Significance
Worldwide, potato is the third most important crop grown for direct human consumption, but breeders have struggled to produce new varieties that outperform those released over a century ago, as evidenced by the most widely grown North American cultivar (Russet Burbank) released in 1876. Despite its importance, potato genetic diversity at the whole-genome level remains largely unexplored. Analysis of cultivated potato and its wild relatives using modern genomics approaches can provide insight into the genomic diversity of extant germplasm, reveal historic introgressions and hybridization events, and identify genes targeted during domestication that control variance for agricultural traits, all critical information to address food security in 21st century agriculture.
Keywords: potato, diversity, domestication, introgression, adaptation
Abstract
Cultivated potatoes (Solanum tuberosum L.), domesticated from wild Solanum species native to the Andes of southern Peru, possess a diverse gene pool representing more than 100 tuber-bearing relatives (Solanum section Petota). A diversity panel of wild species, landraces, and cultivars was sequenced to assess genetic variation within tuber-bearing Solanum and the impact of domestication on genome diversity and identify key loci selected for cultivation in North and South America. Sequence diversity of diploid and tetraploid S. tuberosum exceeded any crop resequencing study to date, in part due to expanded wild introgressions following polyploidy that captured alleles outside of their geographic origin. We identified 2,622 genes as under selection, with only 14–16% shared by North American and Andean cultivars, showing that a limited gene set drove early improvement of cultivated potato, while adaptation of upland (S. tuberosum group Andigena) and lowland (S. tuberosum groups Chilotanum and Tuberosum) populations targeted distinct loci. Signatures of selection were uncovered in genes controlling carbohydrate metabolism, glycoalkaloid biosynthesis, the shikimate pathway, the cell cycle, and circadian rhythm. Reduced sexual fertility that accompanied the shift to asexual reproduction in cultivars was reflected by signatures of selection in genes regulating pollen development/gametogenesis. Exploration of haplotype diversity at potato’s maturity locus (StCDF1) revealed introgression of truncated alleles from wild species, particularly S. microdontum in long-day–adapted cultivars. This study uncovers a historic role of wild Solanum species in the diversification of long-day–adapted tetraploid potatoes, showing that extant natural populations represent an essential source of untapped adaptive potential.
Cultivated potato (Solanum tuberosum L.) was domesticated 8,000–10,000 y ago from wild species (2n = 2x = 24) native to the Andes of southern Peru (Fig. 1 and Fig. S1) (1), becoming a pillar of food security and cultural heritage for ancient societies populating the highlands of Peru, Bolivia, and Ecuador (2, 3). Potatoes were later cultivated under highland equatorial conditions (modern Colombia and Venezuela) and longer summer days in the southern latitudes of Argentina and Chile (4–6), exhibiting an adaptive potential to fulfill regional dietary requirements. Autopolyploidization of early landrace diploids (S. tuberosum groups Stenotomum and Phureja), likely occurring repeatedly due to the common occurrence of 2n gametes in diploid species (7), produced Andean cultivated tetraploids (S. tuberosum group Andigena; 2n = 4x = 48) (Fig. 1 and Fig. S1). Migration from the Andes to coastal Chile yielded a long-day–adapted subspecific group (S. tuberosum group Chilotanum, 2n = 4x = 48) distinct from its upland progenitors (Fig. 1 and Fig. S1) (1, 8), which later contributed much of the genetic background in S. tuberosum commercial cultivars throughout the world. From these ancient origins, potato has been widely adopted into the global diet and is the third most important food crop for direct human consumption (faostat3.fao.org), providing food security in Asia and South America (9, 10).
Fig. 1.
(A) Phenotypic diversity within wild species, cultivated landraces, and cultivars through domestication, improvement, and modern breeding efforts. Exemplar species, landraces, and elite North American cultivars are shown that highlight tuber size, shape, and pigmentation diversity. (B) Phylogeny and population structure of the samples in the domestication panel. The phylogenetic tree is based Nei’s genetic distances calculated from 687,172 fourfold-degenerate sites from conserved potato genes. Population structure is based on 50,000 genome-wide SNPs. The optimal number of subpopulations (K = 5) included wild outgroups (purple), wild Solanum relatives (green), a wild subgroup diverging from the cultivated lineage after most other species (gold), Andean landraces (teal), and S. tuberosum group Tuberosum (navy).
The adaptability of potato to diverse growing conditions stems from a large germplasm base encompassing distinct cultivated groups and over 100 tuber-bearing relatives (Solanum section Petota) (11), with distribution ranging from the southwestern United States to southern Chile (12–14). Potato was domesticated in the Andean highlands (15), an arid region 3,000–4,500 m above sea level characterized by cold temperatures, saline soils, and high solar radiation. Rather than an aboveground reproductive structure, the organ of selection was an underground tuber highly effective at nutrient storage; potato ranks second only to soybean in protein produced per acre among the major crops (16) and ranks first in both energy (5,600 kcal/m3) and protein production (150 g/m3) per unit water (17); a single potato provides 50% of the recommended daily allowance of vitamin C, compared with 0% for rice and wheat, 21% of potassium, 12% of fiber, and balanced protein (18).
The historic importance and versatility of potato for providing nutrition in diverse environments make its continued improvement a priority to meet global food demands. In an era of genomics-enabled breeding, evaluating the genetic diversity of tuber-bearing Solanum species and the impacts of human selection is critical for effective germplasm utilization (19). Potato’s domestication altered the regulation of carbohydrate biosynthesis and transport (20, 21) and antinutritional glycoalkaloid content (22, 23). Locating targets of selection in critical pathways informs breeding strategies to control variance of key agricultural traits. This study highlights the genome diversity of potato and its progenitors, reporting the greatest levels of genetic variation observed in any crop diversity study to date. We present evidence for a role of wild Solanum species in the evolution of a long-day–adapted S. tuberosum subspecific group and report signatures of selection in genes controlling both established domestication traits, such as glycoalkaloid biosynthesis and carbohydrate metabolism, and comparatively unexplored potato traits such as circadian rhythm, the shikimate pathway, cell cycling/endoreduplication, and sexual fertility.
Results and Discussion
Genome Variation and Conservation in Solanum Section Petota.
A panel of 67 genotypes was used to capture a broad extent of genome variation in cultivated potato and its progenitors, including 20 wild diploid species, 20 South American landraces (groups Andigena, Phureja, Stenotomum, and Chilotanum) representing locally adapted primitive selections, 23 North American cultivars (group Tuberosum) from modern (post-1850) breeding programs, and four outgroups (Fig. 1, Fig. S2, and Dataset S1). Wild accessions represented South American species sexually compatible with S. tuberosum, excluding taxa of hybrid origin. Landraces included 10 diploids (primitive groups Phureja and Stenotomum) and 10 autotetraploids (eight in group Andigena and two in group Chilotanum) with habitats ranging from equatorial Colombia to southern Chile. Cultivars represented North American varieties (group Tuberosum) derived from Chilean landraces (group Chilotanum) and released over the last 150 y. Although some taxonomic treatments collapse Andean S. tuberosum groups Andigena (4x), Phureja (2x), and Stenotomum (2x) (24, 25), we invoke the penultimate nomenclature (26) that distinguishes ploidy, referring to these as “Andean landraces” (groups Andigena, Phureja, and Stenotomum) for comparison with cultivars (group Tuberosum) and their Chilean landrace progenitors (group Chilotanum).
Sequence and structural variants in the form of SNPs and copy number variation (CNV) were identified by aligning sequences to the S. tuberosum group Phureja DM v4.04 reference genome (27). Genome conservation was ascertained from CNV, using a maximum 10% deletion threshold in the respective wild, landrace, and cultivar groups to identify 390 Mb of the cultivated genome (53.5%) containing 63% of genes as conserved among sexually compatible potato species. High rates of CNV within individuals and across the panel (Fig. 2A and Dataset S2) expanded previous reports of a structurally heterogeneous genome landscape containing many dispensable loci (Fig. 2B and Fig. S3) (27). Gene-level CNV frequency ranged from 10.9–39.8% (Fig. 2 A and B), averaging 12.8% in diploid landraces and 26.7% in wild species. Within S. tuberosum (all cultivated) genotypes 41.7% of genes were affected by deletion (21.9% homozygous), compared with 53.6% in all nonoutgroup genotypes (35.1% homozygous), indicating a core genome containing 46–65% of genes in the current annotation for cultivars and their wild progenitors. Sequence variants included 68.9 million SNPs, of which 44 million were located within the conserved genome (Table S1). Nucleotide divergence from the reference genome sequence averaged 1.8% in diploid landraces, 2.2% in tetraploid landraces, 2.4% in cultivars, 3.4% among wild (nonoutgroup) species, and 4.2% in reproductively isolated Mexican relatives, reaching 4.5% in the nontuberizing outgroup Solanum etuberosum.
