Significance
14-3-3 proteins are conserved scaffolding proteins expressed in all eukaryotic cells, where they regulate the function of several hundreds of partner proteins by constraining their conformation. Yeast neutral trehalases (Nth), enzymes responsible for the hydrolysis of trehalose, compared with trehalases from other organisms, possess distinct structure and regulation involving phosphorylation followed by binding to 14-3-3. Here we present the crystal structures of yeast Nth1 and its complex with the 14-3-3 protein and propose a molecular mechanism in which 14-3-3 activates Nth1 by stabilizing the flexible part of its active site. Comparison of the 14-3-3:Nth1 complex structure with those of other 14-3-3 complexes suggests the highly conserved nature of 14-3-3 affects the structures of many client proteins.
Keywords: 14-3-3 protein, trehalase, crystal structure, enzyme, allostery
Abstract
The 14-3-3 proteins, a family of highly conserved scaffolding proteins ubiquitously expressed in all eukaryotic cells, interact with and regulate the function of several hundreds of partner proteins. Yeast neutral trehalases (Nth), enzymes responsible for the hydrolysis of trehalose to glucose, compared with trehalases from other organisms, possess distinct structure and regulation involving phosphorylation at multiple sites followed by binding to the 14-3-3 protein. Here we report the crystal structures of yeast Nth1 and its complex with Bmh1 (yeast 14-3-3 isoform), which, together with mutational and fluorescence studies, indicate that the binding of Nth1 by 14-3-3 triggers Nth1’s activity by enabling the proper 3D configuration of Nth1’s catalytic and calcium-binding domains relative to each other, thus stabilizing the flexible part of the active site required for catalysis. The presented structure of the Bmh1:Nth1 complex highlights the ability of 14-3-3 to modulate the structure of a multidomain binding partner and to function as an allosteric effector. Furthermore, comparison of the Bmh1:Nth1 complex structure with those of 14-3-3:serotonin N-acetyltransferase and 14-3-3:heat shock protein beta-6 complexes revealed similarities in the 3D structures of bound partner proteins, suggesting the highly conserved nature of 14-3-3 affects the structures of many client proteins.
The 14-3-3 proteins, a family of highly conserved dimeric proteins ubiquitously expressed in all eukaryotic cells, interact with and regulate the function of several hundred partner proteins by recognizing phosphoserine- (pS) or phosphothreonine (pT)-containing motifs (1, 2). Mechanistically, 14-3-3 proteins act as allosteric regulators and/or molecular scaffolds that constrain the conformation of the binding partner; if the target protein is an enzyme, this can affect its catalytic activity. Many enzymes including c-Raf protein kinase (3), serotonin N-acetyltransferase (AANAT) (4), tyrosine and tryptophan hydroxylases (5, 6), apoptosis signal-regulated protein kinase 1 (7), and Cdc25 phosphatases (8) have been shown to be regulated in such a manner. Nonetheless, the underlying molecular mechanisms are only partially identified, mainly as a result of the lack of structural data. Only one high-resolution structure of the complex between 14-3-3 and a fully active enzyme, AANAT, has been reported so far (9). This structure showed that regions of AANAT that interact with 14-3-3 include those involved in substrate binding, suggesting that 14-3-3 structurally modulates the active site of bound AANAT, and thus increases its substrate binding affinity with an accompanying increase in catalytic activity. Besides the direct structural modulation, 14-3-3 proteins can also stabilize the tertiary and/or quaternary structure of the bound enzyme (10, 11) or change its subcellular localization (8, 12). In addition, 14-3-3 can also mediate the interaction between two different proteins, as has been demonstrated for the florigen activation complex composed of florigen homolog Hd3a, plant 14-3-3 protein GF14c, and a peptide comprising the nine carboxyl-terminal amino acids of phosphorylated transcription factor FD1 (13).
One of the enzymes regulated in the 14-3-3-dependent manner is yeast neutral trehalase 1 (Nth1). Trehalases (EC 3.2.1.28) are glycoside hydrolases that catalyze the hydrolysis of the nonreducing disaccharide trehalose (α-d-glucopyranosyl α-d-glucopyranoside) as the sole substrate. Trehalose is found in many organisms ranging from bacteria and fungi to invertebrates and higher plants, where it fulfills a number of physiological roles (14, 15). In yeast, trehalose is a major storage carbohydrate, carbon source, and stress protectant, and its hydrolysis is catalyzed mainly by neutral trehalase with a pH optimum of about 7, encoded by two genes, NTH1 and NTH2, with NTH1 being more active (16). Compared with trehalases from prokaryotic and higher eukaryotic organisms, yeast neutral trehalases possess an N-terminal extension that contains four cAMP-dependent protein kinase (PKA) phosphorylation sites (S20, S21, S60, and S83 in Saccharomyces cerevisiae Nth1) and the calcium-binding EF-hand-like motif (Fig. 1A), indicating significantly different regulation (17–19). It was subsequently shown that S. cerevisiae Nth1 is indeed activated in a PKA- and calcium-dependent manner in a process that requires interaction with the 14-3-3 protein through motifs containing phosphoserines pS60 and pS83 (20–23).
Fig. 1.
Overview of pNth11–751:Bmh1 structure. (A) Domain structure of yeast Nth1. The N-terminal extension and the calcium-binding domain (Nth1-CaBD) are shown in cyan, the catalytic domain (Nth1-CD) in dark cyan. The positions of PKA phosphorylation sites which are also 14-3-3 binding motifs are indicated with red arrows. The position of the EF-hand-like motif is indicated with its sequence. Residues involved in calcium ion coordination are shown in orange. Numbering is according to Nth1 from S. cerevisiae. (B) Structure of the pNth11–751:Bmh1 complex. The protomers of the Bmh1 homodimer are shown in yellow and brown. The N-terminal extension and the calcium-binding domain (Nth1-CaBD) are shown in cyan, the catalytic domain (Nth1-CD) in dark cyan. The phosphorylated S60 and S83 are shown in red. The calcium ion is shown in orange. (C) Close-up view of calcium ion-binding site.
To understand how the ability to interact with the 14-3-3 protein changed the structure and regulation of yeast neutral trehalase, we determined the crystal structures of phosphorylated full-length S. cerevisiae Nth1 (pNth11–751) bound to yeast 14-3-3 protein Bmh1 and the catalytic domain of Nth1 (Nth1153–751) alone. The crystallographic analysis, together with mutational analysis and activity- and time-resolved fluorescence measurements, provides a useful clue for elucidating the molecular mechanism of the 14-3-3-dependent regulation of yeast Nth1. In addition, comparison of the complex structure with other 14-3-3 complexes suggests that the highly conserved nature of 14-3-3 impacts the 3D structures of many of their binding partners.
Results
Overall Structure of the pNth11–751:Bmh1 Complex.
