Abstract
Background
Compared with normal postmenopausal women, estrogen deficiency and hyperglycemia in postmenopausal women with type 2 diabetes (T2DM) lead to more severe bone property degradation. Liraglutide, a glucagon-like peptide-1 (GLP-1) receptor agonist, has been reported to improve bone condition among people with T2DM but the precise mechanisms remain unclear. Exosomes work as mediators in cell-to-cell communication, delivering functional miRNAs between cells. We aimed to explore the role of exosomes in T2DM-related bone metabolic disorders and the bone protective mechanisms of liraglutide.
Material/Methods
We made comparative analyses of bone marrow-derived exosomal miRNAs from ovariectomized (OVX) control rats, OVX + T2DM rats, and OVX + T2DM + liraglutide-treated rats. miRNA profiles were generated using high-throughput sequencing. Target gene prediction and pathway analysis were performed to investigate the signal pathway alterations. Three miRNAs were randomly chosen to validate their absolute expression levels by real-time quantitative PCR.
Results
Bone marrow-derived exosomal miRNAs were different with respect to miRNA numbers, species, and expression levels. miRNA spectra varied under T2DM condition and after liraglutide treatment. By bioinformatics analysis, we found T2DM and liraglutide administration lead to significant changes in exosomal miRNAs which targeted to insulin secretion and insulin-signaling pathway. Wnt signaling pathway alteration was the critical point regarding bone metabolism.
Conclusions
Our findings show the selective packaging of functional miRNA cargoes into exosomes due to T2DM and liraglutide treatment. Bone marrow exosome-mediated Wnt signaling pathway alteration may play a part in the bone protective effect of liraglutide.
MeSH Keywords: Diabetes Mellitus, Type 2; Exosomes; Glucagon-Like Peptide 1; MicroRNAs; Postmenopause
Background
Postmenopausal women with type 2 diabetes (T2DM) are a group of growing concern. Uncontrolled hyperglycemia combined with estrogen deficiency make their bone metabolic situation even worse. Postmenopausal women with T2DM showed more severe problems in bone quality [1]. This phenomenon can be explained partly by the accumulation of advanced glycation end products (AGEs), vitamin D deficiency, and increased risk of falls due to diabetic neuropathy, retinopathy, and hypoglycemia, but the specific details are unclear [2–4]. Glucagon-like peptide-1 (GLP-1) is an endogenous hormone secreted by intestinal L cells and regulates blood glucose via glucose-dependent insulin secretion [5]. Degraded by dipeptidyl peptidase (DPP)-4, GLP-1 loses its bioactivity rapidly, which makes native GLP-1 unsuitable for T2DM treatment. Liraglutide is a long-acting GLP-1 receptor agonist that has 97% overlapped of amino acid sequence with native GLP-1 [6]. Its amino acid sequence is identical to GLP-1(7–37) with Lys34 replaced by Arg. It has a 16-carbon fatty-acid chain with a glutamic acid spacer extending from the backbone of GLP-1. These modifications prevent liraglutide from degrading and prolong its half-life to approximately 13 h [7]. Liraglutide was approved for T2DM treatment by the Food and Drug Administration (FDA) in 2009. Recently, liraglutide was reported to have anabolic bone effects on bone independent of its anti-diabetic function [8]. Liraglutide administration can reduce the incidence of fractures among patients with T2DM [9]. However, the exact mechanisms are still unclear.
Exosomes are nano-sized vesicles derived from endocytic compartments and work as mediators in cell–cell communication [10]. It has been recently confirmed that exosomes have important roles in the onset of many diseases, as well as drug delivery and treatment [11–14]. Functional cargoes of exosomes, such as proteins, lipids, and nucleic acids, are transported between cells, modifying the biological function of receptor cells, in which exosomal miRNAs play a significant role. miRNAs in exosomes vary in their quantities and species depending on the particular disease and its stage [15]. Further research into the role of exosomal miRNAs should help to clarify the exact mechanisms of many diseases.
Biological behavior of cells in the bone marrow microenvironment is closely related to bone metabolism. In the present study, we established an animal model characterized by hyperglycemia and estrogen deficiency to mimic the pathological changes in postmenopausal T2DM. In order to explore the role of bone marrow exosomes in the bone metabolic disorders of postmenopausal T2DM, as well as the bone protective mechanisms of liraglutide, exosomal miRNA expression profiles and signal pathways were analyzed among ovariectomized (OVX) control rats, OVX + T2DM rats, and OVX + T2DM + liraglutide-treated rats. To the best of our knowledge, this is the first study investigating miRNA expression profiles and signal pathway changes of bone marrow-derived exosomes.
Material and Methods
OVX + T2DM animal model
The animal model in this study was established by 2 steps. Firstly, low-dose streptozotocin (STZ) was given together with a high-fat diet to establish the T2DM model. On this basis, bilateral ovaries were removed to establish the OVX + T2DM rats model. We purchased 27 Sprague-Dawley rats (6-month-old females) from the Slac Laboratory Animal Center (Shanghai, China). They were housed at 23~25°C with a 12-h light/dark cycle and allowed unrestricted cage activity and unlimited access to standard chow and water. All animal procedures were approved by the Animal Care Committee of the Second Xiangya Hospital of Central South University. After 1 month of acclimatization, 7-month-old rats were randomly divided into 3 groups (n=9): (1) the OVX control group (OVX); (2) the OVX + T2DM group (DM); and (3) the OVX + T2DM + Liraglutide-treated group (LIR).