Fig. 2.
Genome diversity in tuber-bearing Solanum species. (A) Fraction of annotated potato genes affected by duplication (blue), heterozygous deletion (light red), or homozygous deletion (red) across landraces, cultivars, tuber-bearing wild-species relatives, and outgroups (OG). (B) Heatmap of gene conservation across tuber-bearing Solanum genotypes on chromosome 9. Genes are ordered along the chromosome y axis, and the x axis contains samples organized into panels (separated by thin black lines) containing (Left to Right) 20 landraces, 23 cultivars, 20 tuber-bearing wild species relatives, and four outgroups. (C) Comparison of nucleotide diversity (π) in domesticated germplasm (cultivars diploid landraces, and tetraploid landraces) and wild relatives for major crop species. Color-coding is shown in the key. Species diversity estimates were obtained from existing resequencing studies: cucumber (35), tomato (35), watermelon (31), rice (32), maize (33), and soybean (34). (D) Heterozygous nucleotide frequency within potato outgroups, wild species relatives, diploid landraces, tetraploid landraces, and cultivars.
Cultivated Potato Genome Diversity.
Pedigrees show North American potato varieties are derived from a restricted base of 19th century Chilean and European founders that survived the 1840s Irish potato famine caused by Phytophthora infestans (28–30). To quantify potato’s genetic bottleneck, we estimated the population diversity (π) of wild species, landraces, and cultivars using SNPs from the conserved genome. Cultivated potato harbored striking levels of diversity: πC = 0.0105 (cultivars), πL = 0.0097 (landraces), and πC+L = 0.0111 (all S. tuberosum), exceeding estimates from previous crop-resequencing studies (Fig. 2C) (31–35) and overturning historic assumptions of low founder diversity. Despite having lower heterozygosity than their tetraploid counterparts (Fig. 2D), diploid potato landraces (πl-2x = 0.0087) exhibited greater diversity than maize landraces (33), upholding theories that abundant genome diversity predated polyploidization (27). The similar genome diversities of the combined S. tuberosum lineages and wild Solanum [πW/πC+L = 1.101 (1.188 coding sequence, CDS)] is remarkable, given that the former represent geographic subgroups of a single cultivated species compared with the 20 distinct species representing wild diversity. Potato autotetraploids, containing four homologous chromosomes that undergo tetravalent pairing (36), more than doubled the heterozygosity of diploid landraces (Fig. 2D) with mean heterozygous nucleotide frequency increasing from 1.05% in diploids to 2.73% in tetraploid cultivars (maximum 3.04%). Extensive allele sharing was confirmed in CDS of wild species, landraces, and North American cultivars, with 73% of wild alleles from 20 species extant in cultivated varieties (53% including rare alleles) (Fig. S4A), suggesting that hybridization with wild populations could have contributed to the heterozygosity and diversification of the cultivated lineage.
Population Analysis.
Phylogenetic analysis of 687,172 fourfold-degenerate sites in conserved genes and analysis of population structure (Fig. 1B and Fig. S5) subdivided the panel into three primary populations: wild species, Andean landraces (groups Phureja, Stenotomum, and Andigena), and long-day–maturing genotypes including group Chilotanum landraces and cultivar descendants. Three Peruvian species (S. medians, S. megistacrolobum, and S. raphanifolium) demonstrated substructure within the wild group, being the wild species closest to the cultivated lineage, while outgroup accessions formed a basal wild subgroup. Crop-species admixture was asymmetric, with few examples of cultivated alleles in wild species and frequent evidence of wild introgression into tetraploid landraces and cultivars (Fig. 1B and Fig. S5). The phylogeny resolved tetraploid group Andigena, Chilotanum, and Tuberosum as more basal to the cultivated lineage than diploids (Fig. 1B and Fig. S5), a surprising result given that potatoes were domesticated as diploids, and extant diploids are more primitive (37). Genetic distances between cultivated tetraploids and wild species were indeed lower than their diploid progenitors (Fig. S4B) due to unequal wild introgression; nine landrace diploids had no evidence of admixture, and the tenth (PI195204) was a misidentified wild-crop hybrid, while 9 of 10 landrace tetraploids and numerous cultivars harbored wild introgression. High rates of wild allele sharing and heterozygosity in tetraploids, combined with evidence of greater admixture, suggest that autopolyploidy in the cultivated potato lineage altered the dynamics of wild species interactions, resulting in the diversification of the cultivated gene pool by the influx of novel wild alleles.
Wild Solanum Introgression in the Cultivated Lineage.
High rates of unreduced (2n) gametes in diploid potatoes reduce barriers to interploidy gene flow, helping explain the frequency of wild alleles within autotetraploid S. tuberosum (7). We evaluated sequence introgression in landraces and cultivars to explore genetic contributions from wild Solanum populations. Genomic regions derived from wild species were identified by comparing local genetic distances (5-kb windows) against both species and the cultivated ancestral genotype derived from a consensus of nonadmixed diploid landraces. This revealed large increases of wild introgression in tetraploid Andigena landraces from diploids and higher rates from distinct species in group Tuberosum (Fig. 3A); average fractions of genomic windows predicted to contain wild alleles were 2.0% in diploid landraces, 20.2% in tetraploid group Andigena, and 31.2% in group Tuberosum.
Fig. 3.
Wild Solanum species introgressions in cultivated potato. (A) Fraction of assessed genome sequences (5-kb windows) with introgressions from individual wild species in diploid landraces, tetraploid landraces, and cultivars. (B) Map of wild species introgressions on potato chromosome 11 for diploid landraces, tetraploid landraces, and cultivars. Color codes for species introgressions are blue, ambiguous/multiple taxa; red, S. microdontum; green, S. candolleanum; orange, Solanum sparsipilum; purple, S. leptophyes; pink, S. raphanifolium; gold, S. brevicaule; brown, S. medians; navy, S. chacoense; dark red, S. berthaultii; and light green, Solanum infundibuliforme. All 12 chromosomes, including names of accessions, can be seen in Fig. S6.
All cultivated groups harbored a significant contribution from the domestication progenitor Solanum candolleanum (Fig. 3 and Fig. S6A) (1), showing crop-species hybridization persisted between cultivated potatoes and local progenitors after domestication and polyploidy. Diploid landrace introgression was mainly limited to S. candolleanum, Solanum medians, and Solanum raphanifolium, species native to Peru. Tetraploid introgressions broadened to include species native to Bolivia (Solanum brevicaule, Solanum leptophyes, and Solanum microdontum) and Argentina (Solanum berthaultii, Solanum chacoense, Solanum gourlayi, Solanum kurtzianum, S. microdontum, Solanum spegazzinii, and Solanum vernei) (Fig. 3A). Except for the Andean progenitor S. candolleanum, introgression was consistently stronger in group Tuberosum than in group Andigenum (Fig. 3A). Critically, no species interaction was unique to Andean landraces, while Chilean-derived group Tuberosum housed all Andean interactions and novel introgressions from S. gourlayi, S. kurtzianum, S. microdontum, and S. spegazzinii, species found only south of the Andean domestication origin. S. microdontum, unrepresented in cultivar pedigrees (29), showed large historic contributions to group Tuberosum: 10 chromosomes (1–8, 11, 12) contained introgressions in a majority of cultivars with residual sequences in heterochromatin around centromeres on chromosomes 7, 11, and 12 (Fig. 3B and Dataset S3).
Eliminating the confounding factor of wild hybridization, we reconstructed a nuclear phylogeny excluding variants in regions of introgression (Fig. S6B), unambiguously resolving group Tuberosum and group Chilotanum as derived from group Andigena. These relationships, with evidence of Chilean-derived cultivar introgressions from the same Peruvian and Bolivian species as Andean landraces, advance theories of a single Peruvian domestication. A likely scenario based on Bolivian and Argentinian species alleles in Chilean-derived Tuberosum is that Andigena tetraploids interbred with wild species en route to their eventual destination in southern Chile where long-day adaptation was required for tuberization (Fig. S1).
