Significance
The formation of infectious HIV-1 virions is triggered by sequential proteolysis of the group-specific antigen (Gag) polyprotein by the viral protease. Using chemical exchange-based NMR spectroscopy, we explore, in the context of Gag polyprotein, the formation of productive complexes between protease and Gag cleavage sites located within the intrinsically disordered linkers that connect the globular domains of Gag. We show that the ability of protease to sequentially cleave distinct Gag cleavage sites, which display little to no sequence identity, primarily originates from conformational dynamics of the protease flaps that cover the active site. The protease flaps are actively involved in substrate recognition and regulate the lifetime of productive complexes, allowing protease to differentiate between cognate and noncognate sequences.
Keywords: invisible states, relaxation dispersion, chemical exchange saturation transfer, substrate selection, conformational dynamics
Abstract
The conversion of immature noninfectious HIV-1 particles to infectious virions is dependent upon the sequential cleavage of the precursor group-specific antigen (Gag) polyprotein by HIV-1 protease. The precise mechanism whereby protease recognizes distinct Gag cleavage sites, located in the intrinsically disordered linkers connecting the globular domains of Gag, remains unclear. Here, we probe the dynamics of the interaction of large fragments of Gag and various variants of protease (including a drug resistant construct) using Carr−Purcell−Meiboom−Gill relaxation dispersion and chemical exchange saturation transfer NMR experiments. We show that the conformational dynamics within the flaps of HIV-1 protease that form the lid over the catalytic cleft play a significant role in substrate specificity and ordered Gag processing. Rapid interconversion between closed and open protease flap conformations facilitates the formation of a transient, sparsely populated productive complex between protease and Gag substrates. Flap closure traps the Gag cleavage sites within the catalytic cleft of protease. Modulation of flap opening through protease−Gag interactions fine-tunes the lifetime of the productive complex and hence the likelihood of Gag proteolysis. A productive complex can also be formed in the presence of a noncognate substrate but is short-lived owing to lack of optimal complementarity between the active site cleft of protease and the substrate, resulting in rapid flap opening and substrate release, thereby allowing protease to differentiate between cognate and noncognate substrates.
Conversion of HIV-1, as well as other retroviruses, from immature viral particles to infectious virions is triggered by sequential proteolytic cleavage of the group-specific antigen (Gag) polyprotein catalyzed by a dimeric viral aspartyl protease (1, 2). Hence, HIV-1 protease inhibitors constitute an integral component of current anti-HIV combination therapies (3). HIV-1 protease (PR) is a 99-residue symmetric dimer comprising a β-strand−rich core, a catalytic site centered on the active site residue Asp25, and glycine-rich flexible flaps that form a lid over the catalytic site (1). The Gag polyprotein comprises three ordered domains, matrix (MA), capsid (CA), and nucleocapsid (NC), connected by linkers and organized as follows: MA–CA–spacer peptide 1 (SP1)–NC–spacer peptide 2 (SP2)–p6. The MA|CA linker, SP1, SP2, and p6 are intrinsically disordered in solution such that the ordered domains behave like beads on a string (4–7).
The five cleavage sites within Gag are hydrolyzed at distinct rates by PR, generating a characteristic Gag cleavage pattern whose rates are ordered as follows: SP1|NC > SP2|p6 ≈ MA|CA > CA|SP1 ≈ NC|SP2 (5, 8–11). The underlying mechanism governing ordered Gag cleavage by PR is unclear. Further, there is little to no sequence identity between the different Gag cleavage sites, making it difficult to assess the determinants of PR substrate specificity or to derive a generalized consensus sequence recognized by PR. The precise control of Gag proteolysis and the apparent promiscuous substrate specificity of PR are seemingly paradoxical. One current explanation lies in the “substrate envelope” hypothesis (12), based on crystal structures of inactive PR variants in complex with peptide substrates (12–14), which posits that PR recognizes the 3D conformation of Gag cleavage sites rather than their primary sequences. As the Gag polyprotein is refractory to crystallization, all crystallographic work on PR−Gag interactions has been carried out using peptide analogs. Such peptides, however, are poor substitutes for the Gag polyprotein, since there are large differences in proteolysis rates of peptides and Gag under identical experimental conditions (6, 15). The latter observations imply the presence of additional factors governing PR−Gag interactions, as well as a role for the ordered domains of Gag. Indeed, paramagnetic relaxation enhancement (PRE) measurements have revealed the existence of transient (lifetimes ≤ 250 μs to 500 μs), sparsely populated encounter complexes between PR and the ordered domains of Gag that may serve to efficiently guide PR to the Gag cleavage junctions (6). Details, however, regarding the formation of productive complexes between Gag and PR have remained elusive.
Here we make use of Carr−Purcell−Meiboom−Gill (CPMG) relaxation dispersion and chemical exchange saturation transfer (CEST) experiments to probe the association of PR with Gag cleavage sites on the millisecond time scale in the context of large fragments of Gag comprising MA−CA−SP1−NC and CA−SP1−NC. We show that the time scale of opening and closing of the PR flaps is modulated by PR−Gag interactions, allowing PR to fine-tune the lifetime of the productive complex and hence differentiate between cognate and noncognate Gag cleavage sites.
Results
Recombinant Gag and PR Constructs.
