteostasis
Abstract
Ubiquitin E3 ligases are crucial for eliminating misfolded proteins before they form cytotoxic aggregates that threaten cell fitness and survival. However, it remains unclear how emerging misfolded proteins in the cytoplasm can be selectively recognized and eliminated by E3 ligases in plants. We found that Misfolded Protein Sensing RING E3 ligase 1 (MPSR1) is an indispensable E3 ligase required for plant survival after protein-damaging stress. Under no stress, MPSR1 is prone to rapid degradation by the 26S proteasome, concealing its protein quality control (PQC) E3 ligase activity. Upon proteotoxic stress, MPSR1 directly senses incipient misfolded proteins and tethers ubiquitins for subsequent degradation. Furthermore, MPSR1 sustains the structural integrity of the proteasome complex at the initial stage of proteotoxic stress. Here, we suggest that the MPSR1 pathway is a constitutive mechanism for proteostasis under protein-damaging stress, as a front-line surveillance system in the cytoplasm.
To prevent cytotoxic aggregation of abnormal proteins, cells employ protein quality control (PQC) pathways to refold, sequestrate, or immediately degrade them (1–3). Many chaperones and cochaperones mediate the triage decisions of misfolded proteins in the PQC pathways (4). The ubiquitin-proteasome system (UPS) and autophagy are the most important proteolytic systems in the PQC pathways (4–6). Autophagy contributes to the bulk removal of misfolded protein aggregates, whereas UPS mainly eliminates individual misfolded proteins, preventing harmful aggregate formation (6). E3 ligases tether ubiquitins to aberrant proteins in the UPS-mediated PQC pathway, mostly by the guidance of chaperones (hereafter referred to as PQC E3 ligases) (7, 8). The cytoplasm is a major scene for PQC employing many molecular chaperones, E3 ligases, and proteolytic components (7–12). Cytoplasmic PQC E3 ligases and the mechanisms by which they distinguish misfolded proteins from properly folded proteins have been extensively studied. In mammalian cells, several cytoplasmic PQC E3 ligases have been identified. C terminus of Hsc70-interacting protein (CHIP), ubiquitin protein ligase E3 component N-recognin 1/2 (UBR1/2), Parkin, and E6-associated protein (E6-AP) specifically recognize chaperone-bound misfolded proteins for 26S proteasome-dependent degradation (9–11). In yeast, HECT ubiquitin ligase 5 (Hul5) directly recognizes misfolded proteins without the aid of chaperones (7). Moreover, Reverses Spt-Phenotype 5 (Rsp5) is involved in both the direct and chaperone-dependent ubiquitination of cytosolic misfolded proteins (8). However, to date, only one cytoplasmic PQC E3 ligase, AtCHIP, an ortholog of the mammalian CHIP, has been identified in plants (13). Our knowledge of cytoplasmic PQC E3 ligases and their functions in plants is very limited, especially the molecular mechanisms underlying the specific recognition and removal of emerging misfolded proteins in the cytoplasm. Furthermore, in yeast, listerin E3 ubiquitin ligase 1 (Ltn1), histone E3 ligase 2 (Hel2), and negative regulator of transcription 4 (Not4) form a complex with the ribosome, which is crucial for the elimination of aberrant polypeptides translated from defective mRNAs (12, 14). Hul5 associates with the 26S proteasome, modulating proteasomal processivity (15). Despite the importance of these E3 ligase–ribosome and E3 ligase–proteasome complexes in the cytoplasmic PQC, no related study has been performed in plants.
Here, we describe a cytoplasmic PQC E3 ligase in plants. Misfolded Protein Sensing RING E3 ligase 1 (MPSR1) is a quick responsive element that rapidly eliminates emergent misfolded proteins and attunes the stress threshold of the 26S proteasome and proteostasis.
Results
MPSR1 Is a Major PQC Component for Plant Survival Under Proteotoxic Stress.
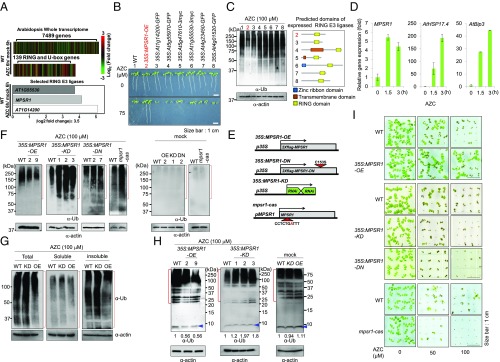
To screen unidentified cytoplasmic PQC E3 ligases, we monitored the expression levels of all E3 ligases following a controlled proteotoxic stress. To systemically modulate the multitude and time of proteotoxic damages, in lieu of various abiotic stresses, we treated 2-wk-old Col-0 seedlings with azetidine-2-carboxylic acid (AZC), an analog of l-proline that causes irreversible protein misfolding when incorporated into the polypeptide chain instead of d-proline (16, 17). By using the RNA-seq method, the transcriptome of AZC-treated samples (6 h) was analyzed and compared with that of a mock control (Fig. 1A, Upper). Gene ontology analysis showed that AZC treatment was sufficient to induce many proteotoxic stress-related genes, such as heat shock proteins (HSPs), confirming the validity of this method (SI Appendix, Table S1). Next, we analyzed the expression levels of CHIP, Hul5, Rsp5/Nedd4, and Ubr1/2 homologous E3 ligase genes in Arabidopsis (SI Appendix, Fig. S1 A and B). Interestingly, we found that the expression of these genes was not significantly increased upon AZC treatment (SI Appendix, Fig. S1A). Instead, we noticed that the expression levels of three RING E3 ligase genes (MPSR1, At1g14200, and At1g55530) were significantly elevated (log2 value > 3.5; P < 0.05) with total expression of at least 200 fragments per exon kilobase per million reads mapped (FKPM) (Fig. 1A, Lower and SI Appendix, Fig. S1 C and D and Dataset S1). To test whether these RING E3 ligases are involved in proteotoxic stress responses, we performed serial genetic and molecular analyses. First, to perform gain-of-function analyses, the three best candidates (MPSR1, At1g14200, and At1g55530), a negatively regulated control (At5g47610), and three mildly induced E3 ligases (At5g47610, At4g23450, and At5g01520) were individually expressed under the control of a 35S promoter in Col-0 background (SI Appendix, Fig. S1 D–F). Most of the transgenic plants showed severely defective growth under continuous treatment with AZC (75 μM) for 2 wk, whereas the transgenic plant harboring At1g26800 (MPSR1) gene grew unaffected (Fig. 1B). We further tested whether the stress-resistant phenotype of the At1g26800 overexpression lines (35S:MPSR1-OE) was related to PQC. Under strong protein-misfolding stress, the overloading of the 26S proteasome typically leads to the accumulation