Analysis of gene functions in wild-introgressed regions supports an adaptive role of crop-species hybridization in tetraploids. The 1,415 genes within introgressions conserved in 70% of tetraploids, showing preferential retention of wild alleles, were highly enriched (fivefold expression increase; P < 0.0001) among potato genes induced by abiotic stress, with 21.3% of wild-donated genes but only 1.5% of nonassociated genes being stress-responsive (Dataset S4). Genes associated with wild species introgressions were also more likely to be highly expressed [fragments per kilobase of transcript per million mapped reads (FPKM) >10] than genes from nonintrogressed regions (P < 0.0001), supporting an impact on cultivated phenotypes. The 407 genes within regions retaining 80% wild sequence contained many loci functioning in disease resistance, heat and drought tolerance, and antioxidant pathways (Dataset S5). These data offer one possible model by which wild Solanum species assisted the spread of cultivated potatoes by transmitting alleles for tolerance of new ecological factors, enabling colonization of nonnative habitats as they migrated south following domestication and polyploidy.
Potato Day-Length Adaptation.
A key event in the history of the cultivated potato lineage was adaptation of long-day–maturing tetraploids able to tuberize under 16-h days in southern Chile, and later in Europe and North America, enabling global cultivation at nonequatorial latitudes. Kloosterman et al. (38) demonstrated that naturally occurring alleles of StCDF1 (positive regulator of tuberization via CONSTANS) harbor transposable element (TE)-induced structural variants resulting in truncated StCDF1 proteins with dominant-allele effects leading to circadian deregulation and, as a consequence, the long-day–maturity phenotype. We resolved StCDF1 haplotypes for 58 samples in the panel along with cultivated tomato (cv. Heinz 1706) to predict alleles conferring long-day tuberization in cultivars, identifying 55 haplotypes encoding 27 peptide variants (Dataset S6). Four haplotype groups contained conserved deletions affecting the structure of the StCDF1 peptide encoded by the short-day reference haplotype (H1) (Figs. S7 and S8A); some encode StCDF1 proteins lacking a C-terminal domain, potentially resulting in higher protein stability. Phylogenetic analysis of StCDF1 haplotypes revealed that nearly all long-day clones (23/25) contained alleles encoding shortened StCDF1 proteins (H35, H45) derived from either S. microdontum or a second wild species (Fig. S7). A single Andigena genotype (PI214421) contained a putative “long-day allele,” while PI258885 was confirmed phylogenetically as Chilotanum. Two Tuberosum cultivars (Missaukee and Yukon Gold) lacked putative long-day alleles based on CDS variation; however, the DM v4.04 annotation contains an alternate StCDF1 transcript isoform (PGSC0003DMT400047371) whose exon boundary (chr5:4,539,678) is flanked by a G→T SNP (left) and a single base-pair deletion (right) impacting adjacent splice motifs in both clone haplotypes (Fig. S8B). Two of three StCDF1 proteins identified by Kloosterman et al. were identified in our panel: StCDF1.1 (short-day allele) was identical to PH1, the predominant short-day protein encoded in the DM reference and most Andigena landraces, while StCDF1.2 (long-day allele) was structurally identical to PH15, PH16, and PH20 and shared 99.75% amino acid identity with PH20 (Dataset S6). StCDF1.3 was not identified. Chilotanum and Tuberosum’s possession of wild alleles impacting the protein-coding structure of StCDF1, potato’s maturity locus and the key quantitative trait locus (QTL) for yield in the northern hemisphere, shows that wild Solanum introgression (particularly from S. microdontum) not only shaped tetraploid potato diversification but may have introduced the adaptive variants enabling S. tuberosum cultivation in Europe and North America. Characterizing a broader set of StCDF1 alleles is required as reports of this locus as a major yield QTL in Tuberosum populations lacking the TE-induced variant (StCDF1.2) support a functional impact of the wild alleles. Discovery of alleles impacting StCDF1 protein structure beyond those previously reported shows that disruption of the predominant equatorial short-day haplotype has occurred repeatedly by TE-induced (StCDF1.2) and non–TE-induced mutations and was introduced into Andean populations from multiple sources following domestication, which may have enabled tuberization outside their earliest geographic range.
Genes Under Selection in Cultivated Potato.
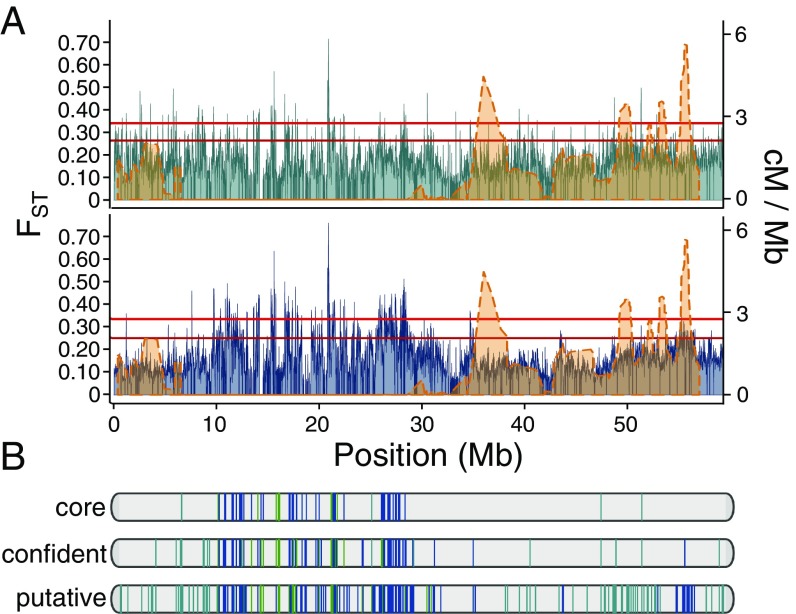
Allele frequencies were used to scan the potato genome for gene-selection signatures based on population metrics comparing North American cultivars against wild species (cultivar selected) and South American Andean landraces against wild species (landrace selected). Selected genes were ranked under three stringency cutoffs: putative, confident, and core, based on scoring in the top 5% (putative), 2% (confident), or 1% (core) values for FST estimates in adjacent genomic windows (20 kb) and at least one per-locus selection metric (Tajima’s D, relative nucleotide diversity, maximum single-variant allele frequency difference) within the gene. This approach identified 2,622 (6.7%) putatively selected genes in cultivated potato, 841 (2.1%) confidently selected genes, and 315 genes (0.8%) under core selection (Fig. 4, Fig. S9, and Dataset S7). Domestication candidates were regarded as genes under selection in both landraces and cultivars, impacting performance regardless of hemisphere or local adaptation. These accounted for only 14.4–16.3% of selected loci across all confidence thresholds, evidence that highland Andigena and lowland Tuberosum populations achieved agricultural performance by distinct selection strategies, relying more on regional adaptation than on conserved developmental processes. Signatures of selection between groups Tuberosum and Andigena (Dataset S7) were comparatively weak, lacking strong evidence of loci approaching fixation of distinct alleles.
Fig. 4.
Genome-wide selection signatures on potato chromosome 6. (A) Chromosome-wide FST estimates (left axis) from 20-kb sliding window analysis (5-kb step size) comparing Andean landraces (teal) and S. tuberosum group Tuberosum (navy) to wild Solanum; plotted with chromosomal recombination rate in centimorgan/Mb (orange; right axis) based on a diploid biparental population (111). Red lines indicate top 1% and 5% cutoffs for FST window estimates. (B) Locations of genes under putative, confident, and core selective pressure in Andean landraces (teal), S. tuberosum group Tuberosum (navy), or both groups (domestication candidates; green).
A range of molecular functions, including regulation of gene expression, was enriched in genes under selection (Dataset S7). Tomato domestication involved modification of transcriptional networks (39), and transcription factors were enriched in potato genes with signatures of selection (152 genes, χ2 test, P ≤ 0.005). Potato domestication gave rise to enlarged tubers with a concomitant increase in leaf carbon fixation and transport, tuber-specific reduction of harmful glycoalkaloids, adaptation to a long-day photoperiod, and reduced sexual fertility. Signatures of selection associated with key structural and regulatory genes involved in the cell cycle, circadian rhythm, carbohydrate metabolism, endoreduplication, glycoalkaloid biosynthesis, shikimate biosynthesis, and pollen development provide insights into the biological processes and molecular mechanisms by which ancient farmers and modern breeders reshaped pathways critical to agronomic traits.