The current work made use of three engineered monomeric Gag constructs, each carrying a double mutation at the CA dimerization interface (W316A/M317A): a larger Gag fragment consisting of the MA, CA, SP1, and NC domains (Fig. 1A), hereafter referred to as (Group M, HXB2 strain, residues 1 to 432, ∼48 kDa); a smaller fragment comprising the CA, SP1, and NC domains, (Group M, pLN43 strain, residues 133 to 432, ∼33 kDa); and a variant of the latter containing two point mutations at the CA|SP1 (L363I) and SP1|NC (M377I) cleavage sites, denoted as . The cleavage pattern and proteolysis rates obtained for monomeric are very similar to those obtained using wild-type ΔGag, which exists in a monomer−dimer equilibrium in solution (5, 6), indicating that Gag proteolysis and hence Gag−PR association are independent of Gag oligomerization state.
Fig. 1.
Kinetic scheme for PR−Gag binding. (A) Schematic representation of architecture. Amino acid sequences of the MA|CA, CA|SP1, and SP1|NC junctions are shown with the sites of cleavage indicated by dashed lines and scissors, and the sequential order of proteolysis indicated by numbers (Fig. S3A). NTD and CTD refer to the N- and C-terminal domains of CA, respectively; N-Zn and C-Zn refer to N- and C-terminal Zn knuckles of NC, respectively. (B) General kinetic scheme for PR−Gag association (also see Fig. S1A). Schematics of the PR dimer and Gag are shown in blue and red, respectively. The species colored in blue (PRclosed) and red (free Gag) are the NMR visible major states, while those in gray are not considered explicitly in the models used to fit the experimental CEST/CPMG data. The asterisk for the bound species signifies that the symmetry of the PR dimer is broken in the bound state (see Overall Kinetic Scheme for PR–Gag Binding). For analysis of the CPMG relaxation dispersion and CEST data, the overall kinetic scheme depicted in B can be simplified to (C) a pseudo three-state model from the perspective of PR (as the chemical shifts of the two subunits are no longer identical in the bound state) and (D) a two-state model for Gag (see Supporting Information for further details).
Cleavage reactions were carried out using protease from HIV-1 Group O denoted as PR, and a multidrug-resistant protease bearing 20 mutations from HIV-1 group M denoted as PR20 (16, 17). The NMR experiments made use of the corresponding inactive variants, PRD25N and PR20D25N, respectively, in which the active site Asp25 was substituted for Asn.
Overall Kinetic Scheme for PR−Gag Binding.
The most general overall scheme for the binding of PRD25N to Gag is shown in Fig. 1B (and Fig. S1A). The flaps of PR that cover the active site adopt a range of conformations in crystal structures: open, semiopen, and closed (1). In solution, however, NMR residual dipolar coupling data indicate that the flaps of PRD25N are predominantly closed (PRclosed) (6, 18) (see also Fig. S2). The exchange lifetime (τex = 1/kex) between the major closed (PRclosed) and minor open (PRopen) states is ≤80 μs at 20 °C as deduced from measurement of backbone 1H and 15N transverse relaxation rates as a function of effective rotating frame field (19). The active site of PR is not accessible to substrate in the closed flap conformation, and therefore the initial binding event of PRD25N to Gag cleavage sites must involve PRopen to generate an initial asymmetric PRopen*−Gag complex (where the asterisk denotes that the two subunits of PR are no longer symmetric with regard to the bound substrate). The flaps in crystal structures of PR−peptide complexes are in the closed state with numerous interactions between the bound peptide and the flaps, as expected for productive complexes (1). One can therefore surmise that the flaps are also closed in the end-state productive PR*−Gag complexes in solution.
No 15N CPMG dispersions are observed for free PRD25N at 30 °C, the conditions used here, indicating that the interconversion rate between the closed (major) and open (minor) states is faster than the limit of detection of CPMG experiments (∼100 μs). The generalized scheme in Fig. 1B can therefore be simplified to exchange between free and bound states, represented by pseudo three-state (Fig. 1C) or two-state (Fig. 1D) models when monitoring the binding of isotopically labeled PRD25N or Gag, respectively. [The pseudo three-state model in the context of PR arises because of asymmetry in the bound state such that the chemical shifts for the two PR subunits are different (20); see Supporting Information]. The binding kinetics in the simplified “two species” scheme are governed by an overall association rate constant (given by , where is the fitted pseudo first-order association rate constant and [S] is the concentration of the free unlabeled species), and an overall dissociation rate . In the context of the full binding scheme shown in Fig. 1B (and Fig. S1A), ≈ (k1/k−1) (k2k3)/(k−2 + k3) under conditions where ; and = k−2k−3/(k−2 + k3) (Supporting Information).
The PRD25N− Complex.