Low-dose STZ injection was given together with a high-fat diet to establish the T2DM model according to previous studies with some minor modifications [16]. Rats in the OVX group were fed standard laboratory animal diet containing 3% fat, 20% protein, and 55% carbohydrate with 310 kJ/kg total caloric value, while rats in the DM and LIR groups were fed high-fat diet containing 15% butter, 10% egg yolk powder, 2% cholesterol, and 73% standard food, providing 412 kJ/kg calories for animals. Both standard diets and high-fat diets were purchased from Guangdong Provincial Medical Experimental Animal Center, China. After 2 months on these diets, rats in the LIR and DM groups were intraperitoneally injected with a freshly prepared solution of STZ (35 mg/kg) (Sigma, St. Louis, MO) in 0.1 M of cold citrate buffer (pH 4.2) to induce T2DM, while rats in the OVX group were injected with an equal volume of 0.1 M citrate buffer. Blood glucose was measured 72 h after STZ injection using a glucometer (ACCU-CHEK, Roche, Basel, Switzerland). Blood samples were collected from tails. Rats with blood glucose levels lower than 16.7 mmol/l were injected with STZ (35 mg/kg) a second time. Rats with random blood glucose levels higher than 16.7 mmol/l were considered to be T2DM and used in the following research. Random blood glucose measurements were made weekly during this study.
After the establishment of the T2DM animal model, all rats were returned to standard laboratory animal diet. Ovariectomy was performed as described in our previous work [17]. Briefly, anesthesia was induced with 3% pentobarbital sodium (1 ml/Kg) (Sigma, St. Louis, MO). Bilateral ovaries were clamped and removed with the fallopian tubes being ligated, and the skin was then sutured. The OVX+T2DM rat model was established and the follow-up experiments began 3 weeks later [17].
Drug administration
After establishment of the OVX+T2DM model, rats in the LIR group were treated with liraglutide (Novo Nordisk, Copenhagen, Denmark) 0.2 mg/kg, i.h., bid for 2 months. The drug dose was determined according to several previous studies [18–20]. Rats in the OVX and DM groups were injected with equal volumes of normal saline.
Isolation of whole bone marrow (WBM)
After 2 months of drug administration, all rats were anesthetized and sacrificed under sterile conditions. Bilateral femurs were rinsed in sterile phosphate-buffered saline (PBS, 137 mM NaCl, 2.68 mM KCl, 8.10 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4) followed by the removal of attached fatty tissues, connective tissues, and periosteum. WBM extraction was performed according to a method previously described [21]. Briefly, after epiphyses were dissected, WBM of each femur was flushed with a syringe filled with 2 ml sterile PBS solution, and repeatedly beaten into a homogeneous suspension. The suspensions were then transferred into RNase-free centrifugal tubes. After concentration at 500×g for 10 min, cell debris was removed. The supernatant was transferred into RNase-free EP tubes and stored at −80°C.
Isolation and purification of exosomes
Exosomes were isolated from bone marrow samples by a differential centrifugation method, as described previously [22]. Samples were centrifuged at 2000×g for 30 min and 12 000×g for 45 min to remove dead cells and cell debris. The supernatant was ultracentrifuged at 110 000×g for 2 h (Beckman Optima L-90K, CA). The pellets were then resuspended in 1 mL PBS and pooled into new tubes by filtering through a 0.22-μm filter sterilizer (Millipore, Billerica, MA). The suspensions were ultracentrifuged at 110 000×g for 70 min, after which the pellets were resuspended for another 70 min of ultracentrifugation. Finally, the supernatant was discarded. The exosome pellets were resuspended in 200 μL PBS and stored at −80°C. All centrifugations were performed at 4°C. Exosomal protein concentration was quantified by use of a BCA protein assay kit (Beyotime Biotechnology, China).