Circadian rhythm.
The plant circadian clock regulates growth, metabolism, and stress responses (40). Clock variation is adaptive, enhancing performance of domesticated species by fine-tuning expression and physiological responses to match fluctuation in their environments (41, 42). Migration of domesticated tomato (Solanum lycopersicum) from the equator to higher latitudes coincided with deceleration of its clock, extending the circadian period (43). As with tomato, potato domestication preceded migration to different latitudes with longer summer days. Strong selection signatures were identified in genes regulating potato circadian rhythm, including REVEILLE 6 (RVE6) and EARLY FLOWERING 4 (ELF4) homologs (Dataset S7). We measured circadian rhythms of wild species, landraces, and cultivars using delayed fluorescence (Fig. 5 A and B) (44), including wild and cultivated tomato checks to validate our results (43), and observed correlation of period length and latitude of origin (R2 = 0.31) only for wild potatoes (Dataset S8). In contrast to their tomato relatives, potato landraces and cultivars have maintained similar short periods independent of latitude (Fig. 5B and Fig. S10), although similar weakening of clock oscillation was observed (Fig. 5B). Correlated period length and latitude have been observed in several plant species (43, 45, 46); longer circadian periods delay daily peaks in gene expression, potentially enhancing growth under longer days by extending the expression of genes controlling growth processes (43, 47). However, in monkey flower, correlation of latitude and period length is observed in annual but not perennial species (48), indicating that modes of reproduction may influence clock adaptation. As potato shifted to rely on belowground asexual structures for annual regrowth, distinct metabolic or developmental processes associated with tuberization and dormancy could have selected against changes in the circadian period, unlike tomato. However, strong selection signatures at regulatory loci of the circadian pathway (Fig. 5C) imply that, despite conservation of a shorter period, potatoes have retuned metabolic or physiological responses to the circadian clock.
Fig. 5.
Circadian rhythms and pollen development phenotypes in wild species, landraces, and cultivars of potato. (A) Example of delayed fluorescence (DF) traces of one potato wild species and landrace. Data are from one representative experiment; values are the average ± SEM of five (wild) or six (landrace) plants. (B) Circadian period length and latitude of origin of potato populations. The origin of modern cultivated lines was determined as the location in which the line was initially bred. Circadian rhythms were measured using delayed fluorescence. Values are the average ± SEM of 2–11 plants from at least two independent experiments. (C) Simplified model of the photoperiod control of tuberization in potato. Elements in white display signatures of selection. Black lines represent transcriptional regulation; blue lines represent posttranslational regulation. (D–G) Pollen grains stained with actetocarmine of S. infundibuliforme PI 472894 at 10× (D) and 40× (E) magnification and S. tuberosum group Tuberosum cv Superior at 10× (F) and 40× (G) magnification; viable pollen grains stain red, and nonviable pollen grains are unstained.
Cell cycle and endoreduplication.
Endoreduplication is a deviation from the standard cell cycle wherein cells forgo cytokinesis, undergoing successive rounds of DNA replication, increasing ploidy and nuclear size. This frequently coincides with increased cellular volume and specialized cellular functions such as nutrient storage, as observed for maize endosperm (49) and tomato pericarp (50). Considerable endoreduplication has been observed within potato tubers: DNA content as high as 16C (16 times the haploid genome) has been observed in cv. Superior and 64C in S. candolleanum, the putative diploid precursor to domestication (51). This, and observations that tuber enlargement is primarily due to cell expansion rather than division (52), suggests that endoreduplication may benefit the tuber’s function as a sink/storage organ by allowing greater cell expansion and increasing the potential for starch biosynthesis and deposition. Given that endoreduplication has the capacity to increase sink organ size (53), selection of endoreduplication-promoting alleles during domestication and improvement may contribute to differences in wild and cultivated tuber size (Fig. 1A). We observed selection of 18 genes regulating cell cycle and endoreduplication (Dataset S9), including the cell-cycle switch 52 gene (CCS52B), showing signatures of selection in both landraces and cultivars. CCS52B homologs are involved in the shift from mitotic to endoreduplication cycles; Arabidopsis CCS52B-overexpressing lines displayed >50% reduction in nonendopolyploid root cells (54), whereas tobacco BY-2 cells overexpressing CCS52B showed a sevenfold increase in cells with >2C DNA content (55). Selected genes also included two homologs of KAKTUS and two cyclins (CYCD3;2 and CYCA3;4), which regulate the transition to endoreduplication during organ development (56, 57). While the 13 additional selected cell-cycle genes do not control endoreduplication, they may have been selected for other processes, such as altering the rate of cell division, rather than expansion during early organ development, as is the case of the tomato fw2.2 domestication locus, which accounts for ∼30% of size variance between wild and cultivated populations (58, 59). This may also be the case for CCS5B, which is preferentially expressed in developing tissues such as immature tubers (60) and fruit (61), with endoreduplication resulting from perturbations in other cell-cycle genes when CCS5B is overexpressed. Estimates of endoreduplication in potato tubers have been limited by its recalcitrance to routine flow cytometry protocols; new methodology may improve our understanding of the process and genes that regulate it.
Sexual reproduction and pollen development.
The low sexual fertility of cultivated potato is a common hurdle to breeding new cultivars, as reliance on asexual propagation minimizes the need for sexual fitness. Pollen from wild species and cultivars typically show wide differences in the frequency of aborted pollen grains (Fig. 5 D–G). A comparison of cultivars with wild species identified 11 genes controlling aspects of pollen development with signatures of selection. Orthologs for several of these genes have been studied in Arabidopsis, where insertional mutants exhibit various degrees of male sterility throughout microspore and pollen development, including faulty transcription factors [AT2G14760; 25% aborted pollen (62)], failure of cell division at the end of male meiosis [AT3G43210 (63)], burst pollen grains [AT3G45040; phosphatidate cytidylyltransferase family protein (64)], incomplete pollen tube growth [AT5G15470; galacturonosyltransferase 14 (65)], and failure to produce functional sperm cells [AT3G60460; DUO1 transcription factor (66)]. Of the six genes that revealed signatures of selection between wild species and landraces, all were expressed across a wide range of tissues, e.g., genes encoding NAC (AT3G10490) or BTB (AT5G63160) domain-containing proteins that affect shoot architecture and nitrogen uptake, respectively. The differences between cultivars and landraces in genes that specifically or indirectly influence pollen development may reflect the diminishing reliance on sexual reproduction as wild species transitioned to landraces and finally to cultivars.
Steroidal glycoalkaloid biosynthesis.
Steroidal glycoalkaloids (SGAs) provide potatoes above- and belowground resistance to insects and pathogens (22). Due to antinutritional properties (67, 68), low SGA concentrations are required for tuber consumption. Domestication and breeding reduced tuber SGAs to acceptable levels (<200 mg/kg), while the key underlying genes remained undetermined. Most genes in the SGA-specific metabolic pathway were unselected (Dataset S10); however, strong signatures were observed in squalene synthase (SQS), the enzyme acting as the gateway for substrates entering the sterol biosynthetic pathway (Fig. 6) (69, 70). SQS was a core selection candidate in landraces and cultivars, with variant sites showing complete allele fixation relative to wild species. Additional selection signatures were associated with a cluster of ethylene response factor (ERF) genes on chromosome 1 harboring GLYCOALKALOID METABOLISM 9 (GAME9) (71), the key transcriptional regulator of GAME genes encoding enzymes in the SGA-specific pathway (Fig. 6, shaded red) (72). Allelic diversity within GAME9 showed broad diversification in wild species. Landraces and cultivars contain almost no GAME9 variation (Fig. S11), implying conserved SGA-pathway regulation by a limited set of alleles. Unlike SGA-pathway genes, SQS and upstream enzymes are not coexpressed or regulated by GAME9, suggesting that cultivated potatoes may exercise multitiered control over SGA biosynthesis, including regulation of substrate flux into the sterol pathway (via SQS) and subsequent activity of downstream genes involved in hydroxylation, oxidation, and glycosylation to synthesize SGAs (via GAME9). Formerly unknown to breeders, this two-locus model for selection against SGAs may offer an ideal marker strategy to screen populations, given renewed interest in the use of wild species for diploid breeding.