Previous work demonstrated that a dimer of Bmh1 binds one molecule of pNth11–751 (23). Thus, for crystallization trials, pNth11–751 and Bmh1 were mixed in a 1:2 molar ratio. The structure of the complex was solved at a resolution of 2.29 Å by molecular replacement, using the structures of the N-terminally truncated Nth1 (Nth1153–751) and 14-3-3ε (24) as search models (SI Appendix, Table S1). The crystal structure of the Bmh1:pNth11–751 complex shows pNth11–751 in a bidentate interaction with the Bmh1 dimer, which simultaneously interacts with both 14-3-3 binding motifs of pNth11–751 (Fig. 1B; the Greek letters identify the helices of Nth1 and capital letters those of Bmh1). The main-chain conformation of the first 14-3-3 binding motif of pNth11–751 (sequence RTRTMpS60VFDN), the pS60 coordination, and other contacts in the 14-3-3 binding groove are similar to those previously seen in other complexes of 14-3-3 proteins (9, 25). The Bmh1 side chains coordinating the pS60 moiety of pNth11–751 include R58, R132, and Y133 (Fig. 2A). An Arg residue of pNth11–751 at the −5 position relative to the phosphorylated residue pS60 is salt-bridged to E136 and E185 of Bmh1 and the phosphate group of pS60, and thus similarly to the Arg residue at positions −4 and −2 in complexes of the phosphopeptide with the “mode 2” consensus sequence and AANAT, respectively (9, 25) (Fig. 3). Interactions between pNth11–751 residues D63 and N64 at the +3 and +4 positions and Bmh1 residues K51 and N52 appear to force a change in the direction of the polypeptide chain, mimicking the role of the Pro residue, which is frequently found in 14-3-3 binding motifs at the +2 position relative to the phosphorylated residue (9, 25–28). The second 14-3-3 binding motif of pNth11–751 (sequence RRGpS83ED) also does not contain a Pro residue at the +2 position relative to the phosphorylated residue (Fig. 2B). Whereas the pS83 moiety is coordinated similar to that of pS60 by the side-chains of Bmh1 residues K51, R58, R132, and Y133, residue R81 of pNth11–751 at the −2 position relative to pS83 is salt-bridged to D85 at the +2 position, and R80 at the −3 position interacts with the side-chain of Bmh1 residue N229. Interestingly, the salt bridge between pNth11–751 residues R81 and D85 is responsible for an abrupt change in the direction of the polypeptide chain (Figs. 2B and 3B), thus again mimicking the role of the Pro residue at the +2 position (9, 25, 27, 28). The intervening linker sequence between the two 14-3-3 binding motifs of pNth11–751 (residues 64−79) is ordered compared with known structures of 14-3-3 protein complexes, most likely a result of numerous interactions with helices A1 and A3 of Bmh1 (Fig. 2C).
Fig. 2.
Contacts between Bmh1 and the 14-3-3 binding motifs of pNth11–751. (A) Contacts between Bmh1 and the first 14-3-3 binding motif of pNth11–751 containing pS60. The Nth1 residues are labeled in cyan, the Bmh1 residues are labeled in black. (B) Contacts between Bmh1 and the second 14-3-3 binding motif of pNth11–751 containing pS83. The Nth1 residues are labeled in cyan, the Bmh1 residues are labeled in black. (C) Contacts between Bmh1 and the intervening linker sequence between the two 14-3-3 binding motifs of pNth11–751. The omit 2F0 − Fc electron density map is contoured at 1σ. The Nth1 residues are labeled in cyan, the Bmh1 residues are labeled in black.
Fig. 3.
Comparison of the Bmh1:pNth11–751, 14-3-3ζ:phosphopeptide, 14-3-3ζ:AANAT, and 14-3-3σ:heat shock protein beta-6 complexes. (A) Comparison of the main-chain conformation of the first 14-3-3 binding motif of pNth1 (sequence RTRTMpS60VFDN, shown in cyan) with a “mode 2” 14-3-3 peptide (sequence RLYHpSLPA, PDB ID 1QJA, shown in green) (25) and the 14-3-3 binding motifs of AANAT (sequence QRRHpTLPA, PDB ID 1IB1, shown in orange) (9) and heat shock protein beta-6 (sequence LRRApSAPL, PDB ID 5LTW, shown in red) (27). The C-terminal portion of the 14-3-3 binding motifs is indicated by black ellipse. (B) Comparison of the main-chain conformation of the second 14-3-3 binding motif of pNth1 (sequence QTRRGpS83EDDT, shown in cyan) with the same complexes as in A. (C and D) Comparison of the first 14-3-3 binding motif of pNth1 (sequence QTRRGpS83EDDT, shown in cyan) with a “mode 2” 14-3-3 peptide [sequence RLYHpSLPA, PDB ID 1QJA (25), shown in green] and the 14-3-3 binding motif of AANAT [sequence QRRHpTLPA, PDB ID 1IB1 (9), shown in orange]. The numbers describe the position relative to pS/pT.
The structure of the calcium-binding domain of pNth11–751 (Nth1-CaBD, residues 96−176) is composed of three α helices and two antiparallel β-sheets, each composed of two strands with the EF-hand-like motif (sequence D114TDKNYQITIED125) located in a loop between α3 and β2 (SI Appendix, Fig. S1). The calcium ion is coordinated in a pentagonal bipyramidal configuration by the side-chains of residues D114, D116, N118, and D125; the main-chain carbonyl group of Q120; and a water molecule (Fig. 1C). The side chains of D114, D116, N118, and D125 and the main chain of Q120 are 2.24, 2.29, 2.38, 2.27, and 2.29 Å, respectively, from the Ca2+ ion. Nth1-CaBD is embedded within the central channel of the Bmh1 dimer, where it borders both phosphorylated motifs and interacts with Bmh1 helices A1, A3, and A9, as well as the loop between A1 and A2 through numerous direct and water-mediated contacts. In terms of Nth1-CaBD, major contributions to this interface arise from the surface of helix α2, strands β2 and β3, and loops between β2 and β3 and between α4 and β4 (Fig. 4A).
Fig. 4.
Other Bmh1:pNth11–751 interactions. (A) Close-up view of interactions between Nth1-CaBD (light cyan) and Bmh1 (yellow and brown). (B) Close-up view of interactions between the C-terminal helices A8 and A9 of Bmh1 (brown) with Nth1-CaBD (light cyan) and Nth1-CD (dark cyan).
The catalytic domain of pNth11–751 (Nth1-CD, residues 180−751) is positioned on the top of the Bmh1 central channel, where it interacts with helices A8 and A9 from the C terminus of one Bmh1 protomer and the helix α4 and the EF-hand-like motif of Nth1-CaBD. Nth1-CD contributes to this region of the interface via the loop preceding the helix α6, the C-terminal end of helix α28, the loop region between α28 and α29, and helices α30 and α31 from the C terminus of the domain (Figs. 1B and 4B). The total solvent-excluded surface area between the Bmh1 dimer and pNth11–751 is 2,955 Å2, of which ∼81% (2,384 Å2) is contributed by the phosphorylated N-terminal segment and Nth1-CaBD (residues 54−176), whereas the rest is contributed by Nth1-CD. Although just ∼19% of the interface between pNth11–751 and Bmh1 arises from Nth1-CD, the extensive interactions between Nth1-CD and the rest of the complex enable a stable and well-defined 3D configuration of this domain relative to Nth1-CaBD and Bmh1.