Western blot analysis
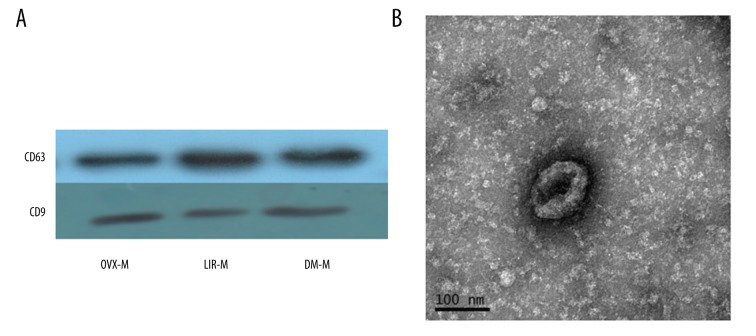
For exosome validation, CD63 and CD9 were chosen as the exosome-specific membrane markers. Western blot analysis was performed as previously described [23,24]. Briefly, total exosomes proteins in each group were extracted and lysed by a reagent (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Protein lysates of exosomes were mixed with sodium dodecyl sulfate (SDS) buffer and boiled at 100°C for 5 min. Proteins were separated by 8–10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) at 70 V for 30 min and 110V for 90 min by a blotting system (Bio-Rad, Hercules, CA). The separated proteins in gel were electrotransferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica) at constant 300 mA for 30 min. PVDF membranes were blocked in 5% skim milk for 1.5 h at room temperature and incubated with primary antibodies CD63 (ab108950; Abcam, Cambridge, UK, 1: 1000), CD9 (ab92726; Abcam, Cambridge, UK, 1: 1000) in 3% skim milk for 12 h at 4°C. After that, the membranes were washed 3 times for 15 min in Tris-buffered saline (TBS)-Tween (10 mM Tris HCl, 150 mM NaCl, 0.02% Tween-20), and reprobed by secondary antibodies (ab6789, ab6721; Abcam, Cambridge, UK, 1: 2000) in 3% skim milk for 1 h at 37°C. The protein bands were developed by chemiluminescence reagents (BestBio, Shanghai, China) according to manufacturer’s instructions and exposed to X-ray films in a darkroom.
Transmission electron microscope (TEM)
Purified exosomes were diluted 1: 100 in PBS and fixed with 2% glutaraldehyde for 12 h at room temperature. We adsorbed 10-μL samples onto charged carbon-coated grids (Quantifoil, Jena, Germany) and dried them overnight at room temperature. After that, exosomes were contrasted with a freshly prepared 2% uranyl acetate solution for 30 min, air-dried, and examined with a transmission electron microscope (TEM, FEI Tecnai G2 Spirit, Eindhoven, Netherlands).
Atomic force microscopy (AFM)
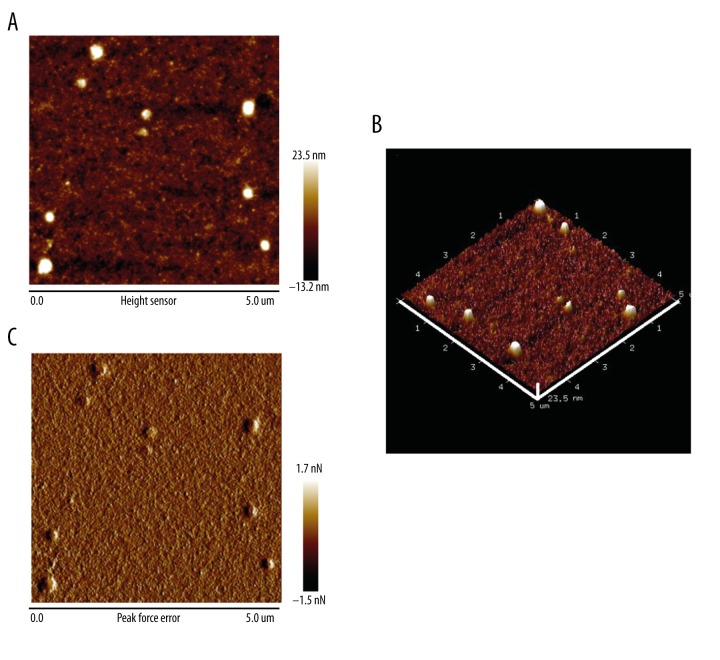
Purified exosomes were diluted 1: 50 in deionized water; 5- to 10-μL samples were spotted onto freshly cleaved mica sheets (V-1, thickness 0.15 mm, size 15×15 mm) and dried with mild nitrogen flow for atomic force microscopy (AFM) scanning at room temperature. An Icon AFM (Dimension Icon, Bruker; Camarillo, CA) was used to obtain exosome topographic images at tapping mode, which generates high-resolution images by detecting the change in the vibration amplitude of the cantilever by silicon probes (SNL-10 and Scanasyst-Air, Bruker, USA). A 5×5 μm sample area was scanned in every microscopic field. Topographic images were recorded at 512×512 pixels at a scan rate of 1 Hz and 100 pN force. After that, samples were analyzed in peak force quantitative nanomechanical mapping (Peak Force QNM) mode to obtain nanomechanical maps of exosomes. In Peak Force QNM mode, the silicon probes acted on samples to obtain force-distance curves which are then used as feedback signals to generate the peak force error images. Image processing was performed with NanoScope Analysis software (Bruker; Camarillo, CA).
Total miRNA extraction
Total RNAs were extracted from exosomes using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Samples were mixed with TRIzol (1: 3) and pipetted several times to lyse the samples for 5 min. Lysates were centrifuged at 12 000×g for 15 min at 4°C. After that, chloroform was added into supernatant liquor (1: 5) and pipetted to homogenize. The mixture was centrifuged at 12 000×g for 15 min at 4°C. The upper aqueous layer was transferred into a new tube and mixed with an equal volume of 70% ethanol. The mixture was centrifuged at 12 000×g for 15 min at 4°C. Sediment RNA was dried at room temperature. Quantity and quality of RNA were estimated by use of a Nano-Drop spectrophotometer (Thermo Scientific, Wilmington, DE) by measuring the absorbance at 260 nm and the purity at 260/280 and 260/230 nm ratios.