of insoluble polyubiquitinated (pUb)-proteins (18). Therefore, we monitored insoluble pUb-proteins in the At1g26800 overexpression lines. For separation of soluble pUb proteins from insoluble pUb-protein fractions, we performed the fractionation assay with 1% Triton X-100 supplemented buffer, which was used in a previous study (36). As shown in Fig. 1C, we found that the accumulation of pUb proteins was dramatically reduced compared with the other six transgenic plants and the Col-0 control, implying that At1g26800 is a positive regulator of PQC. Accordingly, we named the identified RING-type E3 ligase as MPSR1. MPSR1 transcripts were immediately increased by the AZC treatment, showing that MPSR1 is a fast-responsive gene for proteotoxic stress. AtHSP17.4 and AtBip3, encoding an endoplasmic reticulum (ER) stress-specific protein, were used as control genes for proteotoxic stress (Fig. 1D). For loss-of-function analyses, we constructed RNAi-induced knockdown lines (35S:MPSR1-KD), dominant negative (DN) transgenic lines (35S:MPSR1-DN), and a CRISPR-mediated point mutant, mpsr1-cas (Fig. 1E and SI Appendix, Fig. S2). Using these lines, we examined whether MPSR1 deficiency or MPSR1 activity suppression resulted in a defective PQC pathway and compromised plant survival. We treated the seedlings of MPSR1-deficient lines with AZC (100 μM) for 2 wk. Then, the levels of soluble and insoluble pUb proteins were determined with α-Ub antibody. We found that insoluble pUb proteins remarkably accumulated in the KD, DN, and mpsr1-cas lines compared with the wild-type and 35S:MPSR1-OE lines. However, the soluble pUb proteins were notably changed neither by MPSR deficiency nor by MPSR1 excessivity (Fig. 1 F and G). We speculated that in the absence of MPSR1, plants are incapable of eliminating emergent misfolded proteins, resulting in the accumulation of insoluble pUb proteins. In plants, ubiquitin (UBQ) genes are highly up-regulated in response to many abiotic stresses, including proteotoxic stress (19–23) (SI Appendix, Fig. S3). Thus, we examined the levels of free ubiquitins in the tested lines. Compared with the level of free ubiquitins in wild-type, 35S:MPSR1-KD lines showed increased levels of free ubiquitins (20% ∼ 90%) whereas 35S:MPSR1-OE lines showed reduced free ubiquitins under the stress condition (∼45%) (Fig. 1H, Left and Middle). Under nonstress condition, the levels of free ubiquitins were not varied in wild-type, 35S:MPSR1-KD, and 35S:MPSR1-OE lines (>±10%) (Fig. 1H, Right). In accordance with these results, KD, DN, and mpsr1-cas seedlings were hypersensitive to AZC treatment and their growth were severely inhibited (Fig. 1I and SI Appendix, Fig. S4). These results, taken together, indicate that MPSR1 activity is indeed necessary to implement PQC in Arabidopsis and to remove damaged proteins before forming cytotoxic aggregates that threaten survival.
Fig. 1.
MPSR1 is essential for plant survival under proteotoxic stress. (A) Comparative transcriptome analysis in response to proteotoxic stress. (Upper) The heat map illustrates the up-and-down expression profile of genes. (Lower) The induced expression levels of three selected RING E3 ligases (mock vs. AZC). (B) Phenotypic screening of transgenic plants, overexpressing seven different RING E3 ligases, under proteotoxic stress. The numbers stand for: 1, Col-0; 2, 2xflag-At1g26800 (MPSR1); 3, At1g14200-sGFP; 4, At5g55970-sGFP; 5, At5g47610-3xmyc; 6, At1g55530-3xmyc; 7, At4g23450-sGFP (AtAIRP1); 8, At5g01520-sGFP (AtAIRP2). (C) Accumulation of insoluble pUb proteins in seven transgenic plants and wild-type seedlings upon AZC treatment (100 µM). The level of actin was used as a loading control. At Right, the predicted domains of the tested RING E3 ligases are shown. (D) Relative expression of MPSR1 transcript under AZC treatment (5 mM). AtHSP17.4, a small heat-shock protein gene, and AtBip3, an ER stress-responsive gene, were used as controls. The data are the average values of three biological samples ± SD (n = 3). (E) Cartoon showing schematically the structures of transgenic genes and the position of CRISPR-mediated mutation. (F) Accumulation of insoluble pUb proteins in 35S:MPSR1-OE, 35S:MPSR1-KD, 35S:MPSR1-DN, and mpsr1-cas lines under stress or nonstress condition. (G) Levels of ubiquitinated proteins in the soluble fraction, insoluble fraction, and whole samples. Protein samples were resolved by 8% SDS/PAGE. (H) Levels of free ubiquitins in WT, 35S:MPSR1-KD, and 35S:MPSR1-OE lines under proteotoxic stress condition or nonstress condition. Protein samples were resolved by 15% SDS/PAGE. Blue arrowheads indicate free ubiquitins. Red brackets indicate ubiquitinated proteins. (I) Phenotype analysis of 35S:MPSR1-OE, 35S:MPSR1-KD, 35S:MPSR1-DN, and mpsr1-cas lines upon AZC treatment (50, 100 µM).
MPSR1 Functions as a PQC E3 Ligase Independently of Cytoplasmic Chaperones.
The covalent modification of misfolded proteins by ubiquitin for proteasomal degradation is the major function of PQC E3 ligases. To test whether MPSR1 has E3 ligase activity, we performed an in vitro self-ubiquitination assay, which is widely used to monitor E3 ligase activity, and we clearly observed MPSR1 self-ubiquitination (Fig. 2A). To confirm this, we mutated the RING domain of MPSR1 by replacing C153 with S153 (MPSR1-DN) (Fig. 2A, Upper). This one-base substitution eliminated E3 ligase activity, demonstrating that MPSR1 is a bona fide E3 ligase (Fig. 2B). Many RING E3 ligases tend to form self-dimers to achieve full activity; thus, in most cases, chimeric dimerization between the RING domains of mutant and wild-type proteins seriously undermines their E3 ligase functionality (24–28), which could account for the dominant negative phenotype of 35S:MPSR1-DN lines (Fig. 1I). To test this assumption, we performed in vitro pulldown assay using recombinant His-MPSR1-DN and GST-MPSR1. Indeed, His-MPSR1-DN directly interacts with GST-MPSR1 (SI Appendix, Fig. S5). Therefore, we speculate that overexpression of MPSR1-DN led to the malfunction of endogenous MPSR1 by forming a nonfunctional dimer. However, we cannot rule out the possibility that the excessive MPSR1-DN dominantly bound to misfolded proteins, preventing MPSR1-dependent ubiquitination.
Fig. 2.