The structure of the Bmh1 dimer in the complex is very similar to the recently reported structure of Bmh1 from Lachancea thermotolerans (29); the two structures can be aligned over 393 Cα atoms with a r.m.s. deviation of 1.26 Å. The most significant differences involve the C-terminal helices A7–A9, most likely because of the interaction of helices A7 and A9 with pNth11–751.
Structure of the Catalytic Domain of Nth1.
The structure of Nth1-CD consists of an (α/α)6 barrel found in all enzymes belonging to the six-hairpin glycosidases superfamily of an α/α-toroidal fold. The active site is located in a deep pocket formed by the loops of the (α/α)6 barrel (shown in yellow in Fig. 5A) and two subdomains, I and II, which are inserted within the N-terminal half of the (α/α)6 barrel (shown in cyan and green, respectively, in Fig. 5A), and contains a molecule of sucrose (SUC), which was present in the crystallization solution as an additive. The structure of Nth1-CD of Bmh1-bound pNth11–751 is very similar to the structure of the N-terminally truncated Nth1153–751, which is composed of only the catalytic domain (Fig. 5B); the two structures can be aligned over 526 Cα atoms with an r.m.s. deviation of 0.54 Å. The only difference between these two structures involves the long loop region located between α28 and α29, which in complexed pNth11–751 folds back onto the active site and covers it as a “lid” (hereafter referred to as the “lid” loop). In contrast, in the Nth1153–751 structure, the C-terminal portion of this “lid” loop (residues 685−700) is disordered, presumably because of its flexibility. Furthermore, the same part of the “lid” loop is also disordered in the structure of Nth1153–751 with bound trehalose (Nth1153–751:TRE), suggesting it is not caused by the absence of a bound ligand (Fig. 5B). The “lid” loop of Bmh1-bound pNth11–751 is structured likely as a result of interactions with the EF-hand-like motif-containing region of Nth1-CaBD, including the salt bridge between E124 and R686 and hydrogen bonds between the side-chain of Q120 and main-chain carbonyl groups of residues A696 and F698 (Fig. 5C). These interactions appear to stabilize the bent conformation of this loop, thus placing its tip onto the active site pocket. To investigate whether these interactions are also present in the absence of Bmh1, we crystallized an Nth1 construct composed of residues 100−751, thus containing both Nth1-CaBD and Nth1-CD (SI Appendix, Table S1). However, this structure only exhibited a continuous electron density for residues 180−682 and 703−751 of Nth1-CD, and no density was seen for Nth1-CaBD and residues 683−702 from the “lid” loop. This suggests that in the absence of Bmh1, the calcium-binding and catalytic domains of Nth1 are not engaged in a stable protein–protein interaction, and as a result, the “lid” loop is disordered similarly as observed in the structures of Nth1153–751 and Nth1153–751:TRE.
Fig. 5.
Structure of catalytic domain of Nth1. (A) Structure of catalytic domain of Bmh1-bound pNth11–751. The (α/α)6 barrel, subdomains I and II, and the C-terminal helices are shown in yellow, cyan, green, and blue, respectively. SUC is shown in gray. The C-terminal portion of the “lid” loop is shown in red. The rest of the structure is not shown. (B) Superimposition of the catalytic domain of Bmh1-bound pNth11–751 (blue), Nth1153–751 (green), and Nth1153–751:TRE (orange). The C-terminal portion of the “lid” loop of pNth11–751 is shown in red. (C) Close-up view of interactions between the “lid” loop and the EF-hand-like motif of Nth1-CaBD. (D) Sequence alignment of “lid” loops of yeast (S. cerevisiae) Nth1 and trehalase Tre37A from E. coli. The C-terminal portions of the “lid” loops are shown in red. (E) Close-up view of Nth1153–751 active site with bound trehalose (TRE). (F) Close-up view of active site of trehalase Tre37A from E. coli with bound inhibitor VDM (30).
The superimposition of Nth1-CD of the Bmh1 bound pNth11–751 on the crystal structure of the periplasmic trehalase Tre37A from Escherichia coli (Tre37A) with the bound inhibitor validoxylamine A (VDM) (30), which consists of only the catalytic domain and lacks the N-terminal extension, yielded a r.m.s. deviation value of 1.25 Å over 337 Cα atoms and revealed a similar arrangement of helices of the (α/α)6 barrel (SI Appendix, Fig. S2A). In contrast, significant differences exist in both N-terminal subdomain structures as well as the C-terminal domain, which consists of three α-helices in Nth1-CD (α30−32, shown in blue in Fig. 5A), whereas Tre37A possesses no helices in this region (SI Appendix, Figs. S1 and S2A). Interestingly, the “lid” loop of Tre37A is ordered and adopts a similar conformation to that only observed in Bmh1-bound pNth11–751 (SI Appendix, Fig. S2B). The different behaviors of the “lid” loops of Tre37A and yeast Nth1 likely result from their distinct sequences and lengths (Fig. 5D). The “lid” loop of Tre37A is significantly shorter and contains a Gly-rich sequence, and its position is mainly stabilized by contacts with bordering loop regions (SI Appendix, Fig. S2C).
Substrate Binding and the Active Site Structure.
The cocrystal structure of the N-terminally truncated, and thus catalytically inactive, Nth1153–751 with trehalose showed a clear and interpretable electron density for the bound ligand (TRE) in the −1 and +1 subsites of the active site (Fig. 5E). The glucose moieties in the catalytic −1 and the leaving-group +1 subsites adopt 4C1 and 1C4 chair conformations, respectively, and form contacts that are similar to those observed between Tre37A and VDM (Fig. 5F), with trehalose occupying the same position as VDM (30). Comparison of the Nth1153–751:TRE structure with that of Tre37A suggests that Nth1 residues D478 (D312 in Tre37A) and E674 (E496 in Tre37A) function as the catalytic acid and base, respectively. Their side-chains are located 13 Å apart, with D478 being a hydrogen bond distance (3.7 Å) from the glycosidic oxygen atom of trehalose, whereas E674 is located 7.8 Å from the C1 atom of the glucose moiety in the −1 subsite (Fig. 5E). In the structure of Tre37A with bound VDM, this residue (E496) is hydrogen bonded to a water molecule that is responsible for a nucleophilic attack of the glycoside (Fig. 5F). Although the resolution of the Nth1153–751:TRE structure did not enable us to assign water molecules, the sequence and structural similarities between Nth1-CD and Tre37A (SI Appendix, Figs. S1 and S2A) strongly suggest that E674 in Nth1 plays the same role as E496 in Tre37A. This was further corroborated by site-directed mutagenesis, which revealed that both D478A and E674A mutants of pNth11–751 bound to Bmh1 are catalytically inactive (SI Appendix, Fig. S3). The structures of apo and TRE-bound forms of Nth1153–751 are highly similar and can be aligned with an overall r.m.s. deviation at 542 Cα positions of 0.72 Å. The only structural difference between the two forms of Nth1153–751 occurs in the vicinity of the active site, where the substrate binding induces a shift of the surrounding loop regions toward the active site (Fig. 5B). The rest of the catalytic domain exhibited a nearly identical conformation.