Library construction and sequencing
RNAs extracted from exosomes were used for miRNA library construction by use of the TruSeq Small RNA Sample Preparation Kit (Illumina, San Diego, CA) according to the manufacturer’s instruction. Briefly, RNAs were ligated with 3′- and 5′-adapters. Adapter-ligated RNAs were then subjected to low-cycle reverse transcription PCR. PCR products of about 18–40 nt in length were selected by PAGE gel for sequencing. The library was sequenced using HiSeq2500 (Illumina, San Diego, CA) as 50 bp single reads. miRNA library construction and sequencing were performed at RiboBio, China. For sequence analysis, raw reads were filtered, discarding low-quality reads, 3′- and 5′-adapters, and contaminated reads to acquire clean reads. All clean reads were mapped to databases to eliminate tRNA, rRNA, snRNa, snoRNA (Rfam12.1), and piRNA (Pirnabank). For differentially expressed miRNAs, P<0.05 and fold-changes >2 were considered significant. miRNAs expression was normalized by reads per million (RPM) by mapping to miRBase version 21. RPM=(number of reads mapping to miRNA/number of reads in clean data)×106.
Target gene prediction and function analysis
Target genes were predicted by Target Scan 7.0 (http://www.targetscan.org), miRDB (http://www.mirdb.org/miRDB) and miRanda (http://www.microrna.org/microrna/home.do). Predicted target genes were analyzed for enrichment using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (http://www.genome.jp). Enriched gene target pathways with P<0.05 were considered as significant.
Validation of sequencing data
To validate the sequencing data, 3 distinctive miRNAs were selected randomly to test their absolute expression levels by real-time quantitative polymerase chain reaction (qRT-PCR), including miR-322-3p, miR-194-5p, and miR-9a-5p. Primers for the 3 miRNAs were designed and synthesized by RiboBio Co., Ltd. (Guangzhou, China). Total RNA extraction was performed as described above, and reverse transcription reactions were performed using a PrimeScript RT Reagent kit (Takara, Dalian, China). The qRT-PCR was performed using a SYBR Green PCR kit (Bio-Rad, California, USA).
Statistical analysis
SPSS 19.0 (SPSS, Chicago, IL, USA) was used for statistical analysis. Blood glucose data are presented as mean ±SD. Differences among 3 groups were evaluated using analysis of variance (ANOVA). When a significant overall F value (P<0.05) was observed, differences between individual group means were tested using Fisher’s protected least-significant difference (PLSD) post hoc test. P<0.05 was considered significant.
Results
Blood glucose levels of each group
Baseline blood glucose showed no significant difference among the 3 groups. After STZ injection at 0 weeks, blood glucose in the DM group and LIR group increased significantly compared to the OVX group. During the period of liraglutide administration, blood glucose levels in the DM group and LIR group were significantly higher than that in the OVX group (p<0.01). Liraglutide administration decreased the blood glucose compared with DM but it did not return to normal values (Figure 1).
Figure 1.
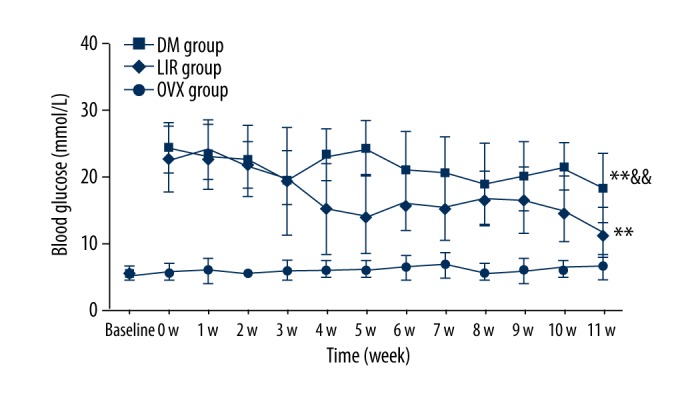
Blood glucose measurement during this study. ** Means p<0.01 vs. OVX group, && means p<0.01 vs. LIR group. OVX – ovariectomized; DM – diabetes mellitus; LIR – liraglutide.
Exosome identification
Western blot analysis confirmed the positive membrane markers CD63 and CD9 of exosomes (Figure 2A). Morphology of exosomes was observed with TEM and AFM. A typical cup-shaped phenotype was observed under TEM (Figure 2B). Two-dimensional (Figure 3A), peak force (Figure 3B), and three-dimensional (Figure 3C) images were observed by AFM.
Figure 2.
Characterization of exosomes. (A) Membrane markers CD63 and CD9 by Western blot. (B) Typical cup-rounded phenotype under transmission electron microscope. Scale bar=100 nm.
Figure 3.
Exosomes images observed by atomic force microscopy. The topography of a 5×5 μm sample area was depicted. (A) Exosome images in 2-dimension. (B) Exosome images in 3-dimension. The maximum altitude of exosomes in this microscopic field was about 23.5 nm. (C) Peak force error images. In peak force error images, microstructure and nanomechanical images of the exosomes can be obtained simultaneously at the same area. Scale bar=5 μm.