MPSR1 is a PQC E3 ligase that targets misfolded proteins independently of chaperones. (A) In vitro self-ubiquitination assay of MPSR1. The GST-MPSR1 protein was incubated with ubiquitination reaction buffer in the presence or absence of ATP, ubiquitin, UBA1, and UBC8 for 1 h, resolved by SDS/PAGE on an 8% gel, and detected using α-GST or α-Ub antibodies. The cartoon shows schematically the structure of wild-type and dominant negative (DN) mutant MPSR1. The inverted red arrowhead indicates the substituted base in the MPSR1 RING domain mutant. (B) Substitution of a conserved cysteine to serine at the RING domain of MPSR1 (MPSR1-DN-GFP) abolished the self-ubiquitination activity of MPSR1-GFP in vitro. (C) Yeast two-hybrid assay of MPSR1 and cytoplasmic chaperones. HSP101, HSP90.1, HSP70.1, HSP40, and HSP17.4 were not associated with MPSR1. (D) In vitro pulldown assay of Δ2GFP (artificial misfolded protein) with GST-MPSR1. Δ2GFP or GFP was incubated with GST-MPSR1 and pulled down with GST-resin. Retrieved proteins were determined with an α-GFP antibody. GST was used as a negative control. (E) Yeast two-hybrid assay using full-length and deletion constructs of MPSR1. C-terminal disordered segment of MPSR1 interacted with Δ2GFP (Left). MPSR1 did not interact with GFP (Right). (F) In vitro ubiquitination of Δ2GFP by GST-MPSR1. Ubiquitinated Δ2GFP was determined with α-GFP an antibody (Left and Middle). GFP used as a negative control (Right). Red arrowhead indicates the nonspecific aggregation of misfolded GFP. (G) In vivo coimmunoprecipitation analysis of flag-tagged MPSR1 (flag-MPSR1) and catalase using 35S:MPSR1-OE seedlings, treated with the indicated chemicals. flag-MPSR1 and associated proteins were precipitated with α-flag resin. Coprecipitated catalase and flag-MPSR1 were determined with an α-catalase antibody and an α-flag antibody, respectively (Left and Middle). Ubiquitinated catalases were immunoprecipitated with an α-catalase antibody and determined with an α-Ub antibody (Right). The blue arrowheads indicate nonubiquitinated flag-MPSR1. The red arrowheads indicate nonubiquitinated catalase. The green arrowheads indicate IgG band. The red square brackets indicate ubiquitinated proteins. (H) Subcellular colocalization of MPSR1 and CAT. 35S:MPSR1-OE seedlings were used for an immunohistochemistry assay using α-flag for MPSR1 and α-CAT for CAT. After the AZC treatment, green fluorescence from MPSR1 and red fluorescence from CAT were colocalized as speckles near the nucleus. White dotted squares show the zoom-in area. White dotted circles indicate the nucleus. The nucleus was detected with DAPI staining.
The first step in all PQC pathways is the sensing of misfolded proteins, which is followed by damage assessment and a fate decision. In many cases, chaperones and cochaperones serve to recognize damaged proteins and to facilitate refolding, or if refolding does not occur, to recruit E3 ligases and the 26S proteasome for degradation (4). For instance, CHIP E3 ligase forms an E3 ligase–chaperone complex with HSP90 and HSP70 to selectively eliminate misfolded proteins in animals and plants (29, 30). Therefore, we examined whether MPSR1 is able to recognize misfolded proteins independently or dependently of chaperones. First, using yeast-two hybrid assay, we investigated whether MPSR1 forms an E3 ligase–chaperone complex with cytoplasmic chaperones in plants. Interestingly, MPSR1 was unable to associate with five typical cytoplasmic chaperones in plants: HSP101, HSP90.1, HSP70.1, HSP40, and HSP17.4 (Fig. 2C). To rule out whether MPSR1 requires uninvestigated minor chaperones to recognize and eliminate misfolded proteins, we treated Col-0 seedlings with sodium arsenite, which interferes with chaperone-mediated protein folding system and induces protein aggregations (31–33). Upon treatment of 2-wk-old seedlings with arsenite (300 μM), the marginal expression of the MPSR1 transcripts was immediately elevated ∼ninefold, while the expression of AtBip3 was somewhat decreased (SI Appendix, Fig. S6A). Treatment with arsenite for 24 h was sufficient to cause chaperone malfunction, leading to insoluble pUb-protein accumulation in wild-type seedlings. In contrast, it was less effective on 35S:MPSR1-OE lines, showing that MPSR1 is able to remove arsenite-bound misfolded proteins or to prevent arsenite-induced protein aggregations (SI Appendix, Fig. S6B). When the wild-type and 35S:MPSR1-OE seedlings were treated under continuous strong arsenite stress for more than a week, wild-type seedlings failed to survive, while 35S:MPSR1-OE seedlings maintained their growth, showing that MPSR1 alleviates arsenite stress leading to plant viability (SI Appendix, Fig. S6C). Next, we monitored the levels of MPSR1 under the influence of sodium arsenite and two specific chaperone inhibitors: pifithrin-μ (PES) for HSP70 and geldanamycin (GDA) for HSP90. MPSR1 was notably accumulated under the treatment of chemicals, showing that MPSR1 senses misfolded proteins irrespective of chaperones, such as HSP90 and HSP70 (SI Appendix, Fig. S6D). Like the arsenite treatment, the inhibition of HSP70 and HSP90 led to the accumulation of MPSR1 transcripts (SI Appendix, Fig. S6E).
As a next step, we constructed an artificial misfolded protein, Δ2GFP, which has a deletion of the 13 N-terminal amino acids (34, 35) (Fig. 2D, Left). Then, we performed an in vitro pulldown assay using GST-MPSR1 as a bait and recombinant Δ2GFP or GFP control as a prey. Δ2GFP was clearly pulled down by GST-MPSR1, whereas the control GFP was not retrieved, showing that MPSR1 prefers to interact with misfolded Δ2GFP (Fig. 2D). To define the interacting domain of MPSR1 for Δ2GFP, we performed yeast-two hybrid assay. The full-length protein and C-terminal fragment of MPSR1 are both able to recognize Δ2GFP, showing that the disordered segment of MPSR1 is responsible for target recognition (Fig. 2E, Left and SI Appendix, Fig. S7). GFP was unable to interact with MPSR1 (Fig. 2E, Right). Further, to determine whether MPSR1 selectively conjugates pUb to misfolded proteins, we conducted in vitro ubiquitination assay with MPSR1, Ub, UBA1 (E1), UBC8 (E2), and Δ2GFP or GFP as a substrate. We found that pUb was efficiently conjugated to Δ2GFP by MPSR1. In contrast, MPSR1 was unable to tether ubiquitins to GFP (Fig. 2F). These results suggest that MPSR is capable of recognizing misfolded proteins selectively and tethering pUb to the proteins directly.
