Abstract
Bacterial pathogenesis frequently requires metal acquisition by specialized, small-molecule metallophores. We hypothesized that the Gram-negative Pseudomonas aeruginosa encodes the enzymes nicotianamine synthase (NAS) and opine dehydrogenase (ODH), biosynthesizing a new class of opine metallophore, previously characterized only in the unrelated Gram-positive organism Staphylococcus aureus. The identity of this metallophore, herein named pseudopaline, was determined through measurements of binding affinity, the in vitro reconstitution of the biosynthetic pathway to screen potential substrates, and the confirmation of product formation by mass spectrometry. Pseudopaline and the S. aureus metallophore staphylopine exhibit opposite stereochemistry for the histidine moiety, indicating unique recognition by NAS. Additionally, we demonstrate SaODH catalysis in the presence of pyruvate, as previously shown, but also oxaloacetate, suggesting the potential for the production of a variant form of staphylopine, while PaODH specifically recognizes α-ketoglutarate. Both the staphylopine and pseudopaline operons have been implicated in the pathogenesis of key infectious disease states and warrant further study.
Bacterial pathogens use small-molecule metallophores to scavenge metals from their hosts.1 This process is frequently critical to pathogenesis.2–4 In Staphylococcus aureus, the metallophores staphyloferrin A and B are well characterized, staphyloferrin B being an established virulence factor.5 Recently, a new S. aureus metallophore named staphylopine has been identified.6 Staphylopine receptor mutants show significantly attenuated virulence in murine bacteremia and urinary tract infection models,7 emphasizing the importance of this metallophore pathway in two infectious disease states that are major threats to human health.8
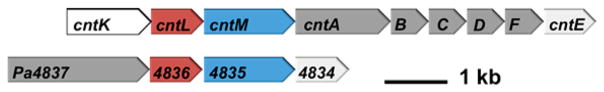
The staphylopine operon encodes three enzymes, a histidine racemase (CntK), a nicotianamine synthase (CntL), and an opine dehydrogenase (CntM)6 (Figure 1). The first enzyme, the histidine racemase, converts L-histidine to D-histidine. The second enzyme shares sequence identity with nicotianamine synthases (NAS), enzymes with S-adenosyl-L-methionine (SAM)-dependent aminoalkyl transferase activity. Nicotianamine is a plant metallophore consisting of three aminobutyrate moieties derived from SAM. NAS enzymes transfer the aminobutyrate moiety in a reaction similar to that performed by spermidine synthase, which uses decarboxy-S-adenosyl-L-methionine as a substrate.9 In S. aureus, NAS links the aminobutyrate moiety of SAM to the primary amine of D-histidine. This product then acts as a substrate for an opine dehydrogenase, the final enzyme in the pathway.
Figure 1.
Map of homologous operons. Gene arrangement in opine metallophore operons: top, S. aureus; bottom, Pseudomonas aeruginosa. CntL and Pa4836 (red): nicotinamine synthase. CntM and Pa4835 (blue): opine dehydrogenase. CntK: histidine racemase. CntA-F and Pa4837: import proteins. CntE and Pa4834: export proteins.
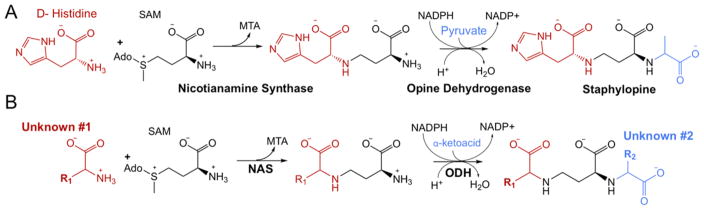
Opine dehydrogenases (ODHs) typically form a secondary amine by condensation of an amino acid with an α-keto acid. Opine products are diverse, as identified in crown gall tumors formed by Agrobacterium tumefaciens. In these infections, bacterial DNA encoding a variety of opine dehydrogenases is incorporated into the plant genome. The plant then produces opines, utilizing amino acids such as leucine and arginine and α-keto acids such as α-ketoglutarate and pyruvate, providing a source of energy for the bacterial pathogen.10 ODHs have also been described in marine molluscs, such as Arctica islandica (king scallop), where they catalyze the reductive condensation of an opine product from pyruvate and L-arginine.11 This reaction regenerates NAD+ in muscle tissue, allowing glycolysis to continue under anaerobic conditions. In S. aureus, ODH uses the primary amine of the D-histidine-aminobutyrate product of NAS as a nucleophile in a condensation with pyruvate followed by hydride transfer from NADPH. Together, these three enzymes biosynthesize the opine metallophore staphylopine (Figure 2A).
Figure 2.
(A) Staphylopine biosynthesis. SAM is S-adenosyl-L-methionine. MTA is methylthioadenosine. Ado is adenosine. (B) Pseudopaline hypothesis.