Overview of exosomal miRNA expression profiles
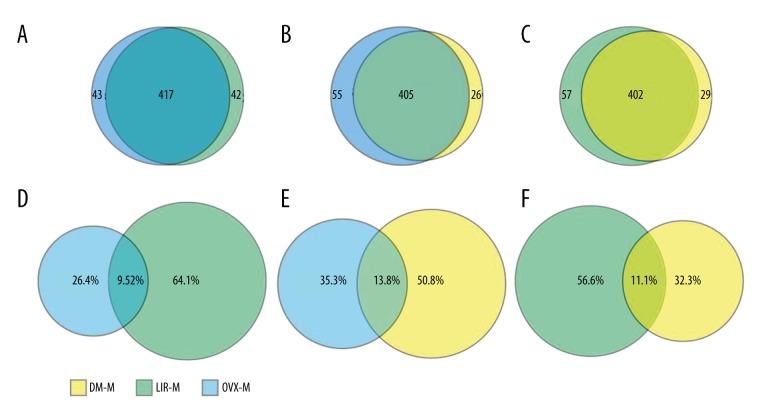
High-throughput sequencing was used to characterize miRNA expression profiles of the 3 groups. A total of 460 exosomal miRNAs were detected in the OVX group, 431 miRNAs for the DM group, and 459 miRNAs for the LIR group. We found 39 exosomal miRNAs expressed differently between the LIR and DM groups, 84 between the OVX and DM groups, and 90 between the OVX and LIR groups. Differentially expressed miRNAs were investigated by edgeR. The number of overlapping miRNA species and ratio of overlapping miRNA reads are shown in Figure 4.
Figure 4.
Number of overlapping miRNA species by pairwise comparison (A–C). Ratio of overlapping miRNA reads (D–F). Ratio of overlapping miRNA reads=number of overlapping miRNA reads/total number of miRNA reads.
miRNA profile analysis
The most highly expressed miRNAs of all 3 groups included 12 miRNAs, of which 3 miRNAs (let-7i-5p, let-7f-5p, and miR-148a-3p) were commonly expressed among the 3 groups. Only 1 miRNA was expressed specially in 1 sample (miR-3557-5p in OVX group) (Table 1).
Table 1.
Top 10 highly expressed miRNAs in each group.
| Rank | OVX group | DM group | LIR group |
|---|---|---|---|
| 1 | miR-148a-3p | miR-1-3p | miR-1-3p |
| 2 | miR-1-3p | let-7i-5p | let-7f-5p |
| 3 | let-7i-5p | let-7f-5p | miR-148a-3p |
| 4 | miR-1b | miR-148a-3p | let-7i-5p |
| 5 | let-7f-5p | let-7g-5p | miR-3571 |
| 6 | miR-21-5p | miR-3571 | miR-126a-3p |
| 7 | let-7g-5p | miR-1b | miR-21-5p |
| 8 | miR-3571 | miR-126a-3p | let-7g-5p |
| 9 | miR-143-3p | miR-206-3p | miR-206-3p |
| 10 | miR-3557-5p | miR-143-3p | miR-1b |
OVX – ovariectomized; DM – diabetes mellitus; LIR – liraglutide.
After comparative analysis, the differentially expressed miRNAs between 2 samples were selected for target gene prediction. KEGG pathway analysis was performed for the target genes of the miRNAs. Differentially expressed miRNAs targeted the function associated with glucose regulation and bone metabolism were analyzed in this study (Table 2, Figure 5, p<0.05).
Table 2.
KEGG pathway analysis of differentially expressed miRNAs among the three groups.
| miRNA species | Group with increased expression | Target pathway | Target gens (encoding proteins) |
|---|---|---|---|
| OVX group vs. LIR group | |||
| let-7c-2-3p | OVX | Wnt signaling pathway | LRP6 (LPR6), TCF3 (TCF) |
| miR-322-3p | OVX | Wnt signaling pathway | CREBBP (CBP), FOSL1 (Fra-1) |
| OVX group vs. DM group | |||
| miR-9a-5p | DM | Insulin signaling pathway | RHOQ (RhoQ) |
| Insulin secretion pathway | SLC2A2 (GLUT2), ADCY5 (ADCY5) | ||
| let-7a-1-3p | DM | Wnt signaling pathway | LRP6 (LPR6), TCF3 (TCF) |
| miR-27b-3p | OVX | Osteoclast differentiation pathway | PPARG (PPAR γ) |
| LIR group vs. DM group | |||
| miR-335 | DM | Insulin secretion | ADCY5 (ADCY5) |
| miR-322-3p | DM | Wnt signaling pathway | CREBBP (CBP), FOSL1 (Fra-1) |
KEGG – Kyoto Encyclopedia of Genes and Genomes; OVX – ovariectomized; DM – diabetes mellitus; LIR – liraglutide; LPR6 – lipoprotein receptor-related protein 6; TCF – T cell factor; CBP – Crebbp binding protein; RhoQ – ras homologue family member Q; Fra-1 – FOS-related antigen-1; GLUT2 – glucose transporter type 2; ADCY5 – adenylate cyclase-5; PPAR γ – peroxisome proliferator-activated receptor gamma.