Implications for Activation.
The previous structural work on Tre37A from E. coli indicated that residues E511 and Y512 from the tip of the “lid” loop might be directly involved in substrate binding and catalysis, respectively (30). The side-chain of E511 is hydrogen bonded to the pseudosugar ring of VDM in the +1 subsite, whereas the side-chain of Y512 was suggested to impede the attacking water molecule, thus directly participating in catalysis (Fig. 5F). Furthermore, this pair of residues (EY) is totally conserved among 101 trehalase sequences available in the UniProt database (SI Appendix, Fig. S4A), further supporting their functional importance. The corresponding residues of yeast Nth1 (E690 and Y691) are, in contrast to the structure of Tre37A, hydrogen-bonded to subdomain II residues T479 and E426, respectively (Fig. 5C). This difference is likely because the active site of pNth11–751 contains SUC instead of its natural substrate TRE. Although the glucose moiety of SUC occupies a similar position as the glucose moiety of TRE in the catalytic −1 subsite, the fructose moiety of SUC is located above the leaving-group +1 subsite where the second glucose moiety of TRE is bound, thus forcing the tip of the “lid” loop to shift further away from the active site (SI Appendix, Fig. S2B). The functional importance of Nth1 residues E690 and Y691 was confirmed by a mutational analysis of pNth11–751, which revealed that the E690A mutant exhibits significantly reduced activity and the Y691A mutant is catalytically inactive (SI Appendix, Fig. S3).
The structure of pNth11–751 bound to Bmh1 suggested that the “lid” loop conformation is stabilized through interactions with the EF-hand-like motif of Nth1-CaBD (Fig. 5C). Moreover, the structure of Nth1100–751 indicated that in the absence of Bmh1, the calcium-binding and catalytic domains of Nth1 are not engaged in a stable protein–protein interaction. This indicates that contacts between the “lid” loop and Nth1-CaBD, which appear to be mediated by Bmh1 binding, might be important for the activation of pNth1. To investigate this hypothesis, we performed site-directed mutagenesis of residues Q120, E124, and R686 involved in these interactions. Indeed, the enzyme activity measurements showed that mutations, especially Q120A and R686A, significantly reduced the catalytic activity of the Bmh1-bound pNth11–751 (SI Appendix, Fig. S3). In addition, the fact that in the presence of calcium, the Bmh1-mediated activation of pNth1 is significantly enhanced further supports this hypothesis, as the calcium binding likely facilitates these interactions by inducing folding of the EF-hand-like motif. Taken together, these results indicate that residues E690 and Y691 from the tip of the “lid” loop, as well as interactions between the “lid” loop and Nth1-CaBD, are essential for the catalytic activity of Bmh1-bound Nth11–751.
Bmh1 Binding Affects the Conformation of the Active Site of pNth1.
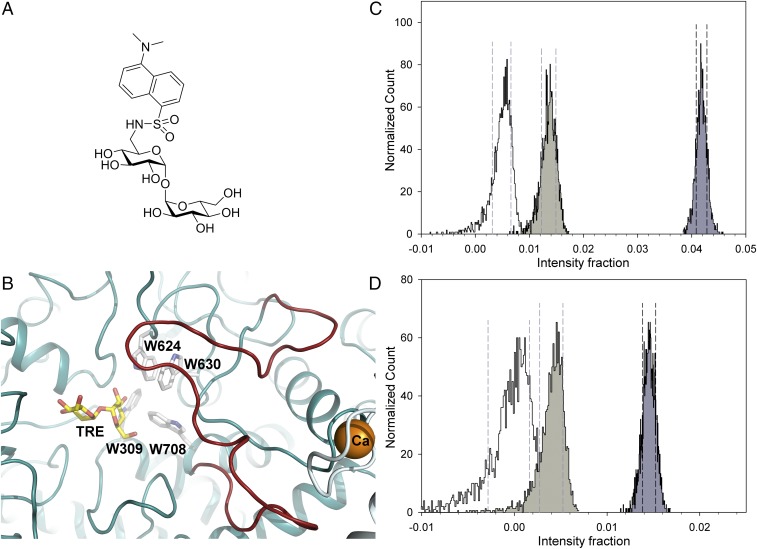
To further investigate the conformational behavior of the “lid” loop of pNth11–751 in the absence and the presence of Bmh1, we synthetized 6-[5′-(dimethylamino)-1′-naphthalenesulfonamido]-6-deoxy-trehalose (DANS-TRE; Fig. 6A), consisting of a dansyl group attached at position 6 of one glucose moiety of TRE, and performed time-resolved fluorescence measurements. We decided to modify the hydroxyl group at position 6 of TRE as it faces outward from the active site pocket toward the “lid” loop (Fig. 6B). The dansyl moiety is highly sensitive to solvent polarity and serves as an excellent fluorescent probe for sensing subtle changes in the local microenvironment by significantly increased quantum yield and emission lifetime in a less polar environment (31). To avoid changes in the substrate concentration during experiments, an enzymatically inactive mutant of pNth11–751(E674A) was chosen for these experiments. A series of time-resolved experiments was performed with excitation at 355 and 290 nm, which correspond to the DANS-TRE and Trp absorption bands, respectively. The latter experiment explored the possibility of sensitized emission from bound DANS-TRE facilitated by Förster resonance energy transfer (FRET) from W309, W624, W630, and W708, located near the binding site (Fig. 6B). These Trp donors are located within a sphere with a radius of 2R0 = 42 Å (32) (R0, critical Förster distance) from the bound DANS-TRE, which makes the FRET feasible. Because FRET efficiency is highly sensitive to the distance and mutual orientation of the donor-acceptor pair (33), Bmh1-induced conformational changes of pNth11–751(E674A) involving the region surrounding the binding site should affect FRET efficiency and DANS-TRE emission.
Fig. 6.