To recapitulate these results in vivo, we investigated whether MPSR1 prefers to associate with misfolding-prone proteins by testing the association of MPSR1 with a misfolding-prone cytoplasmic protein, catalase (CAT), which is highly ubiquitinated under proteotoxic stress (36). 35S:MPSR1-OE (overexpressing flag-MPSR1) seedlings were treated with MG132 to accumulate pUb proteins, or with MG132/AZC to increase misfolded pUb proteins. Using an anti-flag antibody, we immunoprecipitated flag-MPSR1 (Fig. 2G, Middle) and detected MPSR1-associated misfolded pUb-CAT with an α-CAT antibody. Indeed, a greater amount of pUb-CAT was detected in the precipitates from the AZC/MG132 double-treated plantlets than in those from the MG132 single-treated plantlets, although CAT inputs were similar in both samples (Fig. 2G, Left). To confirm the ubiquitination of CAT, we precipitated CAT with α-CAT antibody and determined its ubiquitination using α-Ub antibody. We observed that CAT proteins were more ubiquitinated under AZC treatment condition (Fig. 2G, Right). Furthermore, we visualized the subcellular localization of specific interactions between MPSR1 and aberrant CAT by immunohistochemistry. Under the mock treatment, only red fluorescence from CAT was observed, as individually dispersed speckles in the cytoplasm (Fig. 2H). Upon proteotoxic damage, considerable colocalization between green (MPSR1) and red (CAT) speckles was observed near the nucleus (Pearson correlation coefficient: 0.59 ± 0.16) (Fig. 2H). These results indicate that MPSR1 is a cytoplasmic PQC E3 ligase that targets proteins with damaged configurations due to proteotoxic stress.
MPSR1 Is a Self-Regulatory E3 Ligase That Is Stabilized by Conjugating Ubiquitins on Misfolded Proteins.
As shown in Fig. 2G, we observed that flag-MPSR1 was highly ubiquitinated in the MG132-treated 35S:MPSR1-OE seedling samples (Middle, red bracket), whereas in the MG132/AZC-treated samples, ubiquitinated flag-MPSR1 was significantly diminished and unmodified flag-MPSR1 was accumulated (Middle, red bracket). We also observed by immunohistochemistry that the green speckles of MPSR1 were barely detectable in the mock-treated seedlings (Fig. 2H), implying that MPSR1 is stabilized by proteotoxicity. To confirm the ubiquitination of MPSR1 is efficiently suppressed by proteotoxic stress, we performed an immunoprecipitation assay using the extracts from mock-treated, AZC-treated, MG132, or AZC/MG132 double-treated 35S:MPSR1-OE seedlings. Ubiquitinated MPSR1 proteins in the precipitates were distinctively observed as smeared bands when the activity of the 26S proteasome was blocked upon MG132 treatment (Fig. 3A, see red bracket), whereas upon AZC treatment, ubiquitinated MPSR1 proteins were hardly detectable and unmodified MPSR1 was significantly accumulated (Fig. 3A, see blue arrowhead). In the case of double treatment, although unmodified MPSR1 accumulated more than in the AZC single-treated sample, ubiquitinated MPSR1 was also slightly detected. MPSR1 was highly unstable in the mock treatment (Fig. 3A). Furthermore, using α-Ub antibody, we determined that MPSR1 is indeed modified by ubiquitination (Fig. 3A, Right). Hence, we hypothesized that MPSR1 is rapidly self-ubiquitinated or ubiquitinated by another E3 ligase for destruction, which is inhibited by protein-damaging stress. To test whether MPSR1 is degraded by self-ubiquitination, we investigated MPSR1 stability using two transgenic lines, 35S:MPSR1-OE and 35S:MPSR1-DN. Overexpressed flag-MPSR1 was scarcely seen after the mock treatment, whereas after 6 and 24 h of AZC treatment, the protein levels were significantly elevated. Clearly, flag-MPSR1 transgene transcription under the 35S promoter was not affected by the AZC treatment. These results showed that flag-MPSR1 protein is stabilized by proteotoxic stress (Fig. 3B). In contrast to the wild-type protein, flag-MPSR1-DN was highly accumulated even after the mock treatment, and there were no discernible differences in its expression between the nonstress and stress conditions, indicating that MPSR1 degradation is solely dependent on its own E3 ligase activity (Fig. 3B). MPSR1 self-regulation was also visualized by immunohistochemistry using the 35S:MPSR1-OE lines and protoplast transient expression system (SI Appendix, Fig. S8). To test whether UPS is responsible for MPSR1 destabilization, 35S:MPSR1-OE seedlings were treated with MG132 or a protease inhibitor mixture (PI). MPSR1 levels were dramatically increased only in the MG132-treated samples, implying that MPSR1 is degraded by UPS (Fig. 3C). The MPSR1 half-life was calculated with a cell-free degradation assay; recombinant GST-MPSR1 was incubated with the cell crude extract in a time-dependent manner. Within 1.5 h, 50% of GST-MPSR1 was rapidly destabilized, confirming that MPSR1 is a short-lived protein and is degraded by UPS (Fig. 3D). We therefore conclude that under no stress, MPSR1 becomes highly unstable by its self-ubiquitination activity.
Fig. 3.
Client-induced stabilization of MPSR1 under proteotoxic stress. (A) MPSR1 self-ubiquitination was blocked by AZC treatment. In vivo coimmunoprecipitation assay using 35S:MPSR1-OE plantlets treated with AZC (5 mM), MG132 (20 µM), or AZC (5 mM)/MG132 (20 µM). Ubiquitinated and nonubiquitinated flag-MPSR1 were precipitated with α-flag resin and detected with an α-flag antibody or with an α-Ub antibody. The red square brackets indicate ubiquitinated MPSR1. Blue arrowhead indicates nonubiquitinated MPSR1. Green arrowhead indicates IgG band, which used for immunoprecipitation. (B) flag-MPSR1 and flag-MPSR1-DN expression levels in 35S:MPSR1-OE and 35S:MPSR1-DN seedlings, respectively, treated with or without AZC (5 mM) for the indicated times were determined with an α-flag antibody. RT-PCR analysis showing flag-MPSR1 and flag-MPSR1-DN transcript levels. (C) Effect of proteolysis inhibitor MG132 (10, 20 µM) or protease inhibitor mixture (PI) (0.1, 0.2×) on flag-MPSR1 degradation. RT-PCR analysis showing flag-MPSR1 transcript levels. (D) In vitro cell-free GST-MPSR1 degradation assay using Col-0 cytosolic extracts. Time-dependent GST-MPSR1 degradation (arbitrary units) is plotted versus control MBP levels. The data shown are the averages of three replicates ±SD (n = 3). (E) Transiently expressed Δ2GFP stabilizes MPSR1. GFP or Δ2GFP were expressed in protoplasts from two 35S:MPSR1-OE transgenic seedlings and their expression levels were determined with an α-GFP antibody. MPSR1 levels were determined with an α-MPSR1 antibody. The cartoon shows the structure of the GFP and Δ2GFP expression vectors. (F) Chemically induced Δ2GFP stabilizes endogenous MPSR1. The endogenous MPSR1 levels in WT and XVE:Δ2GFP transgenic plantlets treated with β-estradiol (5 and 20 µM) for a day are shown. MPSR1 levels were determined with an α-MPSR1 antibody. Induced Δ2GFP was determined with an α-GFP antibody. Equal loading of samples was determined with an α-actin antibody. (G) The relative amounts of MPSR1 were calculated by image analysis and plotted (arbitrary units) versus the level of the initial sample (F). The data shown are average values of four immunoblot analyses with ±SD (n = 4). (H) The self-ubiquitination of MPSR1 is hindered by Δ2GFP. (I) The self-ubiquitination of MPSR1 is not hindered by GFP. Ubiquitinated MPSR1 was determined with an α-GST antibody. The red square brackets indicate ubiquitinated proteins.