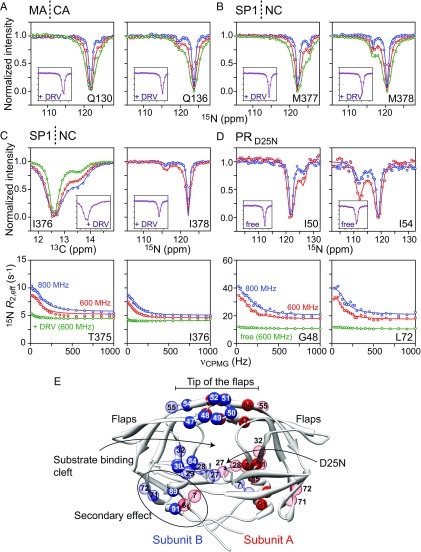
The kinetics of PRD25N binding to the cleavage sites within were investigated by analysis of 15N CEST profiles (21) recorded on 2H/15N-labeled (Fig. 2 A and B). The CEST experiment probes sparsely populated states (as low as ∼0.5%) that are in exchange with the major species on a time scale of 2 ms to 50 ms (22). When the exchanging species have significantly different chemical shifts, application of a weak radiofrequency (RF) field at the resonance position of the minor state results in a loss in intensity of the corresponding resonance of the major species. Fitting CEST profiles, generated by constant wave saturation at a series of frequency offsets with several RF field strengths, to the Bloch−McConnell equations (23) yields kinetic rate constants, populations, and chemical shifts of the minor (bound) state. CEST effects are observed at the MA|CA (Fig. 2A) and SP1|NC junctions (Fig. 2B) which are abolished upon addition of the protease inhibitor darunavir (Fig. 2 A and B, Insets). The data are well fit to a two-site exchange model (Fig. 1D, Table 1, and Table S1). Exchange at the SP1|NC site is about 3 times faster than at the MA|CA site, with values of ∼93 and ∼35 s−1, respectively; under the experimental conditions used here (200 μM 15N-labeled and 50 μM protease dimer), the population of bound to PRD25N at either cleavage site is ∼2.5%, corresponding to values of ∼5 × 104 and ∼2 × 104 M−1⋅s−1, respectively (Table 1). The values of are at least two orders of magnitude lower than those expected for a diffusion-limited association rate constant, in part due to the low population of the open, binding-competent conformation of PRD25N (∼k1/k−1) and possibly due to the dissociation rate constant k−2 in the Fig. 1B scheme being significantly larger than the rate constant k3 for the conversion from bound-open to bound-closed states of PRD25N (such that the expression for is reduced to ∼k1k2k3/k−1k−2; see Supporting Information). As a result, the overall equilibrium dissociation constant () at both the SP1|NC and MA|CA sites is ∼2 mM. No CEST effects are observed at the CA|SP1 cleavage site, indicating that occupancy at the CA|SP1 site is below the limit of detection of the CEST experiment (≤0.5%) consistent with the order of magnitude slower cleavage rate at the CA|SP1 site compared with the SP1|NC and MA|CA sites (Fig. S3A).
Fig. 2.
Kinetic analysis of PR−Gag binding. Examples of 15N CEST profiles observed for 200 μM 2H/15N-labeled in the presence of 50 μM unlabeled PRD25N dimer at the (A) MA|CA and (B) SP1|NC junctions obtained with the constant wave (CW) saturation field applied for TCEST = 200 ms at three RF field strengths (15, 30, and 50 Hz in blue, red, and green, respectively). Insets show the corresponding CEST profiles (TCEST = 200 ms, RF field = 30 Hz) in the presence of the PR inhibitor darunavir (DRV). Examples of CEST (Top) and 15N CPMG relaxation dispersion (Bottom) profiles obtained for (C) 200 μM 2H/15N/13Cmethyl-labeled CA−SP1− in the presence of 50 μM unlabeled PRD25N dimer and (D) 75 μM 2H/15N-labeled PRD25N dimer in the presence of 200 μM unlabeled CA−SP1−. For the 13Cmethyl CEST data, TCEST = 300 ms with RF field strengths of 15 Hz (green) and 30 Hz (blue), and TCEST = 200 ms with an RF field strength of 30 Hz (red). For the 15N CEST, TCEST = 200 ms with RF field strengths of 15 Hz (blue) and 30 Hz (red). Insets in Top of C and D show control CEST profiles in the presence of DRV and for PRD25N alone, respectively. The 15N CPMG relaxation dispersion profiles (Bottom) were recorded at 1H spectrometer frequencies of 600 MHz (red) and 800 MHz (blue). Control relaxation dispersions at 600 MHz obtained in the presence of DRV (C, Bottom) and for free PRD25N (D, Bottom) are shown in green. The experimental data in A–D are displayed as circles, and the solid lines represent the global best fits to the data using the schemes in Fig. 1 C and D for PRD25N and Gag, respectively (Table 1, Table S1, and SI Materials and Methods). For the control data, the green lines are used to guide the eye. (E) Residues of PRD25N that exhibit significant changes in backbone 15N chemical shifts (1 ppm to 3 ppm, semitransparent red and blue; 3 ppm to 8 ppm, solid red and blue) upon binding Gag displayed on a ribbon diagram of the structure of PRD25N (6).
Table 1.