Time-resolved fluorescence experiments with DANS-TRE. (A) DANS-TRE. (B) Four tryptophan residues (W309, W624, W630, and W708) are located in close proximity to the substrate binding site of yeast Nth1. The molecule of trehalose (TRE) was modeled by superimposing the structure of Nth1153–751:TRE with Nth1-CD of Bmh1-bound pNth11–751. (C and D) Statistical confidence-interval analysis of intensity fraction of long fluorescence decay component corresponding to bound DANS-TRE in the presence of pNth11–751(E674A) (white), Bmh1 (light gray), and the pNth11–751(E674A):Bmh1complex (dark gray), with DANS-TRE excited at 290 (C) and 355 nm (D). Histograms represent the probability of recovering a particular intensity fraction from measured data as a result of 2,000 bootstrap fitting cycles (52). Dashed lines border 68% confidence intervals (1 SD). The histograms reveal that the intensity fraction of bound DANS-TRE is significantly higher in the presence of the pNth11–751(E674A):Bmh1complex compared with the control samples. The significantly increased bound-intensity fraction on excitation at 290 nm compared with excitation at 355 nm is likely a result of a combined effect of DANS-TRE FRET facilitated by Trp–DANS-TRE proximity in the binding pocket of the pNth11–751(E674A):Bmh1complex and blue shift of the absorption spectrum of bound DANS-TRE.
Fluorescence decays of DANS-TRE in the presence of pNth11–751(E674A), Bmh1, and the Bmh1:pNth11–751(E674A) complex excited at 290 nm can be found in SI Appendix, Fig. S5A and Table S2. As shown in SI Appendix, Table S2, DANS-TRE alone decays close to a single exponential with a lifetime near 3.3 ns with only minor contamination supposedly caused by impurities. After binding to the Bmh1:pNth11–751(E674A) complex, DANS-TRE decay becomes heterogeneous. Apart from the emission from free sugar, a new lifetime component of 14.3 ns appears. This could be attributed to a bound fraction of DANS-TRE sensing lower solvation and decreased microenvironmental polarity in the binding pocket of the complex. The value is consistent with values commonly found for the dansyl moiety covalently linked to globular proteins (34, 35). The fractional intensity of the 14.3-ns component in the Bmh1:pNth11–751(E674A) complex (4.2%) is significantly higher than in the presence of pNth11–751(E674A) (0.5%) or Bmh1 alone (1.3%), where the presence of the long component likely indicates some nonspecific interaction of DANS-TRE with Bmh1. Importantly, the intensity fraction from DANS-TRE bound to the complex is 2.3× higher than would result from the simple addition of corresponding intensities from pNth11–751(E674A) and Bmh1 that constitute the complex. Because both pNth11–751(E674A) and Bmh1:pNth11–751(E674A) bind TRE with comparable affinities (SI Appendix, Fig. S6), the observed increase is not caused by higher saturation of the complex and might be assigned to a Bmh1-induced conformational change of the pNth11–751(E674A) region surrounding the binding site. The effect is also visible from the raw decays presented in SI Appendix, Fig. S5A. The formation of the Bmh1:pNth11–751(E674A) complex is accompanied by a significant increase in the long-decaying component that is visibly above the noise level. The decay of DANS-TRE alone is omitted for clarity, as it is almost indistinguishable from the decay in the presence of pNth11–751(E674A) (SI Appendix, Fig. S5, green curve).
Fluorescence decays of DANS-TRE excited at 355 nm are presented in SI Appendix, Fig. S5B and Table S3. The observed effects qualitatively agree with the results for 290 nm excitation described earlier. As expected, the measured effect of Bmh1 is smaller without FRET sensitization, and its significance is also lower.
Finally, we rigorously examined the statistical confidence limits of changes extracted for the 14.3-ns intensity fraction with 290 nm (Fig. 6C) and 355 nm excitation (Fig. 6D). Histograms represent the probability of recovering a particular intensity fraction from the measured data resulting from 2,000 bootstrap fitting cycles (36). Dashed lines denote 68.2% confidence intervals (±1 SD). The histograms reveal that for both excitations, the intensity fraction of DANS-TRE bound to the Bmh1:pNth11–751(E674A) complex is significantly higher than in control samples. This further supports our conclusion that the binding of Bmh1 induces conformational changes of pNth11–751(E674A) involving the region close to the substrate binding site.
Discussion
The yeast Nth1, compared with trehalases from prokaryotic and higher eukaryotic organisms, exhibits distinct domain structure and regulation, as its activity is triggered by association with the 14-3-3 protein, which recognizes two phosphorylated motifs within the N-terminal extension of Nth1 (20–23). Previous studies have suggested an allosteric mechanism, whereby 14-3-3 protein binding affects the structure of the catalytic domain of pNth11–751, with Nth1-CaBD (the EF-hand-like motif-containing domain) being the intermediary between 14-3-3 and the catalytic domain (37, 38). Indeed, the crystal structure of the Bmh1:pNth11–751 complex reported in this study revealed that Bmh1 extensively interacts with both Nth1-CaBD and Nth1-CD and enables their proper 3D configuration relative to each other (Fig. 1). This, in turn, appears to facilitate interactions between the EF-hand-like motif of Nth1-CaBD and the flexible “lid” loop, which gets stabilized and completes the active site of pNth1 by providing side-chains important for catalysis. The suggested mechanism of Nth1 activation was corroborated by mutational analysis, which confirmed that residues involved in contacts between the EF-hand-like motif of Nth1-CaBD and the “lid” loop, as well as residues from the tip of the “lid” loop, are required for the activation of pNth1 (Fig. 5C and SI Appendix, Fig. S3). Moreover, time-resolved fluorescence measurements with DANS-TRE further corroborated the suggested mechanism by indicating conformational changes in the vicinity of the active site of pNth1 induced by Bmh1 binding (Fig. 6).
The interaction between pNth11–751 and Bmh1 was previously studied using hydrogen-deuterium exchange coupled to MS, chemical cross-linking, and small-angle X-ray scattering (SAXS) (37, 38). Although the observed changes in deuteration profiles of both pNth11–751 and Bmh1 are in a very good agreement with the crystal structure of the Bmh1:pNth11–751 complex (SI Appendix, Fig. S7A), the previously identified intermolecular cross-links appear to be incompatible with the conformation of the complex in the crystal (SI Appendix, Fig. S7B). However, these intermolecular cross-links were preferentially formed in the absence of calcium and involved Bmh1 residues K76, K127, and K145 and pNth1 residues K214, K393, and K563. In contrast, in the presence of calcium, the mentioned lysine residues of pNth1 preferentially formed intramolecular cross-links within the catalytic domain (cross-links K214–K563 and K258–K393). This might indicate that in the presence of calcium, the EF-hand-like motif of pNth1 is folded (Figs. 1B and 5C), thus providing optimal interactions with the catalytic domain with an accompanying increase in catalytic activity (SI Appendix, Fig. S3). In contrast, in the absence of calcium, when the region containing the EF-hand-like motif is likely unfolded, the interaction between the catalytic domain and the rest of the complex is less stable and the catalytic domain transiently interacts with other surfaces of Bmh1, thus bringing lysine residues to a distance enabling their cross-linking. However, another possibility for the formation of observed intermolecular cross-links might be the presence of weak transient interactions that resemble the crystal-packing contacts, as such interactions would bring Cα atoms of cross-linked lysine residues of Bmh1 and Nth1 at a distance of approximately 30 Å (SI Appendix, Fig. S7 C and D). As a consequence, the previously published structural model of Bmh1 with the catalytic domain of Nth1 is inaccurate (38). The model, which was based on homology models of Bmh1 and Nth1-CD (residues 295–721), distance restraints derived from chemical cross-links discussed earlier and the fitting into the low-resolution SAXS envelope, suggested a clearly incorrect orientation of Nth1-CD with respect to Bmh1. The superimposition of the Bmh1:pNth11–751 crystal structure with the previously published SAXS envelope is shown in SI Appendix, Fig. S8A. Because this SAXS envelope was obtained by refining the averaged bead model, and thus it might not represent a consensus model, we also compared the crystal structure of the complex with the averaged filtered SAXS envelope representing common features of all bead models used in shape reconstruction (39) (SI Appendix, Fig. S8B). The reasonable agreement of the averaged filtered SAXS envelope with the overall shape of the complex structure indicates similarity of the solution and crystal structures of the Bmh1:pNth11–751 complex.