We next asked how MPSR1 is stabilized in response to proteotoxic stress. The most plausible and simple mechanism would be the inhibition of self-ubiquitination by substrates (37). To test this hypothesis, we constructed an artificial misfolded protein, Δ2GFP, which has a deletion of the 13 N-terminal amino acids (34, 35) (Fig. 3E, Upper). Δ2GFP and wild-type GFP were transiently expressed in 35S:MPSR1-OE protoplasts. More flag-MPSR1 protein was accumulated in the cells expressing Δ2GFP than in the cells with normal GFP. Considering the lower expression level of Δ2GFP in comparison with GFP, the effect of the misfolded protein on MPSR1 stabilization was evident (Fig. 3E). To recapitulate this result in stable transgenic plants, we constructed transgenic lines expressing Δ2GFP under the control of the inducible XVE promoter (Fig. 3F, Upper). Addition of β-estradiol specifically induced Δ2GFP protein in the transgenic plants in a concentration-dependent manner. After the expression of Δ2GFP for 24 h, the level of endogenous MPSR1 was notably increased by ∼10-fold compared with that in the control, confirming the stabilization of MPSR1 by misfolded proteins (Fig. 3F). We plotted the correlation between the levels of MPSR1 and the concentrations of treated β-estradiol in a bar graph (Fig. 3G). Furthermore, we found that the self-ubiquitination of MPSR1 was efficiently suppressed by Δ2GFP in vitro. The reduction of MPSR1 ubiquitination was inversely correlated to Δ2GFP ubiquitination (Fig. 3H). By contrast, GFP did not hinder the self-ubiquitination of MPSR1 (Fig. 3I). This inverse correlation between MPSR1 ubiquitination and client ubiquitination was consistent with the ubiquitination pattern of MPSR1 and CAT in vivo as shown in Fig. 2G. These results not only show that MPSR1 is efficiently stabilized by tethering ubiquitins to misfolded proteins but also support that the E3 ligase activity of MPSR1 is irrelevant to chaperones.
MPSR1 Modulates the Activity and Integrity of the 26S Proteasome.
Overloading or dysfunction of the 26S proteasome by prolonged stress, such as heat shock or radical oxygen species, leads to its dissociation, resulting in pUb-protein aggregation into insoluble inclusion bodies (38). Therefore, the dramatic insoluble pUb-protein alterations in MPSR1-overexpressing and MPSR1-lacking lines could reflect: (i) the presence of an MPSR1-independent pathway for aberrant protein ubiquitination and (ii) the essential role of MPSR1 in proteasomal degradation, in addition to its role as a specific E3 ligase for damaged proteins (Fig. 1F). If MPSR1 is exclusively a cytoplasmic PQC E3 ligase, the amassed pUb-protein levels in the 35S:MPSR1-KD, 35S:MPSR1-DN, and mpsr1-cas lines would not be observed, implying the existence of an MPSR1-independent pathway for aberrant protein ubiquitination. However, the significantly reduced level of pUb-misfolded protein aggregates in the 35S:MPSR1-OE lines could be explained if MPSR1 modulates the efficacy of pUb-protein degradation by the 26S proteasome, also accounting for the pUb-protein aggregation in the MPSR1-lacking lines (Fig. 1F).
The first possibility can be easily accepted, owing to the function of other PQC E3 ligases such as AtCHIP (39). However, to date there has been no study that investigated the roles of PQC E3 ligases in modulating the activity of 26S proteasome directly or indirectly in plants. To test the second possibility, we measured the proteasomal activity under proteotoxic stress, with and without MPSR1 activity. Surprisingly, the 26S proteasome enzymatic activity was significantly diminished in the 35S:MPSR1-KD and 35S:MPSR1-DN lines, whereas the 35S:MPSR1-OE line showed a higher proteasomal activity, implying a positive effect of MPSR1 on 26S proteasome activity (Fig. 4A). Because the 26S proteasome components were not significantly varied in the tested lines, the 26S proteasome activity could not be altered owing to differences in its subunits expression levels (Fig. 4B). We therefore tested whether MPSR1 could modulate the integrity of the 26S proteasome complex and, thereby, maintain the proteasomal activity, even under proteotoxic stress. The 26S proteasome complex consists of a barrel-shaped 20S catalytic core particle (CP) and two 19S regulatory particles (RPs) of ∼2,000 kDa. To assess complex integrity by size fractionation, the crude extracts from wild-type, 35S:MPSR1-OE, and 35S:MPSR1-KD seedlings were resolved by gel filtration chromatography, and the elution of the 26S proteasome components, regulatory particle non-ATPase (RPN12), regulatory particle triple-A protein 5 (RPT5), and 20S alpha was monitored by immunoblot analysis. Under nonstress condition, RPN12 was eluted without any distinctive difference among the tested lines (Fig. 4C and SI Appendix, Fig. S9). However, in the wild-type plants under stress, most of RPN12, a subunit of RP, was eluted between fractions 46 and 51 (below ∼2,000 kDa), showing that stress strongly induces 26S proteasome dissociation. In the absence of MPSR1, RPN12 dissociation was accelerated and most of RPN12 was eluted in a protein complex below ∼700 kDa. By contrast, in the 35S:MPSR1-OE line, RPN12 was eluted in fraction 45, indicating that the 26S proteasome was still intact (Fig. 4C and SI Appendix, Fig. S9). To confirm this finding, we monitored the elution of RPT5, an AAA-ATPase subunit of the RP base. In the 35S:MPSR1-KD seedlings, the low level of RPT5 retention in the 26S proteasome was similar to that of the wild-type seedlings (fraction number 45), although there was a minor shift toward smaller molecular complexes (fraction numbers 51–52). Additionally, MPSR1 overexpression aided RPT5 to stay in the 26S proteasome complex (fraction number 45), confirming the positive role of MPSR1 in 26S proteasome stability (Fig. 4C). We further tested the elution pattern of 20S alpha, a subunit of the 20S catalytic core with a molecular mass of ∼750 kDa. As shown in Fig. 4C, the 20S alpha was barely detected in the 2,000-kDa size range in either wild-type or 35S:MPSR1-KD lines, whereas in the 35S:MPSR1-OE line the 20S alpha eluted in fraction 45. These results suggest that upon AZC treatment, in the absence of MPSR1, the lid part of the 19S RP is highly prone to dissociation from the 26S proteasome. Furthermore, to see the structural integrity of the proteasome by its latent activity, we performed native in-gel analysis. We prepared the total cell extracts from wild-type, 35S:MPSR1-KD, and 35S:MPSR1-OE seedlings treated with AZC for 4 and 12 h. Then, we resolved the samples on a native gel and then incubated the gel with Suc-LLVY-AMC as a substrate. We observed greater levels of proteasome activity in the 35S:MPSR1-OE line than in the wild type and the 35S:MPSR1-KD line under the AZC treatments (Fig. 4 D and E). The proteasome activities in the native gel corresponded to double- and single-capped proteasomes (RP2-CP and RP1-CP), showing that MPSR1 is able to retain the higher-order complexes of the proteasome. Using immunoblot analysis, we found that the levels of the proteasome complexes were consistent to the levels of the proteasomal activity in all of the tested lines (Fig. 4 D and E). These results suggest that MPSR1 activity is important for sustaining the 26S proteasome complex under proteotoxic stress.