Kinetic parameters for PR−Gag interactions derived from the fits to the CEST and CPMG relaxation dispersion data
| Complex* | % bound* | , s−1 | ×104, M−1⋅s−1 | , s−1 | KD,† mM |
| PRD25N + | |||||
| (SP1|NC site) | 2.5 ± 0.3 | 2.4 ± 0.1 | 4.8 ± 0.2 | 93 ± 12 | 1.9 ± 0.3 |
| (MA|CA site) | 2.2 ± 1.0 | 0.8 ± 0.1 | 1.6 ± 0.2 | 35 ± 15 | 2.2 ± 1.0 |
| PRD25N + CA−SP1− | |||||
| CA−SP1−‡ | 1.5 ± 0.1 | 4.7 ± 0.3 | 9.4 ± 0.6§ | 315 ± 8¶ | 3.4 ± 0.2 |
| PRD25N | 4.1 ± 0.1 | 13.5 ± 0.3 | 6.8 ± 0.2 | 315 ± 8¶ | 4.6 ± 0.2 |
| PRD25N + CA−SP1− | |||||
| PRD25N | 0.9 ± 0.3 | 9.9 ± 3.0 | 5.0 ± 1.5 | 1,120 ± 130 | 23 ± 7 |
| PR20D25N + CA−SP1− | |||||
| CA−SP1−‡ | 0.3 ± 0.1 | 3.2 ± 0.1 | 6.4 ± 0.2§ | 1,190 ± 40¶ | 19 ± 1 |
| PR20D25N | 1.2 ± 0.1 | 14.1 ± 1.5 | 7.1 ± 0.8 | 1,190 ± 40¶ | 17 ± 2 |
| PR20D25N + CA−SP1− | |||||
| PR20D25N# | 0.3 ± 0.1 | 4.5 ± 1.0 | 2.3 ± 0.5 | 1,330 ± 170 | 58 ± 15 |
NMR experiments were carried out at 30 °C in 20 mM sodium phosphate, pH 6.5, 0.1 mM ZnCl2, and 1 mM TCEP. For solubility considerations, the concentration of NaCl used for the ΔGag and CA−SP1−NC constructs was 300 and 50 mM, respectively. Protein concentrations were as follows: 200 μM isotopically labeled Gag + 50 μM unlabeled PR dimer variants, and 75 μM isotopically labeled PR dimer + 200 μM nonlabeled Gag variants.
The PR–Gag complexes are indicated in bold, and the species listed below are the isotopically-labeled components detected in the NMR experiments; % bound is the population of the isotopically labeled bound species.
KD is given by . KD values cannot be determined directly from titration experiments owing to weak binding coupled to low protein solubility.
At the SP1|NC cleavage junction. No CEST or CPMG relaxation dispersion are observed at the CA|SP1 junction.
The total concentration of PRD25N dimer is used to calculate from .
The is a shared optimized parameter in the fits (see Supporting Information).
The rate constants for exchange between the major open and minor closed species in free PR20D25N (derived from 15N CPMG measurements) are 904 ± 44 s−1 for the closed-to-open transition (k1) and 7.0 ± 0.3 s−1 for the open-to-closed transition (k−1), corresponding to a population of 0.8 ± 0.1% for the closed state in free PR20D25N.
The PRD25N−CA−SP1− Complex.
To probe the interaction from the perspective of both PR and Gag, we made use of the CA−SP1− construct, which effectively only contains a single interaction site at the SP1|NC junction (since binding at the CA|SP1 site lies below the limit of detection; see above).
The catalytic efficiency of PR is known to increase with salt concentration (24). Proteolytic cleavage of CA−SP1− is an order of magnitude slower than that of (compare Fig. S3 A and B) due to the much lower ionic strength employed for the former (50 mM NaCl) than the latter (300 mM NaCl), necessitated by the different solubility properties of the two constructs which self-assemble into cones, tubes, and virus-like particles at high and low salt, respectively (4, 5). Slower proteolysis implies faster exchange rates, bringing the CA−SP1−/PRD25N system into the exchange regime amenable to both CEST (21) and CPMG relaxation dispersion (25) experiments (τex ≈ 0.1 ms to 10 ms).
Fig. 2C shows 15N and 13Cmethyl CEST and 15N CPMG relaxation dispersion profiles observed at the SP1|NC cleavage site for 200 μM 2H/15N/13Cmethyl-labeled CA−SP1− in the presence of 50 μM unlabeled PRD25N dimer. No CEST or CPMG relaxation dispersions are observed at the CA|SP1 junction, indicating that the occupancy at the CA|SP1 junction in complexes with PRD25N, under the experimental conditions employed (50 mM NaCl, 20 mM sodium phosphate, pH 6.5, 30 °C), lies below the limit of detection. Examples of corresponding profiles observed for 75 μM 2H/15N-labeled PRD25N dimer in the presence of 200 μM unlabeled CA−SP1− are shown in Fig. 2D. No CEST or CPMG relaxation dispersions are observed either for CA−SP1− when the PRD25N active site is occupied by darunavir or for PRD25N alone. Simultaneous fitting of all of the relevant CEST and CPMG relaxation dispersion data, as well as observable 15N exchange-induced shifts for PRD25N (Table 1 and Table S1) yields values of ≈ 300 s−1 and ≈ 7–9 × 104 M−1⋅s−1, together with the residue-specific values and signs for the differences in chemical shifts (Δϖ) between the exchanging species (see SI Materials and Methods for details). Significant (>1 ppm) |15N Δϖ| values are observed for residues in and around the catalytic site of PRD25N, with the largest (3 ppm to 8 ppm) |15N Δϖ| values occurring in the flaps (residues 47 to 52 and 54) (Fig. 2E). Six residues (6, 7, 71, 72, 89, and 91) located at the lower outer edge of the PR structure in the view shown in Fig. 2E also display significant changes in backbone 15N chemical shifts (>1 ppm) upon CA−SP1− binding, and likely arise from secondary (allosteric) backbone conformational changes transmitted via residues located in the β-sheet constituting the base of the active site cleft (residues 27 to 32 and 84) (19).