Sequence alignment of neutral trehalases from four different yeast species with trehalases from selected prokaryotic and higher eukaryotic organisms revealed that yeast neutral trehalases possess distinct “lid” loop sequences that are longer and do not contain a Gly-rich region (SI Appendix, Fig. S4C, marked with the red box). Furthermore, only yeast sequences possess the N-terminal extension containing conserved PKA phosphorylation sites, which are also the 14-3-3 binding motifs (23), and the calcium-binding EF-hand-like motif (SI Appendix, Fig. S4B, marked with green, blue and orange boxes). In addition, residues involved in contacts between Nth1-CaBD and the “lid” loop (Q120, E124, R686, marked with red circles) and residues whose side-chains are involved in interactions with Bmh1 (SI Appendix, Fig. S4 B and C, marked with blue circles) are also highly conserved among yeast sequences. This indicates that all yeast neutral trehalases are regulated in the 14-3-3 protein-dependent manner.
Both 14-3-3 binding motifs of pNth1 do not contain a Pro residue at the +2 position relative to the phosphorylated residue (Fig. 2 A and B). This pSer +2 proline is frequently found in 14-3-3 binding motifs (26), and is required for introducing a kink in the peptide chain and thus directing the C-terminal portion back into the central channel (25). In the case of the first pNth1 motif, the change in the direction of the polypeptide chain is achieved by interactions between pNth1 residues at the +3 and +4 positions and Bmh1 (Fig. 2A), whereas in the case of the second motif, the kink is introduced by the intramolecular salt bridge between pNth1 residues at the −2 and +2 positions (Fig. 2B). As a result of these interactions, the C-terminal portions of both pNth1 motifs leave the binding grooves in similar orientation as polypeptide chains of other proteins and peptides including, for example, AANAT (9), the heat shock protein beta-6 (27), and short phosphopeptides (25, 28), that were cocrystalized with 14-3-3 and whose 14-3-3 binding motifs contain proline residue at the +2 position (Fig. 3). The observed similarities in the 3D structures of these 14-3-3 binding partners suggest that 14-3-3 proteins, which are highly conserved both in sequence and structure (40, 41), provide a highly conserved evolutionary force that affects the structures of many client proteins.
In conclusion, the structural analysis of yeast Nth1 alone and its complex with Bmh1, together with mutational analysis and activity and time-resolved fluorescence measurements, provides a structural basis for the 14-3-3 protein-mediated activation of Nth1. The data presented here not only highlight the ability of 14-3-3 to modulate the structure of a multidomain binding partner and to function as an allosteric effector of the bound enzyme but also suggest that the highly conserved nature of 14-3-3 proteins affects the structures of many client proteins.
Experimental Procedures
Protein Preparation and Purification.
DNA encoding full-length and two N-terminally truncated versions of S. cerevisiae Nth1 (residues 1−751, 100−751, and 153−751, respectively) were ligated into the pRSFDuet-1 vector (Novagen), using the NcoI, BamHI, and NotI sites. Nth11–751, Nth1100–751, and Nth1153–751 were expressed as N-terminal 6×His-tagged fusion proteins in E. coli BL21(DE3). Protein expressions were induced by isopropyl β-d-1-thiogalactopyranoside for 18 h at 25 °C, and the proteins were purified using Chelating Sepharose Fast Flow (GE Healthcare) according to a standard protocol. Proteins were dialyzed against buffer 1 [20 mM Tris⋅HCl, 150 mM NaCl, 10 mM EDTA, 2 mM β-mercaptoethanol, 10% (wt/vol) glycerol at pH 7.5]. The affinity tag and GB1 domain were removed by tobacco etch virus (TEV) protease cleavage for 2 h at 30 °C and overnight at 4 °C (250 U TEV/mg recombinant protein). After the cleavage, Nth11–751 was purified by cation-exchange chromatography (HiTrap SP column; GE Healthcare). The protein was eluted using a 50−1,000-mM gradient of NaCl in 50 mM citric buffer (pH 6.0), 2 mM DTT. Four hours before phosphorylation, fractions containing Nth11–751 were dialyzed against buffer 2 [20 mM Tris⋅HCl, 150 mM NaCl, 5 mM DTT, 10% (wt/vol) glycerol at pH 7.5]. Purified Nth11–751 was phosphorylated by incubation at 30 °C for 2 h and then overnight at 4 °C with 80 units PKA (Promega) per milligram protein in the presence of 0.75 mM ATP and 20 mM MgCl2. The final purification step was size-exclusion chromatography (Superdex 200; GE Healthcare) in buffer 2. With the Nth1100–751 and Nth1153–751 variants, the final purification step was size-exclusion chromatography (HiLoad Superdex 75; GE Healthcare) in buffer containing 1 mM ammonium acetate, 150 mM NaCl, 5 mM DTT, and 10% (wt/vol) glycerol at pH 7.3. Selenomethionine-substituted Nth1153–751 was expressed using SelenoMet Medium Base and SelenoMet Nutrient Mix (Molecular Dimensions) and purified according to the same protocol. All mutants were generated using a QuikChange site-directed mutagenesis kit (Stratagene), and mutations were confirmed by sequencing. The stability of prepared Nth1 mutants was checked by measuring the thermally induced protein denaturation, using differential scanning fluorimetry. No significant differences in the temperature of the unfolding transition (Tm) were observed for prepared Nth1 mutants (SI Appendix, Table S4).
Bmh1 (C-terminally truncated version composed of residues 1−236) from S. cerevisiae was expressed and purified as described previously (42). The N-terminal 6×His-tag was removed using TEV protease, and final size-exclusion chromatography was performed in a HiLoad Superdex 75 column (GE Healthcare).