Fig. 4.
MPSR1 sustains the integrity of the 26S proteasome complex under proteotoxic stress. (A) In vitro proteasome activity assay using the cytosolic fraction of WT, 35S:MPSR1-OE (OE), 35S:MPSR1-KD (KD), and 35S:MPSR1-DN (DN), and short peptide Suc-LLVY-AMC as substrate. Log2 values of OE/WT, KD/WT, or DN/WT activity ratio are shown on the graph. The data shown are the averages of three independent assays ±SD (*P < 0.05, **P < 0.01 Student’s t test, n = 12). (B) The expression levels of 26S proteasomal subunits in wild-type and two transgenic plants. The 20S core complex was detected with an α-20S alpha antibody. The 19S lid and base complexes were detected with an α-RPN12 antibody and an α-RPT5 antibody, respectively. Equal loading of samples was determined with an α-actin antibody. (C) Size-fractionation analysis of the 26S proteasome using the cytosolic fractions of WT, 35S: MPSR1-OE (OE), and 35S:MPSR1-KD (KD) under nonstress and stress condition. The size-fractionation profile of the 19S lid, the base, and 20S core complex in each plant was determined with an α-RPN12, α-RPT5, and α-20S alpha antibody, respectively. (D and E) Native in-gel analysis of WT, 35S:MPSR1-OE, and 35S:MPSR1-KD to reveal the latent activity of the proteasome complexes. Proteasome activity was visualized using Suc-LLVY-AMC as substrate. The protein levels of proteasome complexes were determined with an α-RPN12 antibody. Equal loading of samples was confirmed with the levels of Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo) using SDS/PAGE. (F) In vivo degradation assay of Δ2GFP. The levels of Δ2GFP in WT/XVE-Δ2GFP and 35S:MPSR1-KD/XVE:Δ2GFP transgenic plants that were treated with β-estradiol (5 µM) for the indicated times are shown. The induced levels of Δ2GFP were monitored with an α-GFP antibody. (G) Deficiency of MPSR1 results in the accumulation of insoluble pUb-catalase in vivo. AZC (5 mM)-treated WT and 35S:MPSR1-KD plantlets were fractionated into soluble extracts and insoluble pellets. The amount of catalase in each fraction was determined with an α-catalase antibody. In all tests, equal loading of samples was determined with an α-actin antibody. Red square bracket indicates unmodified CAT proteins.
Furthermore, using cell-free degradation assay, we demonstrated that the activity of the 26S proteasome was modulated by MPSR1 (SI Appendix, Fig. S10 and SI Appendix). Then, the function of MPSR1 was further determined in vivo by using two transgenic lines: WT/XVE-Δ2GFP and 35S:MPSR1-KD/XVE-Δ2GFP. By adding 5 μM β-estradiol, we monitored the Δ2GFP protein expression levels in both transgenic plant lines. In the wild-type background, there was no significant Δ2GFP accumulation, whereas Δ2GFP was significantly accumulated in the 35S:MPSR1-KD line, indicating that Δ2GFP removal was largely dependent on MPSR1 activity (Fig. 4F). To examine the effect of MPSR1 on other proteins, we compared CAT expression levels in wild-type and 35S:MPSR1-KD lines. Using a cell-fractionation method, we separated the insoluble aggregates from the soluble proteins and found that the levels of aggregated insoluble pUb-CAT in the pellet were significantly increased in 35S:MPSR1-KD after AZC treatment (5 mM) (Fig. 4G). These results suggest that MPSR1 is important to promote the degradation of misfolded proteins under proteotoxic stresses.
MPSR1 Directly Interacts with the 26S Proteasome in the Cytoplasm.
Next, we investigated how MPSR1 maintains the integrity of the 26S proteasome complex under proteotoxic stress. Many molecular factors, including chaperones, proteases, phosphatases, metabolites, and some PQC E3 ligases, tune the integrity and/or activity of the 26S proteasome via direct interactions (38, 40–43). For instance, in yeast, Not4 contributes to proteasome assembly and functional integrity at least partially through Ecm29, a proteasome-interacting chaperone (44). Hul5 directly modulates proteasome processivity (15). Thus, we visualized MPSR1–proteasome complex formation by immunohistochemistry. In the mock-treated seedlings, green fluorescence from MPSR1 was barely detected, whereas red fluorescence speckles from RPN12 were clearly seen (Fig. 5A, Upper). After 6 h of treatment with AZC, the ratio of green fluorescent speckles from MPSR1 dramatically increased and more importantly, the green speckles that colocalized with red speckles of RPN12 were mainly found around the nucleus (Pearson correlation coefficient: 0.47 ± 0.11) (Fig. 5A, Middle and Lower). Using an anti-RPT5 antibody, we also observed circumferentially colocalized speckles of MPSR1 and RPT5, a base subunit of the 19S complex, near the nucleus (Pearson correlation coefficient: 0.46 ± 0.07) (Fig. 5B, Middle and Lower). In combination with the results showing the subcellular localization of misfolded CAT and MPSR1 around the nucleus (Fig. 2H), these results strongly suggest the possible localization of MPSR1, misfolded proteins, and the 26S proteasome within distinctive speckles to the periphery of the nucleus. Next, we confirmed the association of MPSR1 with the 26S proteasome by coimmunoprecipitation. As shown in Fig. 5C, MPSR1 apparently forms a complex with the RP subunits of the 26S proteasome in vivo. These results clearly indicate that MPSR1 associates with the 26S proteasome.
Fig. 5.