Role of PR Flaps in Substrate Recognition.
To investigate the role of the PR flaps in substrate recognition, we made use of a monomeric Gag variant, CA−SP1−, containing point mutations in the CA|SP1 (L363I) and SP1|NC (M377I) cleavage junctions. This Gag construct is resistant to proteolysis by active PR (Fig. S3C), consistent with previous observations that a β-branched amino acid at the P1 position of the cleavage junction restricts PR-mediated hydrolysis (26). Practically no 15N CPMG relaxation dispersions were observed for 200 μM 2H/15N-labeled CA−SP1− in the presence of 50 μM unlabeled PRD25N dimer, either because the population of the bound state or the differences in chemical shifts between free and bound states are too small. However, several residues of the 2H/15N-labeled PRD25N dimer (75 μM) show significant 15N CPMG relaxation dispersions in the presence of 200 μM unlabeled CA−SP1− (Fig. 3A and Fig. S4 and Table S1), and two residues, I54 and I84 located in the flap and base of the active site cleft, respectively, display 15N CEST effects as well (Fig. S4). While the value of (∼5 × 104 M−1⋅s−1) is comparable to that obtained with CA−SP1−, (∼1,200 s−1) increases by about fourfold with a concomitant increase in KD (∼20 mM) (Table 1). From the scheme in Fig. 1B, we would predict that the increase in , given by k−2k−3/(k−2 + k3), can be largely attributed to an increase in k−3 and a decrease in k3, the rate constants for the conversion from closed to open and from open to closed flap conformations in the bound state of PRD25N, rather than any change in the dissociation rate constant k−2 from bound open conformation to free PRD25N (Supporting Information). The weaker binding, and hence lower occupancy of the bound state, is consistent with the reduced rate of proteolysis (Fig. S3C). The values of |15N Δϖmax| obtained with the two CA−SP1−NC variants are highly correlated (Fig. 3B), implying that the flaps of bound PRD25N are closed in both instances. (Note that |15N Δϖmax| is the larger of the two |15N Δϖ| values and therefore has the smallest errors; nondegeneracy of chemical shifts in the bound state is due to breaking of symmetry of the PR dimer upon substrate binding.)
Fig. 3.
Kinetic analysis of PRD25N−CA−SP1− binding. (A) Examples of 15N CPMG relaxation dispersions for 75 μM 2H/15N-labeled PRD25N dimer in the presence of 200 μM unlabeled CA−SP1− recorded at 600 MHz (red) and 800 MHz (blue). The experimental data are shown as circles, and the global best fits to a pseudo three-state exchange model (Fig. 1C) are shown as solid lines. Control relaxation dispersions at 600 MHz recorded on PRD25N alone are shown in green. (B) Correlation plot of the maximal (largest) 15N chemical shift differences, |15N Δϖmax|, between free and bound states of PRD25N in the presence of CA−SP1− (y axis) and CA−SP1− (x axis). (R is the Pearson correlation coefficient.)
Impact of PR Drug-Resistant Mutations.
To gain further insight into the impact of PR flap conformational dynamics on substrate recognition, we carried out 15N CPMG relaxation dispersion experiments using a multidrug-resistant PR variant from HIV-1 group M bearing 20 mutations (in addition to D25N), known as PR20D25N (16). At 300 mM NaCl, PR20 cleaves ΔGag approximately fourfold slower than the corresponding wild-type PR (15); at 50 mM NaCl, the rates of cleavage of CA−SP1− by PR and PR20 are comparable (Fig. S3B). In contrast to PRD25N in which the flaps are predominantly closed (6, 18) (Fig. S2A), the flaps of PR20D25N are largely open in both solution (18) (Fig. S2B) and crystal states (27). Exchange between open and closed states of PR20D25N (τex ≈ 1.1 ms; see Table 1) is an order of magnitude slower than for PRD25N (19), such that 15N CPMG relaxation dispersions are observed for free PR20D25N (with a population of ∼0.8% for the closed state), which have to be taken into account in the kinetic analysis of the relaxation dispersion data in the presence of CA−SP1− (Fig. 4A and Fig. S1B). Examples of 15N CPMG relaxation dispersions for 200 μM 2H/15N-labeled CA−SP1− in the presence of 50 μM unlabeled PR20D25N dimer, and of 75 μM 2H/15N-labeled PR20D25N in the presence of 200 μM CA−SP1− are shown in Fig. 4 B and C, respectively. Simultaneous fitting of all of the CPMG relaxation dispersion data (Table S1) using a two-state model for 2H/15N-labeled CA−SP1− and a pseudo four-state model for 2H/15N-labeled PR20D25N (Fig. S1B) yields ≈ 1,200 s−1, about 4 times faster than that with PRD25N (Table 1). The for the binding of PR20D25N to Gag is given by (k2k3)/(k−2 + k3) in the Fig. 4A scheme and has a value of ∼7 × 104 M−1⋅s−1 (Table 1), comparable to that obtained with PRD25N where ≈ (k1/k−1) (k2k3)/(k−2 + k3) (see Fig. 1B scheme). If we assume that the rate constants for association (k2) and dissociation (k−2) are approximately the same for PR20D25N (Fig. 4A) and PRD25N (Fig. 1B), this result implies that the rate of conversion (k3) from open to closed flap conformations of bound PR20D25N is slower than that for bound PRD25N by a factor approximately equal to the ratio of closed to open states of free PRD25N (), while the reverse process (k−3) is enhanced by about fourfold (Supporting Information).