Crystallization and X-Ray Data Collection.
All crystallizations were performed by the hanging-drop vapor-diffusion method at 291 K. Crystals of native and selenomethionine-labeled Nth1153–751 were grown from drops consisting of 2 μL of 100 μM protein and 1 μL of 100 mM sodium citrate (pH 5.6), 1 M lithium sulfate, and 400 mM ammonium sulfate. Crystals of the Nth1153–751:TRE complex were prepared by cocrystallization with 200 mM trehalose. Crystals of native Nth1100–751 were grown from drops consisting of 2 μL of 95 μM protein and 1 μL of 100 mM sodium citrate (pH 5.6), 1 M lithium sulfate, and 400 mM ammonium sulfate. Initial crystals of the pNth11–751:Bmh1 complex (pNth11–751 and Bmh1 were mixed in a 1:2 molar ratio) were obtained in condition #53 of the JCSG III screen (Qiagen) consisting of 0.16 M calcium acetate, 80 mM sodium cacodylate (pH 6.5), 14.4% (wt/vol) PEG 8000, and 20% (vol/vol) glycerol. This condition was further optimized by using the Additive Screen (Hampton Research). Diffraction-quality crystals were grown from drops consisting of 2 μL of 53 μM complex and 1 μL of 100 mM sodium cacodylate (pH 6.5), 200 mM calcium acetate, 18% (wt/vol) PEG 8000, and 12% (wt/vol) sucrose.
All crystals were cryoprotected using 30% (vol/vol) glycerol and flash frozen in liquid nitrogen before data collection in oscillation mode at beamlines P13 of the DESY synchrotron and 14.1 of the BESSY synchrotron. Diffraction data processing was carried out with the packages XDS and XDSAPP (43, 44). The crystal structure of the pNth11–751:Bmh1 complex was solved by molecular replacement in MOLREP (45), using the structures of Nth1153–751 (described here) and PDB 2BR9 (24) as search models, and refined at a resolution of 2.29 Å with PHENIX (46). Data collection and refinement statistics are summarized in SI Appendix, Table S1. No electron density was present for residues 1−26, 38−53, and 437−447 of pNth1, and residues 209−215 of one Bmh1 protomer, and these residues were not included in the final model. The refined model had a MolProbity (47) score of 1.35.
The crystal structure of Nth1153–751 was solved by single-wavelength anomalous dispersion phasing with the data from a selenomethionine-labeled protein crystal diffracting to a resolution 3.13 Å. Single-wavelength anomalous dispersion phasing and initial model building were performed with PHENIX (46). The initial model consisting of residues 180−751 was then used to determine the Nth1153–751 structure by molecular replacement of a higher-resolution native dataset (SI Appendix, Table S1), using MOLREP (45). The structure of apo Nth1153–751 was refined at 2.72 Å, using REFMAC (48), and the refined model had a MolProbity (47) score of 2.13. No electron density was present for residues 153−178 and 680−701, and these residues were not included in the final model.
The Nth1153–751:TRE and Nth1100–751 structures were solved by molecular replacement in MOLREP (45), using the structure of Nth1153–751 as the search model and refined at 2.9 and 3.15 Å, respectively, using PHENIX (46) (SI Appendix, Table S1). The refined models had MolProbity (47) scores of 1.55 and 1.99, respectively. No electron density was present for residues 153−179, 613−620, and 685−700 in the Nth1153–751:TRE structure and residues 100−179, 617−618, and 683−702 in the Nth1100–751 structure, and these residues were not included in the final model. All structural figures were prepared with PyMOL (https://pymol.org/2/).
Enzyme Activity Measurements.
The trehalase activity of Nth1 and its mutants were measured by estimating the glucose produced by the hydrolysis of trehalose, using a stopped assay, as described earlier (23, 49). The assay was performed at 30 °C in buffer 3 [20 mM Tris⋅HCl, 150 mM NaCl, 10 mM CaCl2 (where needed), 10% (wt/vol) glycerol at pH 7.5] and 30 mM trehalose. The final concentrations of Nth1 and Bmh1 (where needed) were 100 nM and 15 μM, respectively. The specific trehalase activity of Nth1 was determined as micromol glucose liberated/min/mg protein.
Microscale Thermophoresis.
The microscale thermophoresis method was described in detail elsewhere (50). The binding affinities of uncomplexed and complexed pNth11–751(E674A) for trehalose were measured using the Monolith NT.LabelFree instrument (NanoTemper Technologies). The solution of trehalose was serially diluted from 250 mM to 8 μM in the presence of 1 μM Nth11–751(E674A) or 0.5 μM pNth11–751(E674A):Bmh1 complex (molar ratio 1:2). All samples were loaded into standard treated zero background capillaries (NanoTemper Technologies) after incubation at 23 °C for 20 min. Measurements were performed at 23 °C in buffer 3 by using 20% LED power and 40–60% IR-laser power. Three independent measurements were performed for each sample, and the obtained data were analyzed using Thermophoresis and the T-Jump signal, using the software MO.Affinity Analysis (version 2.1.2; NanoTemper Technologies).
Differential Scanning Fluorimetry.
The differential scanning fluorimetry experiments were performed using a real-time PCR LightCycler 480 II (Roche Applied Science) according to the standard protocol, as described previously (51). The pNth11–751 concentrations were 2 μM in buffer 3.
Synthesis of DANS-TRE.
The synthesis of DANS-TRE and all corresponding NMR spectra are described in SI Appendix, SI Material and Methods.
Time-Resolved Fluorescence Measurements and Data Analysis.
A full description of method for time-resolved fluorescence measurements and data analysis is available in SI Appendix, SI Material and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Czech Science Foundation (Projects 16-02739S and 17-00726S), the Czech Academy of Sciences (Research Projects RVO: 67985823 of the Institute of Physiology) and project BIOCEV (Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University in Vestec; CZ.1.05/1.1.00.02.0109), a project from the European Regional Development Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. F.D. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5N6N, 5JTA, 5M4A, and 5NIS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714491114/-/DCSupplemental.