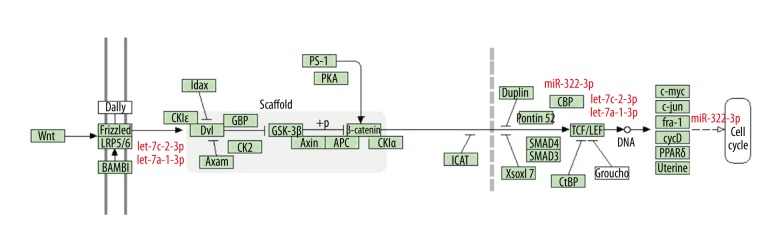
Figure 5.
Wnt/β-catenin pathway. Three differentially expressed miRNAs (including let-7c-2-3p, let-7a-1-3p, and miR-322-3p) were predicted to participate in the Wnt/β-catenin pathway by regulating their potential target genes.
miRNAs validation
qRT-PCR expression data of the 3 selected miRNAs generally agreed with the sequencing data (p<0.05). This result confirmed the reliability of the sequencing data (Table 3).
Table 3.
miRNAs validation by qRT-PCR compared with sequencing data.
| miRNA species | Samples | Absolute expression level by qRT-PCR(μM) | RPM detected by sequencing |
|---|---|---|---|
| miR-322-3p | LIR | 3.89E-08±1.70837E-09 | 15.472 |
| DM | 1.43E-08±6.42968E-10 | 1.7822 | |
| miR-194-5p | DM | 4.72798E-08±5.92621E-09 | 9.8505 |
| OVX | 3.27459E-07±3.87998E-09 | 1.5392 | |
| miR-9a-5p | LIR | 9.83764E-08±8.69045E-09 | 71.5827 |
| OVX | 4.54503E-08±1.68551E-09 | 10.3122 |
qRT-PCR – quantitative polymerase chain reaction; RPM – reads per million; OVX – ovariectomized; DM – diabetes mellitus; LIR – liraglutide.
Discussion
Exosomes are involved in the exchange of genetic information within microenvironments. Particularly, miRNAs play a critical role in exosome-mediated information transportation [25]. We analyzed bone marrow-derived exosomal miRNAs in 3 groups of rats: (1) OVX. (2) OVX + T2DM, and (3) OVX + T2DM + liraglutide-treated. By miRNA sequencing, we found that the exosomal miRNA profiles in these groups differed, which may provide some useful information about the disorders associated with type 2 diabetic postmenopausal subjects. In addition, these findings may assist in understanding the bone-protective mechanism of liraglutide.
Exosomal miRNA expression profiles can be altered by different diseases and drug administration [26,27]. Variance in miRNA expression profiles is involved in disease progression, drug action, and drug resistance [14,15]. In our study, we found 83.1%, 83.3%, and 82.4% overlapping miRNA species among the 3 groups, and among the top 10 most highly expressed miRNAs of each group, only 1 miRNA was expressed just in 1 group. These findings indicate a high degree of similarity in total and dominant miRNA species of bone marrow-derived exosomes among the 3 groups. The pathological state of T2DM and liraglutide treatment did not change the main miRNA species. However, the overlapping ratio of miRNAs reads was only about 9.52% to 13.8% according to pairwise comparisons. Thus, we could infer that, although the miRNA species were similar, the abundance of each miRNA was quite different, which may result in biological differences in bone marrow-derived exosomes.
To understand the different biological functions of exosomal miRNAs during T2DM and with liraglutide treatment, we used pathway analyses among the 3 groups. Our study found that, in bone metabolism, the Wnt signaling pathway was the only pathway that differed among the groups. The Wnt signaling pathway is an important signal transduction pathway in regulating cell growth, differentiation, and tissue morphogenesis, of which the canonical Wnt pathway (Wnt/β-catenin pathway) plays a central role in osteogenesis [28]. In the absence of Wnt ligand, β-catenin is recruited into the destruction complex comprising adenomatous polyposis coli (APC), axin, and glycogen synthase kinase-3β (GSK-3β), which induces phosphorylation of β-catenin, thus targeting it for ubiquitination and proteasomal degradation. The pathway is activated when Wnt ligands bind to Frizzleds (Fzs) and low-density lipoprotein receptor-related protein 5/6 (LRP-5/6) co-receptor proteins, which leads the activation of the intracellular protein, Dishevelled (Dvl). Activation of Dvl results in the collapse of the β-catenin degradation complex. Hence, β-catenin cannot be degraded and it translocates to the nucleus, where it binds to the T cell factor/lymphoid enhancer factor (TCF/LEF) family, and activates the transcription of numerous target genes including runt-related transcription factor 2 (Runx2) and osterix (OSX), thereby strongly stimulating osteogenesis. Recently, miRNA-mediated regulation of Wnt signaling pathway has been described [26]. We found that let-7c-2-3p, let-7a-1-3p, and miR-322-3p are expressed differently among the 3 groups. Bioinformatics analysis showed that all 3 miRNAs targeted and blocked the expression of Wnt/β-catenin pathway genes. They were more expressed in diabetes and less expressed after liraglutide treatment compared with controls, indicating that T2DM may have an inhibiting effect on the Wnt/β-catenin pathway, while liraglutide may exert its bone-protective effect by negating this inhibiting effect.