Fig. 4.
Kinetic analysis of PR20D25N− CA−SP1− binding. (A) Overall kinetic scheme for PR20D25N−Gag association. The open flap conformation of bound PR20D25N (shown in gray) is not used explicitly in the simplified model (Fig. S1B) used to fit the experimental data. The arrows between the free open and closed states are shown in red to indicate that the exchange rate is reduced by an order of magnitude and the equilibrium is reversed relative to PRD25N. Examples of 15N CPMG relaxation dispersion profiles recorded at 600 MHz (red) and 800 MHz (blue) for (B) 200 μM 2H/15N-labeled CA−SP1− in the presence of 50 μM unlabeled PR20D25N and (C) 75 μM 2H/15N-labeled PR20D25N dimer in the presence of 200 μM unlabeled CA−SP1−. Reference dispersions recorded on (B) CA−SP1− and (C) PR20D25N alone are shown in green. The latter show small dispersions arising from exchange between the major open and minor closed flap conformations of PR20D25N. The experimental data are shown as circles, and the global best fits (Table 1 and Table S1) to two-state (for labeled CA−SP1− in the presence of PR20D25N, and for PR20D25N alone) and pseudo four-state (for labeled PR20D25N in the presence of CA−SP1−; see Fig. S1B) models are shown as continuous lines. (D) Correlation plot of |15N Δϖave| values between open free and bound states of PR20D25N (y axis) versus |15N Δϖ| values between open and closed states of free PR20D25N (x axis). Residues G51 and L54, depicted by open circles, do not display a correlation and are not included in the calculation of the correlation coefficient. (E) Examples of 15N CPMG relaxation dispersion profiles at 600 MHz (red) and 800 MHz (blue) for 75 μM 2H/15N-labeled PR20D25N in the presence of 200 μM unlabeled CA−SP1−. The experimental data are shown as circles, and the global best fits to pseudo four-state model are shown as solid lines. (F) Correlation plot of |15N Δϖmax| values between free open and bound states of PR20D25N in the presence of the two monomeric CA−SP1−NC variants.
The average chemical shift differences, |15N Δϖave|, between free open and bound states of PR20D25N, with the exception of two residues, are reasonably well correlated to the |15N Δϖ| values between the open (major) and closed (minor) states of free PR20D25N, with a slope of ∼1 (Fig. 4D), providing good evidence that the flaps of PR20D25N are predominantly closed in the presence of substrate. The absence of correlation for Gly51 and Leu54 suggests additional large contributions from interactions with the bound substrate for these two residues which are not averaged out in |15N Δϖave|.
The 15N CPMG relaxation dispersion experiments recorded on 75 μM 2H/15N-labeled PR20D25N in the presence of the proteolysis-resistant mutant CA−SP1− (Fig. 4E and Table S1) indicate that KD (∼60 mM) is increased about threefold with respect to that obtained with CA−SP1− (∼18 mM) due to a threefold reduction in from ∼7 × 104 M−1⋅s−1 to ∼2 × 104 M−1⋅s−1 (Table 1), presumably due to an increase in the dissociation rate constant k−2 (see Fig. 4A scheme). The |15N Δϖmax| values between the free and bound states of PR20D25N in the presence of the two CA−SP1−NC variants are highly correlated (Fig. 4F) indicating that the flaps of PR20D25N are likely closed in both cognate and noncognate productive complexes.
Concluding Remarks.
The current CEST and CPMG relaxation dispersion data, in combination with our previous intermolecular PRE experiments (6) and real-time NMR experiments used to follow proteolysis of Gag by PR (5), lead us to propose the following overall scheme for the various steps involved in PR−Gag association (Fig. 5). Transient encounter complexes with lifetimes ≤250 μs to 500 μs, detected by intermolecular PRE measurements, involve the globular domains of Gag and serve to guide PR toward the cleavage sites located in the linkers connecting the globular domains. The latter are clearly important, as PR cleaves intact Gag and peptides corresponding to Gag cleavage sites at very different rates (6, 15). The formation of productive PR−Gag complexes at the Gag cleavage sites occurs on a slower time scale (τex ≈ 3 ms to 30 ms), modulated by conformational exchange between open and closed flap conformations of PR, both in the free and bound states. Finally, proteolytic cleavage occurs on a much slower time scale of minutes to hours.
Fig. 5.