Fig. 6.
Selection impacts within the glycoalkaloid biosynthetic pathway. Plant mevalonate pathway [modified from figure 1 in Ginzberg et al. (69) with permission from Springer] with branches into terpenoid, lanosterol, steroidal glycoalkaloid (SGA-specific pathway shaded in red), brassinosteroid, and phytosterol biosynthesis. Red arrows show pathways directly regulated by SQS and GAME9. Blue arrows show pathways controlled indirectly via GAME9 regulation of enzymes guiding substrates into the SGA-specific pathway.
Carbohydrate metabolism.
Potatoes are valued for their high-quality starch derived from leaf sugars transported to the tuber, cytosolic sucrose (Suc) being a key determinant for establishing tuber sink status (73, 74). Of 232 genes functioning in sugar transport/metabolism and starch biosynthesis, a single invertase inhibitor was selected in both Andean landraces and cultivars, demonstrating predominant lineage-specific selection of genes controlling potato carbohydrate metabolism (Dataset S11). In Andean landraces, selection was strongest in genes modulating Suc transport and mobilization, including sucrose-phosphate synthase (SPS), a regulator of diurnal changes in leaf Suc and flux to tubers (75), a sugar transporter, and fructokinase, each functioning upstream of starch synthesis (76–78). Cultivar selection was strongest for genes encoding inorganic pyrophosphatase proteins. During tuber bulking, sucrose mobilization shifts from invertase to sucrose synthase (Susy) activity (79), a pathway supported by pyrophosphate (PPi). PPi has been shown to play a role in cytosolic tuber metabolism (80) and phloem transport (81, 82).
Sucrose nonfermenting-1 (SNF1)–related proteins are global regulators of energy and carbon metabolism in eukaryotes (83), linking metabolism to developmental shifts and stress response (84–87), and both landraces and cultivars have selection signatures within the SNF1-related kinase (SnRK1) protein complex. The plant SnRK1 complex directly regulates SPS (88), is required for Suc-mediated induction of Susy (89), and up-regulates ADP-glucose phyrophosphorylase (AGPase), the key enzyme for starch biosynthesis, in response to Suc availability (90). SnRK1 expression peaks in stolons at tuber initiation, gradually declining in maturing tubers (91), and plays a key role in establishing Suc-mediated sink status in young tubers. The potato gene encoding the plant-specific β3 subunit of the SnRK1 complex was selected in landraces and cultivars, showing that this global regulator of plant-energy homeostasis (85, 86) may be a conserved factor supporting the development of larger cultivated tubers.
Shikimate pathway and disease resistance.
Cultivar selection was highly enriched in genes controlling multiple levels of the shikimate pathway, including 3-dehydroquinate dehydratase, chorismate synthase, and shikimate 3-dehydrogenase activity (Dataset S7). The solanaceous shikimate pathway is wound-induced (92) and generates a tryptophan catabolic sink in potato, altering the phenylpropanoid pathway and susceptibility to P. infestans (93, 94), its most destructive pathogen (28). Supporting this tryptophan catabolic sink, the most highly enriched biological process in cultivar-selected genes was tryptophan catabolism to kynurenine (P = 4.83 × 10−8). Potatoes were recently discovered to be a rich source of kynurenic acid (95), a tryptophan derivative with antioxidant, neuroprotective, and potential antiinflammatory/antiproliferative qualities (95, 96), suggesting that indirect selection in the Solanum shikimate pathway, altering tuber composition, arose while breeding for horizontal P. infestans resistance in cultivars over the last two centuries following the Irish potato famine.
Conclusions
The potato genome remains relatively unexplored, given the crop’s major role in supporting global food security. Using a Solanum section Petota diversity panel, we show that extant germplasm possess heterogeneous genomes littered with CNV, expanding on previous studies of cultivated genotypes (27, 97). Potato contains the highest genomic diversity of any sequenced crop species to date, partly due to autopolyploidy and wild species introgression. Despite the remarkably similar genetic variation within cultivars and wild species (πW/πC = 1.157), the average genetic distance among cultivars (0.026) was 3.27-fold lower than among wild accessions (0.085), confirming that significant allelic diversity existed in 19th century founders, but their small number ensured a group of individually heterozygous but closely related descendants. This is consistent with hypotheses that breeding populations are without significant structure due to the cost of genetic load on inbreeding and continuous reshuffling of alleles from the same heterozygous sites to increase gene interaction while masking deleterious mutations (98). While this creates challenges for exploiting heterosis, our discovery that over 80% of the selected genes in S. tuberosum cultivars (Tuberosum) or Andean landraces (Andigena) are population-specific shows there may be beneficial alleles in South American landraces representing untapped heterotic potential. Recently developed self-compatible S. tuberosum diploids (99) have given breeders an effective sieve to separate alleles of wider agricultural value across latitude or upland/lowland habitats from what is likely a more extensive pool of genes selected for regional adaptation.
Meyer and Purugganan (100) predict 15 changes in traits accompanying the various stages in domestication of root and tuber crops. These include increased yield, modified resource allocation, reduced toxicity, stress tolerance, and reduced sexual fertility—all of which possess signatures of selection in potato. They also anticipate an increase in heterozygosity as a means to leverage heterosis, and we observed an increase in heterozygosity during the progression from wild species to landraces to elite cultivars, likely made possible by the circumstances of potato domestication: substantial nucleotide diversity within the gene pool, 2n gametes allowing for diploid introgressions, additional alleles per locus due to polyploidy, and retention of alleles due to asexual reproduction.
With access to cataloged sequence diversity and annotation of loci controlling key agronomic traits, breeders now have a molecular framework and genome toolbox to develop cultivars outperforming century-old dominant varieties, with adaptation to a wide range of climates and conditions needed to meet global food demands in the 21st century. Given the uncovered contribution of wild Solanum species to cultivated diversity and the recent development of self-compatible diploid cultivars (99), primitive South American populations now offer a rich source of alleles for varietal improvement.
Materials and Methods
Sample Preparation and Sequencing.
Samples were obtained from the US Department of Agriculture potato gene bank and included South American wild species and landrace accessions germinated from seed and North American cultivars as in vitro clones (Dataset S1). Single individuals were selected from accessions to represent populations. DNA was purified from leaves using the Qiagen DNeasy Plant Tissue Kit. Illumina-compatible paired-end sequencing libraries (500-nt fragment size) were prepared and sequenced in paired-end mode (125 nt) to 8× genome coverage (diploids) and 16× coverage (tetraploids) on an Illumina HiSeq 2500 system at the Michigan State University Research Technology Support Facility Genomics Core; one library (Superior) was sequenced in paired-end mode to 150 nt. A subset of libraries was sequenced in paired-end mode (100 nt) on an Illumina HiSeq 2000 system (Dataset S1).
Read Alignment and Variant Calling.
Sequence reads were processed with Trimmomatic (v0.32) (MINLEN = 50, LEADING = 20, TRAILING = 20, SLIDINGWINDOW = 5,20) (101) to remove low-quality bases, adapters, and primers. Clean reads were aligned to the S. tuberosum group Phureja DM reference genome (v4.04) (27) using BWA-mem (v0.7.11) (102). Alignments were processed with Picard tools (v2.1.1) (broadinstitute.github.io/picard) to mark duplicates, and indel realignment was performed using the Genome Analysis Toolkit (v3.3.0) (103). Processed alignments were used for variant genotyping in FreeBayes (v0.9.21.19), requiring minimum 4× coverage in diploids and 8× coverage in tetraploids. Alignments with a MapQ score <20 or base Phred qualities <20 were excluded, and only properly oriented read pairs mapping to the same chromosome were used. Variants were filtered to remove low-quality sites (QUAL <20), mean mapping quality of reference (MQMR) or alternate (MQM) alleles <20, and sites where mean reference allele quality differed from alternate allele quality by 10. Variant sites with >80% strand bias for any allele were excluded. Variant calling ignored regions in the reference genome within 150 bp of gaps to avoid false positives in poorly resolved sequences. Singleton allele variants were removed from downstream analyses. Sample genotypes were filtered for genotype quality (GQ >20). Genome-wide CNV was calculated by comparison of median read coverage in 5-kb windows and within genes to genome-wide median coverage, with copy number (CN) reported as copies per monoploid genome [CN = (region median/genome median)/ploidy]. Sequences were classified as homozygous deletion/absent (CN < 0.1), partial/heterozygous deletion (0.1 ≤ CN < 0.6), conserved (0.6 ≤ CN ≤ 2.0), or duplicated (CN > 2).