Besides bone metabolism, target gene prediction and KEGG pathway analysis indicated that some differently expressed miRNAs are involved in specific target genes in the insulin-signaling and secretion pathways. These miRNAs were more expressed in diabetes compared with controls and drug-treated groups, of which, miR-9a-5p was predicted to be involved in the Cbl/TC10 (also known as RhoQ) pathway, an insulin-signaling cascade implicated in glucose transport. Its target gene RhoQ encodes Rho family protein TC10, which is an important regulator of insulin-stimulated glucose disposal in adipocytes [29]. Also, other target genes, including SLC2A2, encoding glucose transporter type 2 (GLUT2), and Adcy5, encoding adenylate cyclase 5 (ADCY5), are involved in glucose-mediated insulin secretion [30,31]. Negative regulation of these target genes may result in insulin deficiency in T2DM.
Our study has some limitations. According to published studies, GLP-1 receptor agonists have different mechanisms of action in humans, animals, and cell lines. For example, research has shown that GLP-1 receptor agonists can stimulate proliferation and differentiation of pancreatic β-cell, increasing the mass of β-cell in animal models and cell lines [32–34], but these effects have not been confirmed in humans. Thus, human studies are required to validate our findings. We screened out the differentially expressed miRNAs involved in the Wnt/β-catenin pathway and predicted their potential target genes, but the detailed action mechanisms of these miRNAs remain to be investigated in further studies.
Conclusions
Bone marrow exosome-mediated miRNA signal pathways were altered under T2DM status and after liraglutide treatment. Pathways associated with bone metabolism in ovariectomized rats with T2DM showed significant differences from non-diabetic animals. Bone marrow exosome-mediated Wnt/β-catenin pathway change may be a critical node for bone metabolic disorders in T2DM and the bone protective effect of liraglutide. Further studies are needed to clarify the exact mechanisms in humans.
Footnotes
Conflict of interest
None.
Source of support: This work was supported by the China International Medical Foundation China Diabetes Young Scientific Talent Research Fund, the National Natural Science Foundation of China (NSFC, 81670804, 81070695, and 81270960), the Major Research and Development Project of Hunan province (2016WK2020), and the National Clinical Key Subject Construction Project (Endocrinology)
References
- 1.Dytfeld J, Michalak M. Type 2 diabetes and risk of low-energy fractures in postmenopausal women: Meta-analysis of observational studies. Aging Clin Exp Res. 2017;29(2):301–9. doi: 10.1007/s40520-016-0562-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goh SY, Cooper ME. Clinical review: The role of advanced glycation end products in progression and complications of diabetes. J Clin Endocrinol Metab. 2008;93:1143–52. doi: 10.1210/jc.2007-1817. [DOI] [PubMed] [Google Scholar]
- 3.Calvo-Romero JM, Ramiro-Lozano JM. Vitamin D levels in patients with type 2 diabetes mellitus. J Investig Med. 2015;63:921–23. doi: 10.1097/JIM.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 4.Roman de Mettelinge T, Cambier D, Calders P, et al. Understanding the relationship between type 2 diabetes mellitus and falls in older adults: A prospective cohort study. PLoS One. 2013;8:e67055. doi: 10.1371/journal.pone.0067055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renner S, Blutke A, Streckel E, et al. Incretin actions and consequences of incretin-based therapies: Lessons from complementary animal models. J Pathol. 2016;238:345–58. doi: 10.1002/path.4655. [DOI] [PubMed] [Google Scholar]
- 6.Tran KL, Park YI, Pandya S, et al. Overview of glucagon-like peptide-1 receptor agonists for the treatment of patients with type 2 diabetes. Am Health Drug Benefits. 2017;10:178–88. [PMC free article] [PubMed] [Google Scholar]
- 7.Malm-Erjefalt M, Bjornsdottir I, Vanggaard JH, et al. Metabolism and excretion of the once-daily human glucagon-like peptide-1 analog liraglutide in healthy male subjects and its in vitro degradation by dipeptidyl peptidase IV and neutral endopeptidase. Drug Metab Dispos. 2010;38:44–53. doi: 10.1124/dmd.110.034066. [DOI] [PubMed] [Google Scholar]
- 8.Lu N, Sun H, Yu J, et al. Glucagon-like peptide-1 receptor agonist Liraglutide has anabolic bone effects in ovariectomized rats without diabetes. PLoS One. 2015;10:e0132744. doi: 10.1371/journal.pone.0132744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su B, Sheng H, Zhang M, et al. Risk of bone fractures associated with glucagon-like peptide-1 receptor agonists’ treatment: A meta-analysis of randomized controlled trials. Endocrine. 2015;48:107–15. doi: 10.1007/s12020-014-0361-4. [DOI] [PubMed] [Google Scholar]