Schematic of the sequence of events involved in protease−Gag interactions. Sparsely populated encounter complexes, detected by intermolecular PRE measurements (6), are initially formed between Gag domains and PR, τex ≤ 250 μs to 500 μs. A productive complex is assembled through a series of interconnected steps (τex ≈ 3 ms to 30 ms) derived from CEST and CPMG relaxation dispersion measurements (current work); an expanded view is enclosed in a dashed square. Gag proteolysis occurs on the time scale of minutes to hours, amenable to real-time NMR (5). In the presence of Gag, the sparsely populated PROpen state forms an intermediate PRopen*−Gag complex on the path to the productive PRclosed*−Gag complex. With a noncognate substrate, a productive complex can still form, albeit with a significantly smaller population due to the lack of adequate complementarity between the PR and the substrate, resulting in rapid flap opening and, therefore, faster dissociation.
Exchange between the major closed and minor open flap conformations in free protease [PRD25N (6, 18), and Fig. S2A] is very rapid (τex ≤ 80 μs) (19) facilitating the formation of a productive Gag−PRclosed complex which can only occur via the initial formation of a Gag−PRopen complex. Flap opening in the bound state is substantially slower (τex ≈ 3 ms to 30 ms) due to a network of complementary intermolecular interactions between the flaps/catalytic cleft and the substrate. Lack of optimal complementarity, for example as a consequence of the introduction of a β-branched Ile side chain at the P1 position of Gag cleavage sites (as in the case of the CA−SP1−) (26), leads to faster dissociation due to enhanced flap opening in the bound state and subsequent substrate release, with a concomitant decrease in the rate of proteolysis (Fig. S3C). In the case of the drug-resistant PR mutant, PR20, which is catalytically less efficient than its wild-type counterpart (15, 16), the flaps are predominantly in the open conformation in the free state (18) (Fig. S2B), and the interconversion rate between the open and closed free states is comparable to the overall dissociation rate from Gag (Table 1), implying that trapping the substrate by virtue of flap closing plays a key role in substrate recognition.
Thus, rapid opening−closing of the flaps in the absence of substrate and modulation of flap conformational dynamics due to PR−substrate interactions are critical for both the promiscuity and precision of PR and other retroviral proteases. Fit complementarity and multiple intermolecular contacts between the substrate and the catalytic cleft and flaps of PR increase the lifetime of the productive complex, significantly increasing the likelihood of proteolysis; on the other hand, a reduction in these intermolecular interactions for a noncognate substrate leads to rapid substrate release, thereby allowing retroviral proteases to sample multiple binding partners in a short time span.
Materials and Methods
Protein Expression and Purification.
Full details of cloning, expression, site-directed mutagenesis, isotope labeling (2H/15N/13C, 2H/15N, and selective methyl labeling), and purification are provided in SI Materials and Methods. NMR samples were prepared in a buffer containing 20 mM sodium phosphate, pH 6.5, 0.1 mM ZnCl2, and 1 mM TCEP, with 300 and 50 mM NaCl for ΔGag and CA−SP1−NC constructs, respectively.
NMR Spectroscopy.
All heteronuclear NMR experiments were carried out at 30 °C on Bruker 600- and 800-MHz spectrometers equipped with z-gradient triple resonance cryoprobes. Full details of the CEST and CPMG experiments, NMR spectral processing, analysis, and data fits are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Ghirlando and A. Szabo for useful discussions; A. Aniana, J. Baber, and D. Garrett for technical assistance; and J. Lloyd of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Advanced Mass Spectrometry Core for technical support. Darunavir used in this study was obtained through the AIDS Reagent Program, Division of AIDS, National Institute of Allergies and Infectious Diseases, NIH. This work was supported by the intramural program of NIDDK/NIH and the AIDS Targeted Antiviral Program of the Office of the NIH Director (G.M.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716098114/-/DCSupplemental.