Population Analysis and Phylogenetics.
Population structure was calculated using FastStructure (v1.0) (104), testing replicates of five at K = 2–10 with a set of 50,000 biallelic SNPs randomly sampled at equal counts from genome-wide 5-kb windows. The optimal number of model components reflecting structure was selected by the internal fastStructure script chooseK.py. Phylogenetic analysis and estimates of genetic distance (Nei) were performed using PHYLIP (evolution.genetics.washington.edu/phylip.html). A coding-sequence SNP phylogeny was generated using 687,172 fourfold degenerate sites from conserved genes. Neighbor-joining trees were constructed using a consensus derived from 1,000 bootstrap datasets, with branch lengths from the original dataset fitted over the topology of the consensus tree.
Wild Introgressions.
The ancestral potato consensus genotype was reconstructed from diploid landrace accessions (excluding the PI195204 hybrid) by selecting the most common allele composition across variant sites. Genetic-distance matrices were calculated within 5-kb windows and were used to identify windows in which genetic distances between cultivated genotypes and wild species were smaller than between cultivated genotypes and the ancestral cultivated diploid ancestor. Consecutive windows at least 20 kb in length and of the shortest distance to a single wild species were collapsed into putative introgressions from the corresponding species, and those of shortest distance to multiple species were assigned as ambiguous wild introgressions accounting for the heterozygosity of various wild/cultivated haplotypes.
Selection Analysis.
Population statistics used as indicators of selection included FST, Tajima’s D, and nucleotide diversity relative to the founder population (πwild/πcultivated). FST was calculated using Hudson’s estimator (105) for biallelic sites with minor allele frequency (MAF) >0.05. Tajima’s D and population nucleotide diversity were calculated in BioPerl. FST was calculated in 20-kb windows (5-kb steps) using a combining approach described by Bhatia et al. (106). Genes were categorized under three levels of selection: putative, confident, and core selection. Genes under putative selection were identified as those intersecting genomic windows ranked within the top 5% FST values and ranking in the top 5% per-gene estimates of maximum single variant allele frequency difference, Tajima’s D, or reduced nucleotide diversity (π wild/π cultivated). Genes within the top 5% of all three single-gene metric values were considered selected regardless of FST window overlap to allow for erosion of linkage disequilibrium. Genes under confident and core selection were identified using the same criteria with 2% and 1% cutoffs, respectively.
Maturity Locus Analysis.
A universal StCDF1 (PGSC0003DMG400018408) primer pair was designed using Primer3 (biotools.umassmed.edu) and a FASTA sequence extracted from the DM v4.04 reference genome containing the gene plus 1-kb flanking regions. Positions containing variants with MAF >0.10 were masked to reduce amplification failure from sequence variation within primer-binding sites. StCDF1 PCR amplicons were generated with the Q5 High-Fidelity DNA Polymerase kit (New England BioLabs) using 10 ng genomic DNA. Reactions were cycled at 98 °C for 1 min followed by 30 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 90 s, with a final extension at 72 °C for 2 min and were sheared to a 400-bp average insert size using a Covaris S2 ultrasonicator. Illumina-compatible paired-end libraries were prepared as previously described (27) using StCDF1 DNA amplicons and were sequenced in paired-end mode on the Illumina MiSeq, generating 250-nt reads. Sequences were processed as described above, and overlapping reads were merged using FLASH (-m 10 -M 100 -× 0.1) (107). Due to excessive heterozygosity, the haplotypes could not be properly assembled and were manually phased in Integrated Genomics Viewer (IGV) (108). Peptide sequences were derived from the StCDF1 representative transcript isoform (PGSC0003DMT400047370) using TransDecoder (https://github.com/TransDecoder/TransDecoder/wiki), and both DNA and peptide sequences were aligned with ClustalW (109).
Circadian Rhythm.
In vitro plantlets were acclimated in a growth chamber in Redi-Earth soil mix under a 12-h photoperiod (500 μmol⋅m−2⋅s−1 light intensity) at 22 °C for 2 wk. For imaging, plants in 0.12-L plastic pots were transferred to 22 °C and constant 70 μmol⋅m−2⋅s−1 light provided by Heliospectra RX-30 (Heliospectra). The light spectrum was set up to mimic the initial growth chamber: 1% 400 nm, 2% 420 nm, 10% 450 nm, 66% 530 nm, 20% 620 nm, 0.5% 660 nm, and 0.6% 735 nm. Delayed fluorescence was detected using an Andor iKon-M DU-934N-BV camera. Images (1-min exposure time) were collected 2 s after the light was turned off every hour for 4–5 d. Raw fluorescence data were detrended using a linear polynomial function of degree 2, and single-plant measurements were normalized between 0 and 1. Period length was quantified using BRASS (110). Period values with a relative amplitude error larger than 0.6 (<5% total) were discarded. Tomato genotypes Heinz 1706 (S. lycopersicum) and LA0716 (Solanum pennellii) were included for comparing estimates of previous studies using a leaf-movement assay.
Supplementary Material
Acknowledgments
We thank Koichi Sugimoto of the Michigan State University Plant Research Laboratory for kindly providing the tomato plants used to assess circadian rhythm and Joseph Coombs and Walter Amoros for photographs.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.v.d.K. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the BioProject database (ID PRJNA378971).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714380114/-/DCSupplemental.
References
- 1.Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc Natl Acad Sci USA. 2005;102:14694–14699. doi: 10.1073/pnas.0507400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brush SB, Carney HJ, Humán Z. Dynamics of Andean potato agriculture. Econ Bot. 1981;35:70–88. [Google Scholar]
- 3.Pearsall DM. Plant domestication and the shift to agriculture in the Andes. In: Silverman H, Isbell WH, editors. The Handbook of South American Archaeology. Springer; New York: 2008. pp. 105–120. [Google Scholar]
- 4.Hosaka K. Evolutionary pathway of T-type chloroplast DNA in potato. Am J Potato Res. 2004;81:153–158. [Google Scholar]
- 5.Raker CM, Spooner DM. Chilean tetraploid cultivated potato is distinct from the Andean populations. Crop Sci. 2002;42:1451–1458. [Google Scholar]
- 6.Spooner D, Jansky S, Clausen A, del Rosario Herrera M, Ghislain M. The enigma of Solanum maglia in the origin of the Chilean cultivated potato, Solanum tuberosum Chilotanum group. Econ Bot. 2012;66:12–21. [Google Scholar]
- 7.Watanabe K, Peloquin SJ. Occurrence of 2n pollen and ps gene frequencies in cultivated groups and their related wild species in tuber-bearing Solanums. Theor Appl Genet. 1989;78:329–336. doi: 10.1007/BF00265292. [DOI] [PubMed] [Google Scholar]
- 8.Hosaka K. T-type chloroplast DNA in Solanum tuberosum L. ssp. tuberosum was conferred from some populations of S. tarijense Hawkes. Am J Potato Res. 2003;80:21–32. [Google Scholar]
- 9.Birch PR, et al. Crops that feed the world 8: Potato: Are the trends of increased global production sustainable? Food Secur. 2012;4:477–508. [Google Scholar]
- 10.Scott G, Suarez V. The rise of Asia as the centre of global potato production and some implications for industry. Potato J. 2012;39:1–22. [Google Scholar]
- 11.Spooner DM. DNA barcoding will frequently fail in complicated groups: An example in wild potatoes. Am J Bot. 2009;96:1177–1189. doi: 10.3732/ajb.0800246. [DOI] [PubMed] [Google Scholar]
- 12.Hawkes JG. The Potato: Evolution, Biodiversity and Genetic Resources. Smithsonian Institution; Washington, DC: 1990. p. 259. [Google Scholar]
- 13.Ochoa CM. The Potatoes of South America: Bolivia. Cambridge Univ Press; Cambridge, UK: 1990. [Google Scholar]
- 14.Spooner D, Salas A. Handbook of Potato Production, Improvement and Post-Harvest Management. Haworth; Philadelphia: 2006. Structure, biosystematics, and genetic resources of potato; pp. 1–39. [Google Scholar]