References
- 1.Wang W, Shakes DC. Molecular evolution of the 14-3-3 protein family. J Mol Evol. 1996;43:384–398. doi: 10.1007/BF02339012. [DOI] [PubMed] [Google Scholar]
- 2.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 3.Freed E, Symons M, Macdonald SG, McCormick F, Ruggieri R. Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science. 1994;265:1713–1716. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- 4.Ganguly S, et al. Role of a pineal cAMP-operated arylalkylamine N-acetyltransferase/14-3-3-binding switch in melatonin synthesis. Proc Natl Acad Sci USA. 2001;98:8083–8088. doi: 10.1073/pnas.141118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichimura T, Isobe T, Okuyama T, Yamauchi T, Fujisawa H. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+, calmodulin-dependent protein kinase II. FEBS Lett. 1987;219:79–82. doi: 10.1016/0014-5793(87)81194-8. [DOI] [PubMed] [Google Scholar]
- 6.Banik U, Wang GA, Wagner PD, Kaufman S. Interaction of phosphorylated tryptophan hydroxylase with 14-3-3 proteins. J Biol Chem. 1997;272:26219–26225. doi: 10.1074/jbc.272.42.26219. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc Natl Acad Sci USA. 1999;96:8511–8515. doi: 10.1073/pnas.96.15.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumagai A, Dunphy WG. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell. 2001;105:257–267. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- 10.Ottmann C, et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol Cell. 2007;25:427–440. doi: 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Yip-Schneider MT, et al. Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem J. 2000;351:151–159. doi: 10.1042/0264-6021:3510151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng CY, et al. Mitotic and G2 checkpoint control: Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 13.Taoka K, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- 14.Crowe JH, Crowe LM, Chapman D. Preservation of membranes in anhydrobiotic organisms: The role of trehalose. Science. 1984;223:701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- 15.Avonce N, Leyman B, Thevelein J, Iturriaga G. Trehalose metabolism and glucose sensing in plants. Biochem Soc Trans. 2005;33:276–279. doi: 10.1042/BST0330276. [DOI] [PubMed] [Google Scholar]
- 16.Wilson WA, et al. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev. 2010;34:952–985. doi: 10.1111/j.1574-6976.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uno I, Matsumoto K, Adachi K, Ishikawa T. Genetic and biochemical evidence that trehalase is a substrate of cAMP-dependent protein kinase in yeast. J Biol Chem. 1983;258:10867–10872. [PubMed] [Google Scholar]
- 18.Kopp M, Müller H, Holzer H. Molecular analysis of the neutral trehalase gene from Saccharomyces cerevisiae. J Biol Chem. 1993;268:4766–4774. [PubMed] [Google Scholar]
- 19.Franco A, et al. A role for calcium in the regulation of neutral trehalase activity in the fission yeast Schizosaccharomyces pombe. Biochem J. 2003;376:209–217. doi: 10.1042/BJ20030825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wera S, De Schrijver E, Geyskens I, Nwaka S, Thevelein JM. Opposite roles of trehalase activity in heat-shock recovery and heat-shock survival in Saccharomyces cerevisiae. Biochem J. 1999;343:621–626. [PMC free article] [PubMed] [Google Scholar]
- 21.Panni S, Landgraf C, Volkmer-Engert R, Cesareni G, Castagnoli L. Role of 14-3-3 proteins in the regulation of neutral trehalase in the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:53–63. doi: 10.1111/j.1567-1364.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- 22.Schepers W, Van Zeebroeck G, Pinkse M, Verhaert P, Thevelein JM. In vivo phosphorylation of Ser21 and Ser83 during nutrient-induced activation of the yeast protein kinase A (PKA) target trehalase. J Biol Chem. 2012;287:44130–44142. doi: 10.1074/jbc.M112.421503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veisova D, et al. Role of individual phosphorylation sites for the 14-3-3-protein-dependent activation of yeast neutral trehalase Nth1. Biochem J. 2012;443:663–670. doi: 10.1042/BJ20111615. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci USA. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rittinger K, et al. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 26.Johnson C, et al. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sluchanko NN, et al. Structural basis for the interaction of a human small heat shock protein with the 14-3-3 universal signaling regulator. Structure. 2017;25:305–316. doi: 10.1016/j.str.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molzan M, et al. Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Mol Cell Biol. 2010;30:4698–4711. doi: 10.1128/MCB.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenreichova A, Klima M, Boura E. Crystal structures of a yeast 14-3-3 protein from Lachancea thermotolerans in the unliganded form and bound to a human lipid kinase PI4KB-derived peptide reveal high evolutionary conservation. Acta Crystallogr F Struct Biol Commun. 2016;72:799–803. doi: 10.1107/S2053230X16015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson RP, et al. Molecular basis for trehalase inhibition revealed by the structure of trehalase in complex with potent inhibitors. Angew Chem Int Ed Engl. 2007;46:4115–4119. doi: 10.1002/anie.200604825. [DOI] [PubMed] [Google Scholar]
- 31.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd Ed. Springer; New York: 2006. p. 954. [Google Scholar]
- 32.Valeur B. Molecular Fluorescence: Principles and Applications. Wiley-VCH; New York: 2001. p. 402. [Google Scholar]
- 33.Förster Th. Zwischenmolekulare energiewanderung und fluoreszenz. Ann Phys. 1948;437:55–75. [Google Scholar]
- 34.Silhan J, et al. 14-3-3 protein masks the DNA binding interface of forkhead transcription factor FOXO4. J Biol Chem. 2009;284:19349–19360. doi: 10.1074/jbc.M109.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rezabkova L, et al. Structural basis for the 14-3-3 protein-dependent inhibition of the regulator of G protein signaling 3 (RGS3) function. J Biol Chem. 2011;286:43527–43536. doi: 10.1074/jbc.M111.273573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davison AC, Hinkley DV. Bootstrap Methods and Their Application. Cambridge Univ Press; Cambridge, UK: 1997. [Google Scholar]
- 37.Macakova E, et al. Structural basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1. Biochim Biophys Acta. 2013;1830:4491–4499. doi: 10.1016/j.bbagen.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 38.Kopecka M, et al. Role of the EF-hand-like motif in the 14-3-3 protein-mediated activation of yeast neutral trehalase Nth1. J Biol Chem. 2014;289:13948–13961. doi: 10.1074/jbc.M113.544551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Heusden GP. 14-3-3 proteins: Regulators of numerous eukaryotic proteins. IUBMB Life. 2005;57:623–629. doi: 10.1080/15216540500252666. [DOI] [PubMed] [Google Scholar]
- 41.Obsil T, Obsilova V. Structural basis of 14-3-3 protein functions. Semin Cell Dev Biol. 2011;22:663–672. doi: 10.1016/j.semcdb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Veisova D, et al. The C-terminal segment of yeast BMH proteins exhibits different structure compared to other 14-3-3 protein isoforms. Biochemistry. 2010;49:3853–3861. doi: 10.1021/bi100273k. [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sparta KM, Krug M, Heinemann U, Mueller U, Weiss MS. XDSAPP2.0. J Appl Crystallogr. 2016;49:1085–1092. [Google Scholar]
- 45.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 46.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Pernambuco MB, et al. Glucose-triggered signalling in Saccharomyces cerevisiae: Different requirements for sugar phosphorylation between cells grown on glucose and those grown on non-fermentable carbon sources. Microbiology. 1996;142:1775–1782. doi: 10.1099/13500872-142-7-1775. [DOI] [PubMed] [Google Scholar]
- 50.Jerabek-Willemsen M, et al. MicroScale thermophoresis: Interaction analysis and beyond. J Mol Struct. 2014;1077:101–113. [Google Scholar]
- 51.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 52.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman and Hall; New York: 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.