Misfolded protein-mediated association between MPSR1 and the 26S proteasome. (A) Subcellular colocalization of MPSR1 and the 26S proteasome in 35S:MPSR1-OE seedlings determined by immunohistochemistry using α-flag for MPSR1 and α-RPN12 for the 26S proteasome. After AZC treatment, green fluorescence from MPSR1 and red fluorescence from RPN12 colocalized as speckles around the nucleus. The nucleus was detected with DAPI staining. (B) Subcellular colocalization of MPSR1 and RPT5 in 35S:MPSR1-OE seedlings determined by immunohistochemistry using α-flag for MPSR1 and α-RPT5 for the 26S proteasome. Under the AZC treatment only, green fluorescence from MPSR1 and red fluorescence from RPT5 granulated as speckles. White squares show the zoomed-in area. White dotted circles indicate the nucleus. The nucleus was detected with DAPI staining. (C) In vivo coimmunoprecipitation analysis of flag-MPSR1 and the 26S proteasome using 35S:MPSR1-OE and wild-type seedlings treated with AZC (5 mM). flag-MPSR1 and associated proteins were precipitated with an α-flag antibody and the presence of 26S proteasome was determined with α-RPN12, α-RPT5, and α-20S alpha antibodies. (D) In vitro pull-down assay of GST-MPSR1 and purified 26S proteasome complex. GST-MPSR1 was used as the bait and the 26S proteasome as the prey in the presence or absence of His-Δ2GFP. Retrieved proteins were determined with an α-RPT1 antibody. (E) In vitro pull-down assay of GST-MPSR1 and purified 26S proteasome complex. GST-MPSR1 was used as the bait and the 26S proteasome as the prey in the presence or absence of His-GFP. Retrieved proteins were determined with an α-RPT1 antibody. (F) In vitro pulldown assay of GST-MPSR1 and His-RPT1-3HA. GST-MPSR1 was used as the bait and His-RPT1-3HA as the prey in the presence or absence of His-Δ2GFP. Retrieved proteins were determined with an α-HA antibody.
A recent study reported that several yeast E3 ligases are able to interact directly with mammalian 26S proteasome subunits upon proteasome inhibition. We found one of them, Rnf181 RING E3 ligase, which seemingly binds to RPT1, a base subunit of RP, and is partly homologous to MPSR1 (45). Rnf181 has two disordered segments at the both N- and C- terminal regions, which are homologous to those of MPSR1 around ∼33%. Based on this observation, we tested the existence of direct interactions between MPSR1 and the 26S proteasome by an in vitro pulldown assay using GST-MPSR1 as the bait and purified human 26S proteasome as the prey. Contrary to our expectations, GST-MPSR1 was unable to recognize any subunits of the 26S proteasome in vitro. To further investigate the inconsistency between the in vivo and in vitro results, we asked whether misfolded proteins are integral to the interaction. Indeed, the interaction between MPSR1 and RPT1 was induced by the addition of Δ2GFP (Fig. 5D). On the contrary, addition of GFP did not induce the association of MPSR1 with RPT1 (Fig. 5E). Using recombinant AtRPT1, the RPT1 ortholog in Arabidopsis, we confirmed the misfolded protein-mediated interaction between MPSR1 and RPT1 (Fig. 5F). Taken together, our results demonstrate that under proteotoxic stress, MPSR1 specifically interacts with RPT1, mainly through misfolded proteins, and that such interaction contributes to the stability and optimal activity of the 26S proteasome complex. Furthermore, we recently reported that salinity-responsive E3 ligase Arabidopsis ABA-insensitive RING protein 2 (AIRP2) forms a large and single aggresome, or a JUNQ-like subcellular compartment around the nucleus, when it is expressed in tobacco leaf epidermal cells (46). Although not identical to the aggresome, we found that the presence of perinuclear speckles in Arabidopsis, containing the 26S proteasome, misfolded proteins, and MPSR1 for cytoplasmic PQC pathway. To confirm the spatial specificity of MPSR1 in the cytoplasm, we further tested whether MPSR1 is involved in ER-associated protein degradation (ERAD) pathway. Our results showed that the role of MPSR1 is not directly related to ERAD pathway (SI Appendix, Fig. S11 and SI Appendix).
Discussion
Despite the importance of E3 ligases in the cytoplasmic PQC pathway, only AtCHIP E3 ligase has been identified in plants (13, 30, 36). We have therefore focused on discovering novel cytoplasmic PQC E3 ligases in plants. We identified the MPSR1 RING E3 ligase that is essential for the elimination of misfolded proteins in the cytoplasm. MPSR1 has many unique features of a PQC E3 ligase. First, MPSR1 senses misfolded proteins through a client-induced stabilization mechanism. Under no stress, MPSR1 is prone to rapid degradation by the 26S proteasome, concealing its PQC E3 ligase activity. However, upon stress that leads to protein misfolding, MPSR1 escapes self-degradation and instead ubiquitinates misfolded proteins. An advantage of such self-degradation may provide a tunable-oscillatory mechanism to efficiently sense the intensity of protein-misfolding stress. Second, independent of chaperones, MPSR1 efficiently distinguishes misfolded proteins from normal proteins by its disordered segments, as can be similarly observed with San1, a chaperone-independent PQC E3 ligase in yeast that can bind a wide range of misfolded proteins by its multiple disordered segments (47). MPSR1 consists of a RING-type E3 ligase domain and six disordered segments at both N- and C-termini, and their predicted overall disorder rate is over 44% (SI Appendix, Fig. S7). Indeed, we found that the intrinsically disordered segments of MPSR1 could behave similarly as the disordered segments of San1 in maintaining target specificity (Fig. 2E). Third, MPSR1 is an identified cytoplasmic PQC E3 ligase in plants that retains the integrity of the 26S proteasome under proteotoxic stress. Compared with many other cytoplasmic PQC E3 ligases in eukaryotes, MPSR1 not only fosters the properties of Hul5—the chaperone-independent target recognition and the modulation of the 26S proteasome activity—but it also has its unique properties, which are yet to be found in yeast and mammalian PQC E3 ligases. The properties are (i) the client-induced association of MPSR1 with the 26S proteasome, (ii) the formation of speckles with the clients and the 26S proteasome around the nucleus, and (iii) the self-regulatory sensor mechanism. In conclusion, we suggest that MPSR1 is essential to remove emergent misfolded proteins by sensing and tethering pUbs, and by sustaining the activity of 26S proteasome in plants. Hence, in the absence of MRSR1, plants lose their capability to adequately remove damaged proteins, leading to the premature malfunction of the 26S proteasome throughout their growth and development under proteotoxic stress.
Materials and Methods
Plant Materials.
Arabidospis thaliana wild-type (Col-0) was obtained from Arabidopsis Resource Center. 35S:MPSR1-OE, 35S:MPSR1-KD, 35S:MPSR1-DN, and mpsr1-cas transgenic plants were generated via agrobacterium-mediated transformation method, which were described in SI Appendix, SI Materials and Methods.
Plant Growth and Phenotypic Analysis Conditions.