References
- 1.Konvalinka J, Kräusslich HG, Müller B. Retroviral proteases and their roles in virion maturation. Virology. 2015;479-480:403–417. doi: 10.1016/j.virol.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Freed EO. HIV-1 assembly, release and maturation. Nat Rev Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshmukh L, Ghirlando R, Clore GM. Investigation of the structure and dynamics of the capsid-spacer peptide 1-nucleocapsid fragment of the HIV-1 Gag polyprotein by solution NMR spectroscopy. Angew Chem Int Ed Engl. 2014;53:1025–1028. doi: 10.1002/anie.201309127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshmukh L, Ghirlando R, Clore GM. Conformation and dynamics of the Gag polyprotein of the human immunodeficiency virus 1 studied by NMR spectroscopy. Proc Natl Acad Sci USA. 2015;112:3374–3379. doi: 10.1073/pnas.1501985112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshmukh L, Louis JM, Ghirlando R, Clore GM. Transient HIV-1 Gag-protease interactions revealed by paramagnetic NMR suggest origins of compensatory drug resistance mutations. Proc Natl Acad Sci USA. 2016;113:12456–12461. doi: 10.1073/pnas.1615342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshmukh L, Schwieters CD, Grishaev A, Clore GM. Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR, X-ray scattering and protein engineering. ChemPhysChem. 2016;17:1548–1552. doi: 10.1002/cphc.201600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kräusslich HG, et al. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc Natl Acad Sci USA. 1989;86:807–811. doi: 10.1073/pnas.86.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ericksonviitanen S, et al. Cleavage of HIV-1 Gag polyprotein synthesized in vitro: Sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 10.Pettit SC, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiegers K, et al. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Substrate shape determines specificity of recognition for HIV-1 protease: Analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 13.Mahalingam B, Louis JM, Hung J, Harrison RW, Weber IT. Structural implications of drug-resistant mutants of HIV-1 protease: High-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins. 2001;43:455–464. doi: 10.1002/prot.1057. [DOI] [PubMed] [Google Scholar]
- 14.Tie Y, et al. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J. 2005;272:5265–5277. doi: 10.1111/j.1742-4658.2005.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis JM, Deshmukh L, Sayer JM, Aniana A, Clore GM. Mutations proximal to sites of autoproteolysis and the α-helix that co-evolve under drug pressure modulate the autoprocessing and vitality of HIV-1 protease. Biochemistry. 2015;54:5414–5424. doi: 10.1021/acs.biochem.5b00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dierynck I, et al. Binding kinetics of darunavir to human immunodeficiency virus type 1 protease explain the potent antiviral activity and high genetic barrier. J Virol. 2007;81:13845–13851. doi: 10.1128/JVI.01184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis JM, Aniana A, Weber IT, Sayer JM. Inhibition of autoprocessing of natural variants and multidrug resistant mutant precursors of HIV-1 protease by clinical inhibitors. Proc Natl Acad Sci USA. 2011;108:9072–9077. doi: 10.1073/pnas.1102278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roche J, Louis JM, Bax A. Conformation of inhibitor-free HIV-1 protease derived from NMR spectroscopy in a weakly oriented solution. ChemBioChem. 2015;16:214–218. doi: 10.1002/cbic.201402585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishima R, Freedberg DI, Wang YX, Louis JM, Torchia DA. Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Structure. 1999;7:1047–1055. doi: 10.1016/s0969-2126(99)80172-5. [DOI] [PubMed] [Google Scholar]
- 20.Cai M, et al. Solution NMR structure of the barrier-to-autointegration factor-Emerin complex. J Biol Chem. 2007;282:14525–14535. doi: 10.1074/jbc.M700576200. [DOI] [PubMed] [Google Scholar]
- 21.Vallurupalli P, Bouvignies G, Kay LE. Studying “invisible” excited protein states in slow exchange with a major state conformation. J Am Chem Soc. 2012;134:8148–8161. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 22.Vallurupalli P, Sekhar A, Yuwen T, Kay LE. Probing conformational dynamics in biomolecules via chemical exchange saturation transfer: A primer. J Biomol NMR. 2017;67:243–271. doi: 10.1007/s10858-017-0099-4. [DOI] [PubMed] [Google Scholar]
- 23.McConnell HM. Reaction rates by nuclear magnetic resonance. J Chem Phys. 1958;28:430–431. [Google Scholar]
- 24.Tropea JE, Nashed NT, Louis JM, Sayer JM, Jerina DM. Effect of salt on the kinetic parameters of retroviral and mammalian aspartic acid proteases. Bioorg Chem. 1992;20:67–76. [Google Scholar]
- 25.Palmer AG., 3rd Chemical exchange in biomacromolecules: Past, present, and future. J Magn Reson. 2014;241:3–17. doi: 10.1016/j.jmr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettit SC, et al. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 27.Agniswamy J, et al. HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements. Biochemistry. 2012;51:2819–2828. doi: 10.1021/bi2018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshmukh L, et al. Structure and dynamics of full-length HIV-1 capsid protein in solution. J Am Chem Soc. 2013;135:16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 30.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 32.Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 33.Lakomek NA, Ying J, Bax A. Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libich DS, Fawzi NL, Ying J, Clore GM. Probing the transient dark state of substrate binding to GroEL by relaxation-based solution NMR. Proc Natl Acad Sci USA. 2013;110:11361–11366. doi: 10.1073/pnas.1305715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 36.Bouvignies G, Kay LE. A 2D 13C-CEST experiment for studying slowly exchanging protein systems using methyl probes: An application to protein folding. J Biomol NMR. 2012;53:303–310. doi: 10.1007/s10858-012-9640-7. [DOI] [PubMed] [Google Scholar]
- 37.Guenneugues M, Berthault P, Desvaux H. A method for determining B1 field inhomogeneity. Are the biases assumed in heteronuclear relaxation experiments usually underestimated? J Magn Reson. 1999;136:118–126. doi: 10.1006/jmre.1998.1590. [DOI] [PubMed] [Google Scholar]
- 38.Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B. 2008;112:5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- 39.Katoh E, et al. A solution NMR study of the binding kinetics and the internal dynamics of an HIV-1 protease-substrate complex. Protein Sci. 2003;12:1376–1385. doi: 10.1110/ps.0300703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clore GM, Garrett D. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J Am Chem Soc. 1999;121:9008–9012. [Google Scholar]
- 41.Schwieters CD, Bermejo GA, Clore GM. Xplor-NIH for molecular structure determination from NMR and other data sources. Protein Sci. 2017 doi: 10.1002/pro.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 43.Schur FK, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 44.Wagner JM, et al. Crystal structure of an HIV assembly and maturation switch. Elife. 2016;5:e17063. doi: 10.7554/eLife.17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.