- 15.Zimmerer KS. The ecogeography of Andean potatoes. Bioscience. 1998;48:445–454. [Google Scholar]
- 16.Kaldy MS. Protein yield of various crops as related to protein value. Econ Bot. 1972;26:142–144. [Google Scholar]
- 17.Renault D, Wallender W. Nutritional water productivity and diets. Agric Water Manag. 2000;45:275–296. [Google Scholar]
- 18.Kolasa KM. The potato and human nutrition. Am Potato J. 1993;70:375–384. [Google Scholar]
- 19.Jansky S, et al. A case for crop wild relative preservation and use in potato. Crop Sci. 2013;53:746–754. [Google Scholar]
- 20.Bradshaw J, Bryan G, Ramsay G. Genetic resources (including wild and cultivated Solanum species) and progress in their utilisation in potato breeding. Potato Res. 2006;49:49–65. [Google Scholar]
- 21.Jansen G, Flamme W, Schüler K, Vandrey M. Tuber and starch quality of wild and cultivated potato species and cultivars. Potato Res. 2001;44:137–146. [Google Scholar]
- 22.Friedman M. Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. J Agric Food Chem. 2006;54:8655–8681. doi: 10.1021/jf061471t. [DOI] [PubMed] [Google Scholar]
- 23.Johns T, Alonso JG. Glycoalkaloid change during the domestication of the potato, Solanum Section Petota. Euphytica. 1990;50:203–210. [Google Scholar]
- 24.Ovchinnikova A, et al. Taxonomy of cultivated potatoes (Solanum section Petota: Solanaceae) Bot J Linn Soc. 2011;165:107–155. [Google Scholar]
- 25.Spooner DM, et al. Extensive simple sequence repeat genotyping of potato landraces supports a major reevaluation of their gene pool structure and classification. Proc Natl Acad Sci USA. 2007;104:19398–19403. doi: 10.1073/pnas.0709796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huamán Z, Spooner DM. Reclassification of landrace populations of cultivated potatoes (Solanum sect. Petota) Am J Bot. 2002;89:947–965. doi: 10.3732/ajb.89.6.947. [DOI] [PubMed] [Google Scholar]
- 27.Hardigan MA, et al. Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell. 2016;28:388–405. doi: 10.1105/tpc.15.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 29.Love SL. Founding clones, major contributing ancestors, and exotic progenitors of prominent North American potato cultivars. Am J Potato Res. 1999;76:263–272. [Google Scholar]
- 30.Plaisted R, Hoopes R. The past record and future prospects for the use of exotic potato germplasm. Am J Potato Res. 1989;66:603–627. [Google Scholar]
- 31.Guo S, et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013;45:51–58. doi: 10.1038/ng.2470. [DOI] [PubMed] [Google Scholar]
- 32.Huang X, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2011;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 33.Hufford MB, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44:808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam H-M, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 35.Qi J, et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet. 2013;45:1510–1515. doi: 10.1038/ng.2801. [DOI] [PubMed] [Google Scholar]
- 36.Swaminathan MS. Nature of polyploidy in some 48-chromosome species of the genus Solanum, Section Tuberarium. Genetics. 1954;39:59–76. doi: 10.1093/genetics/39.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sukhotu T, Kamijima O, Hosaka K. Chloroplast DNA variation in the most primitive cultivated diploid potato species Solanum stenotomum Juz. et Buk. and its putative wild ancestral species using high-resolution markers. Genet Resour Crop Evol. 2006;53:53–63. [Google Scholar]
- 38.Kloosterman B, et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature. 2013;495:246–250. doi: 10.1038/nature11912. [DOI] [PubMed] [Google Scholar]
- 39.Koenig D, et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci USA. 2013;110:E2655–E2662. doi: 10.1073/pnas.1309606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenham K, McClung CR. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet. 2015;16:598–610. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- 41.Bendix C, Marshall CM, Harmon FG. Circadian clock genes universally control key agricultural traits. Mol Plant. 2015;8:1135–1152. doi: 10.1016/j.molp.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Shor E, Green RM. The impact of domestication on the circadian clock. Trends Plant Sci. 2016;21:281–283. doi: 10.1016/j.tplants.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 43.Müller NA, et al. Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat Genet. 2016;48:89–93. doi: 10.1038/ng.3447. [DOI] [PubMed] [Google Scholar]
- 44.Gould PD, et al. Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 2009;58:893–901. doi: 10.1111/j.1365-313X.2009.03819.x. [DOI] [PubMed] [Google Scholar]
- 45.Greenham K, et al. Geographic variation of plant circadian clock function in natural and agricultural settings. J Biol Rhythms. 2017;32:26–34. doi: 10.1177/0748730416679307. [DOI] [PubMed] [Google Scholar]
- 46.Michael TP, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 47.de Montaigu A, et al. Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc Natl Acad Sci USA. 2015;112:905–910. doi: 10.1073/pnas.1422242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salmela MJ, et al. Variation in circadian rhythms is maintained among and within populations in Boechera stricta. Plant Cell Environ. 2016;39:1293–1303. doi: 10.1111/pce.12670. [DOI] [PubMed] [Google Scholar]
- 49.Grafi G, Larkins BA. Endoreduplication in maize endosperm: Involvement of m phase–Promoting factor inhibition and induction of s phase–Related kinases. Science. 1995;269:1262–1264. doi: 10.1126/science.269.5228.1262. [DOI] [PubMed] [Google Scholar]
- 50.Cheniclet C, et al. Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant Physiol. 2005;139:1984–1994. doi: 10.1104/pp.105.068767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laimbeer FPE, et al. Protoplast isolation prior to flow cytometry reveals clear patterns of endoreduplication in potato tubers, related species, and some starchy root crops. Plant Methods. 2017;13:27. doi: 10.1186/s13007-017-0177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson L, Barker WG, Howarth MJ. Development and structure of tubers. In: Li PH, editor. Potato Physiol. Academic; Orlando, FL: 1985. [Google Scholar]
- 53.Chevalier C, et al. Endoreduplication and fruit growth in tomato: Evidence in favour of the karyoplasmic ratio theory. J Exp Bot. 2014;65:2731–2746. doi: 10.1093/jxb/ert366. [DOI] [PubMed] [Google Scholar]
- 54.de Almeida Engler J, et al. CCS52 and DEL1 genes are key components of the endocycle in nematode-induced feeding sites. Plant J. 2012;72:185–198. doi: 10.1111/j.1365-313X.2012.05054.x. [DOI] [PubMed] [Google Scholar]
- 55.Tarayre S, Vinardell JM, Cebolla A, Kondorosi A, Kondorosi E. Two classes of the CDh1-type activators of the anaphase-promoting complex in plants: Novel functional domains and distinct regulation. Plant Cell. 2004;16:422–434. doi: 10.1105/tpc.018952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.El Refy A, et al. The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol Genet Genomics. 2003;270:403–414. doi: 10.1007/s00438-003-0932-1. [DOI] [PubMed] [Google Scholar]
- 57.Dewitte W, et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA. 2007;104:14537–14542. doi: 10.1073/pnas.0704166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alpert KB, Grandillo S, Tanksley SD. fw 2.2: A major QTL controlling fruit weight is common to both red- and green-fruited tomato species. Theor Appl Genet. 1995;91:994–1000. doi: 10.1007/BF00223911. [DOI] [PubMed] [Google Scholar]
- 59.Cong B, Liu J, Tanksley SD. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc Natl Acad Sci USA. 2002;99:13606–13611. doi: 10.1073/pnas.172520999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Potato Genome Sequencing Consortium Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 61.Mathieu-Rivet E, et al. Functional analysis of the anaphase promoting complex activator CCS52A highlights the crucial role of endo-reduplication for fruit growth in tomato. Plant J. 2010;62:727–741. doi: 10.1111/j.1365-313X.2010.04198.x. [DOI] [PubMed] [Google Scholar]