- 10.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–15. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delic D, Eisele C, Schmid R, et al. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS One. 2016;11:e0150154. doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bang C, Batkai S, Dangwal S, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124:2136–46. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye SB, Li ZL, Luo DH, et al. Tumor-derived exosomes promote tumor progression and T cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–52. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao X, Yu S, Li S, et al. Exosomes: Decreased sensitivity of lung cancer A549 cells to cisplatin. PLoS One. 2014;9:e89534. doi: 10.1371/journal.pone.0089534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin AJ, Guo C, Cook JA, et al. Plasma levels of microRNA are altered with the development of shock in human sepsis: An observational study. Crit Care. 2015;19:440. doi: 10.1186/s13054-015-1162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian C, Zhu C, Yu W, et al. High-fat diet/low-dose streptozotocin-induced type 2 diabetes in rats impacts osteogenesis and Wnt signaling in bone marrow stromal cells. PLoS One. 2015;10:e0136390. doi: 10.1371/journal.pone.0136390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu B, Zhang C, Chen B, et al. Self-repair of rat cortical bone microdamage after fatigue loading in vivo. Int J Endocrinol. 2013:321074. doi: 10.1155/2013/321074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo N, Sun J, Chen H, et al. Liraglutide prevents diabetes progression in prediabetic OLETF rats. Endocr J. 2013;60:15–28. doi: 10.1507/endocrj.ej12-0094. [DOI] [PubMed] [Google Scholar]
- 19.Pereira M, Jeyabalan J, Jorgensen CS, et al. Chronic administration of Glucagon-like peptide-1 receptor agonists improves trabecular bone mass and architecture in ovariectomized mice. Bone. 2015;81:459–67. doi: 10.1016/j.bone.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Zhang J, Ma D, et al. Subcutaneous administration of liraglutide ameliorates Alzheimer-associated tau hyperphosphorylation in rats with type 2 diabetes. J Alzheimer’s Dis. 2013;37:637–48. doi: 10.3233/JAD-130491. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Menocal L, Shareef S, Salgado M, et al. Role of whole bone marrow, whole bone marrow cultured cells, and mesenchymal stem cells in chronic wound healing. Stem Cell Res Ther. 2015;6:24. doi: 10.1186/s13287-015-0001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3.22) doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 23.He F, Du T, Jiang Q, et al. Synergistic effect of Notch-3-specific inhibition and paclitaxel in non-small cell lung cancer (NSCLC) cells via activation of the intrinsic apoptosis pathway. Med Sci Monit. 2017;23:3760–69. doi: 10.12659/MSM.902641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long LM, He BF, Huang GQ, et al. microRNA-214 functions as a tumor suppressor in human colon cancer via the suppression of ADP-ribosylation factor-like protein 2. Oncol Lett. 2015;9:645–50. doi: 10.3892/ol.2014.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bang C, Thum T. Exosomes: New players in cell-cell communication. Int J Biochem Cell Biol. 2012;44:2060–64. doi: 10.1016/j.biocel.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Hiratsuka I, Yamada H, Munetsuna E, et al. Circulating MicroRNAs in Graves’ disease in relation to clinical activity. Thyroid. 2016;26:1431–40. doi: 10.1089/thy.2016.0062. [DOI] [PubMed] [Google Scholar]
- 27.Qin X, Yu S, Xu X, et al. Comparative analysis of microRNA expression profiles between A549, A549/DDP and their respective exosomes. Oncotarget. 2017;8:42125–35. doi: 10.18632/oncotarget.15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Li YP, Paulson C, et al. Wnt and the Wnt signaling pathway in bone development and disease. Front Biosci. 2014;19:379–407. doi: 10.2741/4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao JJ, Lin J, Zhu D, et al. miR-30-5p functions as a tumor suppressor and novel therapeutic tool by targeting the oncogenic Wnt/beta-catenin/BCL9 pathway. Cancer Res. 2014;74:1801–13. doi: 10.1158/0008-5472.CAN-13-3311-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajiaghaalipour F, Khalilpourfarshbafi M, Arya A. Modulation of glucose transporter protein by dietary flavonoids in type 2 diabetes mellitus. Int J Biol Sci. 2015;11:508–24. doi: 10.7150/ijbs.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodson DJ, Mitchell RK, Marselli L, et al. ADCY5 couples glucose to insulin secretion in human islets. Diabetes. 2014;63:3009–21. doi: 10.2337/db13-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoffers DA, Desai BM, DeLeon DD, et al. Neonatal exendin-4 prevents the development of diabetes in the intrauterine growth retarded rat. Diabetes. 2003;52:734–40. doi: 10.2337/diabetes.52.3.734. [DOI] [PubMed] [Google Scholar]
- 33.Abraham EJ, Leech CA, Lin JC, et al. Insulinotropic hormone glucagon-like peptide-1 differentiation of human pancreatic islet-derived progenitor cells into insulin-producing cells. Endocrinology. 2002;143:3152–61. doi: 10.1210/endo.143.8.8973. [DOI] [PubMed] [Google Scholar]
- 34.Xu G, Stoffers DA, Habener JF, et al. Exendin-4 stimulates both beta-cell replication and neogenesis, resulting in increased beta-cell mass and improved glucose tolerance in diabetic rats. Diabetes. 1999;48:2270–76. doi: 10.2337/diabetes.48.12.2270. [DOI] [PubMed] [Google Scholar]