Plants were grown under a long-day photoperiod (16 h light/8 h dark, 100 μmol·m−2·s−1) for the phenotype assay and the generation of wild-type and transgenic plants. Seeds of wild-type, 35S:MPSR1-OE, 35S:MPSR1-DN, and 35S:MPSR1-KD transgenic plants were sterilized by washing with bleach solution (30%) and sown on MS medium with or without Azetidine-2-carboxylic acid (Santa Cruz Biotechnology), tunicamycin (Sigma-Aldrich), and sodium arsenite (Sigma-Aldrich) for 2 wk. Times and concentrations are specified in each figure. To determine the accumulations of soluble, insoluble pUb proteins in wild-type and transgenic plants, seedlings were grown on MS medium with AZC (100 μM) for 2 wk.
DNA Constructions, Recombinant Protein Expression and Purifications, Protein–Protein Interaction Assays, Immunoblot, and qRT-PCR.
Plasmid constructions, recombinant protein purification, assays for protein–protein interaction including yeast two-hybrid, co-IP, in vitro pulldown, qRT-PCR, and immunoblot analyses are described in SI Appendix, SI Materials and Methods.
RNA Sequencing Analysis.
Construction of RNA libraries, sequencing, and bioinformatics analyses were performed by LA Science (www.lascience.co.kr/).
Fractionation of Detergent-Soluble and Insoluble pUb-Proteins.
For separation of soluble pUb-proteins from insoluble pUb-protein fractions, we followed the fractionation method used in a previous study with minor modifications (36). Detailed methods are described in SI Appendix, SI Materials and Methods.
Immunohistochemistry and Confocal Microscopy.
The whole-mount immunostaining method was used as described in Sauer et al. (48) with minor modifications. The assay conditions and detailed protocols are described in SI Appendix, SI Materials and Methods.
Proteasome Activity and Complex Analysis.
In vitro proteasome activity analysis, gel filtration assay, and proteasome analysis using native gel electrophoresis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Research Foundation Projects 2014R1A2A2A01003891 and NRF-2017R1A2B2006750, a Republic of Korea grant (to W.T.K.), and by Radiation Technology R&D Program Project NRF-2015M2A2B2066120 through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (S.W.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713574114/-/DCSupplemental.
References
- 1.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: Folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 3.McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: Chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- 4.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 5.Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 6.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 7.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang NN, et al. Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat Cell Biol. 2014;16:1227–1237. doi: 10.1038/ncb3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummings CJ, et al. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 10.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J Mol Med (Berl) 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 12.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan J, et al. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 2003;132:861–869. doi: 10.1104/pp.103.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol. 2010;30:985–994. doi: 10.1128/MCB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelley PM, Schlesinger MJ. The effect of amino acid analogues and heat shock on gene expression in chicken embryo fibroblasts. Cell. 1978;15:1277–1286. doi: 10.1016/0092-8674(78)90053-3. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg AL. Correlation between rates of degradation of bacterial proteins in vivo and their sensitivity to proteases. Proc Natl Acad Sci USA. 1972;69:2640–2644. doi: 10.1073/pnas.69.9.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 19.Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- 20.Genschik P, et al. Ubiquitin genes are differentially regulated in protoplast-derived cultures of Nicotiana sylvestris and in response to various stresses. Plant Mol Biol. 1992;20:897–910. doi: 10.1007/BF00027161. [DOI] [PubMed] [Google Scholar]
- 21.Sun CW, Callis J. Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J. 1997;11:1017–1027. doi: 10.1046/j.1365-313x.1997.11051017.x. [DOI] [PubMed] [Google Scholar]
- 22.Garbarino JE, Rockhold DR, Belknap WR. Expression of stress-responsive ubiquitin genes in potato tubers. Plant Mol Biol. 1992;20:235–244. doi: 10.1007/BF00014491. [DOI] [PubMed] [Google Scholar]
- 23.Guo Q, et al. Drought tolerance through overexpression of monoubiquitin in transgenic tobacco. J Plant Physiol. 2008;165:1745–1755. doi: 10.1016/j.jplph.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liew CW, Sun H, Hunter T, Day CL. RING domain dimerization is essential for RNF4 function. Biochem J. 2010;431:23–29. doi: 10.1042/BJ20100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plechanovová A, et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol. 2011;18:1052–1059. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas-Fernandez A, et al. SUMO chain-induced dimerization activates RNF4. Mol Cell. 2014;53:880–892. doi: 10.1016/j.molcel.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho SK, Ben Chaabane S, Shah P, Poulsen CP, Yang SW. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat Commun. 2014;5:5867. doi: 10.1038/ncomms6867. [DOI] [PubMed] [Google Scholar]
- 29.Ballinger CA, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, et al. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell. 2009;21:3984–4001. doi: 10.1105/tpc.109.071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobson T, et al. Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J Cell Sci. 2012;125:5073–5083. doi: 10.1242/jcs.107029. [DOI] [PubMed] [Google Scholar]
- 32.Ibstedt S, Sideri TC, Grant CM, Tamás MJ. Global analysis of protein aggregation in yeast during physiological conditions and arsenite stress. Biol Open. 2014;3:913–923. doi: 10.1242/bio.20148938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan X, et al. Trivalent arsenic inhibits the functions of chaperonin complex. Genetics. 2010;186:725–734. doi: 10.1534/genetics.110.117655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JG, Takahama S, Zhang G, Tomarev SI, Ye Y. Unconventional secretion of misfolded proteins promotes adaptation to proteasome dysfunction in mammalian cells. Nat Cell Biol. 2016;18:765–776. doi: 10.1038/ncb3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10:e1004116. doi: 10.1371/journal.pgen.1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bie P, Ciechanover A. Ubiquitination of E3 ligases: Self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011;18:1393–1402. doi: 10.1038/cdd.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonough H, Patterson C. CHIP: A link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kikuchi J, et al. Co- and post-translational modifications of the 26S proteasome in yeast. Proteomics. 2010;10:2769–2779. doi: 10.1002/pmic.200900283. [DOI] [PubMed] [Google Scholar]
- 41.Hanssum A, et al. An inducible chaperone adapts proteasome assembly to stress. Mol Cell. 2014;55:566–577. doi: 10.1016/j.molcel.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X, et al. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol. 2016;18:202–212. doi: 10.1038/ncb3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirano H, Kimura Y, Kimura A. Biological significance of co- and post-translational modifications of the yeast 26S proteasome. J Proteomics. 2016;134:37–46. doi: 10.1016/j.jprot.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Panasenko OO, Collart MA. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol Cell Biol. 2011;31:1610–1623. doi: 10.1128/MCB.01210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besche HC, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh TR, et al. AtAIRP2 E3 ligase affects ABA and high-salinity responses by stimulating its ATP1/SDIRIP1 substrate turnover. Plant Physiol. 2017;174:2515–2531. doi: 10.1104/pp.17.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenbaum JC, et al. Disorder targets misorder in nuclear quality control degradation: A disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer M, Paciorek T, Benková E, Friml J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc. 2006;1:98–103. doi: 10.1038/nprot.2006.15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.