- 62.Reňák D, Dupl’áková N, Honys D. Wide-scale screening of T-DNA lines for transcription factor genes affecting male gametophyte development in Arabidopsis. Sex Plant Reprod. 2012;25:39–60. doi: 10.1007/s00497-011-0178-8. [DOI] [PubMed] [Google Scholar]
- 63.Oh SA, Bourdon V, Dickinson HG, Twell D, Park SK. Arabidopsis Fused kinase TWO-IN-ONE dominantly inhibits male meiotic cytokinesis. Plant Reprod. 2014;27:7–17. doi: 10.1007/s00497-013-0235-6. [DOI] [PubMed] [Google Scholar]
- 64.Lindner H, et al. TURAN and EVAN mediate pollen tube reception in Arabidopsis synergids through protein glycosylation. PLoS Biol. 2015;13:e1002139. doi: 10.1371/journal.pbio.1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, et al. Arabidopsis galacturonosyltransferase (GAUT) 13 and GAUT14 have redundant functions in pollen tube growth. Mol Plant. 2013;6:1131–1148. doi: 10.1093/mp/sst084. [DOI] [PubMed] [Google Scholar]
- 66.Brownfield L, et al. A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 2009;5:e1000430. doi: 10.1371/journal.pgen.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maga JA. Potato glycoalkaloids. Crit Rev Food Sci Nutr. 1980;12:371–405. doi: 10.1080/10408398009527281. [DOI] [PubMed] [Google Scholar]
- 68.Sinden S, Sanford L, Webb R. Genetic and environmental control of potato glycoalkaloids. Am J Potato Res. 1984;61:141–156. [Google Scholar]
- 69.Ginzberg I, et al. Induction of potato steroidal glycoalkaloid biosynthetic pathway by overexpression of cDNA encoding primary metabolism HMG-CoA reductase and squalene synthase. Planta. 2012;235:1341–1353. doi: 10.1007/s00425-011-1578-6. [DOI] [PubMed] [Google Scholar]
- 70.Ginzberg I, Tokuhisa JG, Veilleux RE. Potato steroidal glycoalkaloids: Biosynthesis and genetic manipulation. Potato Res. 2009;52:1–15. [Google Scholar]
- 71.Cárdenas PD, et al. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun. 2016;7:10654. doi: 10.1038/ncomms10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Itkin M, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- 73.Fernie AR, Willmitzer L. Molecular and biochemical triggers of potato tuber development. Plant Physiol. 2001;127:1459–1465. [PMC free article] [PubMed] [Google Scholar]
- 74.Sonnewald U, et al. Expression of a yeast invertase in the apoplast of potato tubers increases tuber size. Nat Biotechnol. 1997;15:794–798. doi: 10.1038/nbt0897-794. [DOI] [PubMed] [Google Scholar]
- 75.Geigenberger P, Stitt M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 2000;23:795–806. doi: 10.1046/j.1365-313x.2000.00848.x. [DOI] [PubMed] [Google Scholar]
- 76.Geigenberger P, Stitt M, Fernie A. Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ. 2004;27:655–673. [Google Scholar]
- 77.Huber SC, Huber JL. Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:431–444. doi: 10.1146/annurev.arplant.47.1.431. [DOI] [PubMed] [Google Scholar]
- 78.Williams LE, Lemoine R, Sauer N. Sugar transporters in higher plants–A diversity of roles and complex regulation. Trends Plant Sci. 2000;5:283–290. doi: 10.1016/s1360-1385(00)01681-2. [DOI] [PubMed] [Google Scholar]
- 79.Appeldoorn NJ, et al. Developmental changes of enzymes involved in conversion of sucrose to hexose-phosphate during early tuberisation of potato. Planta. 1997;202:220–226. [Google Scholar]
- 80.Farré EM, Tech S, Trethewey RN, Fernie AR, Willmitzer L. Subcellular pyrophosphate metabolism in developing tubers of potato (Solanum tuberosum) Plant Mol Biol. 2006;62:165–179. doi: 10.1007/s11103-006-9011-4. [DOI] [PubMed] [Google Scholar]
- 81.Khadilkar AS, et al. Constitutive and companion cell-specific overexpression of AVP1, encoding a proton-pumping pyrophosphatase, enhances biomass accumulation, phloem loading, and long-distance transport. Plant Physiol. 2016;170:401–414. doi: 10.1104/pp.15.01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lerchl J, Geigenberger P, Stitt M, Sonnewald U. Impaired photoassimilate partitioning caused by phloem-specific removal of pyrophosphate can be complemented by a phloem-specific cytosolic yeast-derived invertase in transgenic plants. Plant Cell. 1995;7:259–270. doi: 10.1105/tpc.7.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halford N, Boulyz J-P, Thomas M. SNF1-related protein kinases (SnRKs)—Regulators at the heart of the control of carbon metabolism and partitioning. Adv Bot Res. 2000;32:405–434. [Google Scholar]
- 84.Halford NG, et al. Metabolic signalling and carbon partitioning: Role of Snf1-related (SnRK1) protein kinase. J Exp Bot. 2003;54:467–475. doi: 10.1093/jxb/erg038. [DOI] [PubMed] [Google Scholar]
- 85.Hulsmans S, Rodriguez M, De Coninck B, Rolland F. The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci. 2016;21:648–661. doi: 10.1016/j.tplants.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 86.Lastdrager J, Hanson J, Smeekens S. Sugar signals and the control of plant growth and development. J Exp Bot. 2014;65:799–807. doi: 10.1093/jxb/ert474. [DOI] [PubMed] [Google Scholar]
- 87.Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12:20–28. doi: 10.1016/j.tplants.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 88.Sugden C, Donaghy PG, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Purcell PC, Smith AM, Halford NG. Antisense expression of a sucrose non‐fermenting‐1‐related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose‐inducibility of sucrose synthase transcripts in leaves. Plant J. 1998;14:195–202. [Google Scholar]
- 90.Tiessen A, et al. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 2003;35:490–500. doi: 10.1046/j.1365-313x.2003.01823.x. [DOI] [PubMed] [Google Scholar]
- 91.Man AL, Purcell PC, Hannappel U, Halford NG. Potato SNF1-related protein kinase: Molecular cloning, expression analysis and peptide kinase activity measurements. Plant Mol Biol. 1997;34:31–43. doi: 10.1023/a:1005765719873. [DOI] [PubMed] [Google Scholar]
- 92.Dyer WE, Henstrand JM, Handa AK, Herrmann KM. Wounding induces the first enzyme of the shikimate pathway in Solanaceae. Proc Natl Acad Sci USA. 1989;86:7370–7373. doi: 10.1073/pnas.86.19.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herrmann KM. The shikimate pathway: Early steps in the biosynthesis of aromatic compounds. Plant Cell. 1995;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao K, De Luca V, Brisson N. Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. Plant Cell. 1995;7:1787–1799. doi: 10.1105/tpc.7.11.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Turski MP, Kamiński P, Zgrajka W, Turska M, Turski WA. Potato- an important source of nutritional kynurenic acid. Plant Foods Hum Nutr. 2012;67:17–23. doi: 10.1007/s11130-012-0283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turski MP, Turska M, Zgrajka W, Kuc D, Turski WA. Presence of kynurenic acid in food and honeybee products. Amino Acids. 2009;36:75–80. doi: 10.1007/s00726-008-0031-z. [DOI] [PubMed] [Google Scholar]
- 97.Iovene M, Zhang T, Lou Q, Buell CR, Jiang J. Copy number variation in potato–An asexually propagated autotetraploid species. Plant J. 2013;75:80–89. doi: 10.1111/tpj.12200. [DOI] [PubMed] [Google Scholar]
- 98.Hirsch CN, et al. Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 (Bethesda) 2013;3:1003–1013. doi: 10.1534/g3.113.005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jansky S, et al. Reinventing potato as a diploid inbred line–based crop. Crop Sci. 2016;56:1412–1422. [Google Scholar]
- 100.Meyer RS, Purugganan MD. Evolution of crop species: Genetics of domestication and diversification. Nat Rev Genet. 2013;14:840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- 101.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. arXiv:1303.3997v2.
- 103.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: The impact of rare variants. Genome Res. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thompson JD, Gibson T, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002:2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 110.Plautz JD, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 111.Manrique-Carpintero NC, et al. Genetic map and quantitative trait locus analysis of agronomic traits in a diploid potato population using single nucleotide polymorphism markers. Crop Sci. 2015;55:2566–2579. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.