Pseudomonas aeruginosa produces two well-characterized siderophores, pyoverdin and pyochelin, but a recent report suggests that an uncharacterized P. aeruginosa operon may be generating a novel nicotianamine-like metallophore12 (Figure 1). This four-gene operon encodes Pa4837, predicted to be an outer membrane TonB-dependent receptor, and Pa4834, predicted to be an EamA-like export transporter.13 Notably, genes encoding inner membrane import function and outer membrane export function are absent, suggesting that they are encoded elsewhere and/or may be shared with another pathway. This operon is also predicted to encode a NAS and ODH that are 24 and 30% identical, respectively, with S. aureus as determined by Clustal Omega.14 In addition, the S. aureus and P. aeruginosa NAS enzymes have active site residues conserved with the Methanothermobacter thermautotrophicus NAS,15 and both ODHs have GXGXXG/A NAD(P)H binding motifs. The entire operon is upregulated in P. aeruginosa clinical isolates from human burn wound and cystic fibrosis lung infections16,17 as well as in an airway mucus secretion growth model. In addition, transport mutants (ΔPa4834) show significantly attenuated growth in murine airway and burn wound infection models.12 Taken together, these data suggest that P. aeruginosa may be producing a metallophore similar to staphylopine and that this metallophore plays an important role in pathogenesis, which is surprising given the expanse of evolution separating the two species. We hypothesized that PaNAS would produce a nicotianamine product using SAM exclusively or in concert with an amino acid substrate (unknown #1, Figure 2B) and that PaODH would act on the NAS product by incorporating an α-keto acid (unknown #2, Figure 2B).
To test this hypothesis, the P. aeruginosa and S. aureus nicotianamine synthase (PaNAS and SaNAS, respectively) and opine dehydrogenase (PaODH and SaODH, respectively) enzymes were heterologously expressed and purified (Figure S1; supplementary figures and all methods can be found in the Supporting Information). The affinity of PaNAS for SAM was measured by intrinsic tryptophan fluorescence while titrating SAM. The resulting dissociation constant was 1.2 ± 0.3 μM, which is 57-fold tighter than the value of 68 μM previously reported for SaNAS6 (Figure S2A,B). The affinity of PaODH for NADPH was determined by measuring NADPH fluorescence, as the enzyme was titrated. The resulting dissociation constant was 11 ± 1 μM, 5-fold tighter than the value of 50 μM reported for SaODH6 (Figure S2C). A dissociation constant for NADH could not be determined in the concentration range used in this method, indicating a lower affinity of PaODH for NADH. These results confirm that PaNAS binds SAM and PaODH binds NADPH.
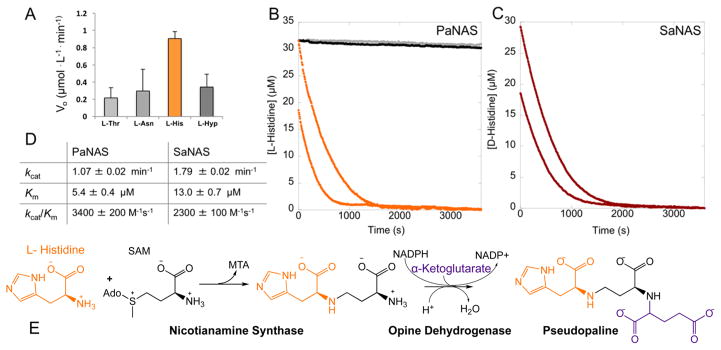
To identify the unknown P. aeruginosa NAS and ODH substrates, we reconstituted the biosynthetic pathway in vitro using purified protein. We expected to observe a decrease in the absorbance at 340 nm as the NADPH was oxidized to NADP+ by the PaODH in the presence of PaNAS, SAM, and the correct amino acid and α-keto acid substrates (Figure 2B). Pyruvate, oxaloacetate, and α-ketoglutarate were screened with SAM, but no oxidation was observed, suggesting that SAM alone is not sufficient as a substrate for PaNAS. We screened 42 L- and D-amino acid substrates (Table S1) in combination with pyruvate, oxaloacetate, or α-ketoglutarate. Several amino acids, L-threonine, L-asparagine, and L-hydroxyproline (L-Hyp), appeared to show limited turnover in this screen, while the combination of L-histidine and α-ketoglutarate resulted in significant oxidation of NADPH by PaODH (Figure 3A and Figure S3B–D).
Figure 3.
(A) P. aeruginosa screening. Each enzyme (2 μM), NADPH (75 μM), an amino acid (200 μM each), α-ketoglutarate, and SAM were combined in 50 mM KPi pH 8 buffer (Hyp is L-hydroxyproline). (B) P. aeruginosa NAS full progress curves. No amino acid (0 μM, black), 25 μM L-Thr, L-Asn, or L-Hyp (gray), or 25 μM (orange, top) or 12.5 μM L-histidine (orange, bottom) was combined with 2 μM PaNAS, 20 μM PaODH, 200 μM NADPH, 400 μM SAM, and 400 μM α-ketoglutarate in 50 mM KPi pH 8 buffer. L-Thr and L-Asn were omitted for the sake of clarity as they overlay with the 0 μM control. (C) S. aureus NAS full progress curves. D-Histidine at 25 μM (red, top) or 12.5 μM (red, bottom) was combined with 2 μM SaNAS, 20 μM SaODH, 200 μM NADPH, 400 μM SAM, and 400 μM pyruvate in 50 mM Tris pH 8 buffer. (D) NAS kinetic parameters. Determined by fitting the Michaelis–Menten equation to a secondary plot of initial rates for varying concentrations of L-His (PaNAS) or D-His (SaNAS). Error propagated on the basis of the standard deviation of four trials. (E) Pseudopaline biosynthesis.
To confirm PaNAS catalysis with these substrates, we measured full progress curves on a TgK stopped flow spectrometer. Using this more precise method, catalysis was observed for only L-histidine, indicating that the limited NADPH oxidation measured for L-Thr, L-Asn, and L-Hyp in our amino acid screening was the result of variance in our plate reader measurements (Figure 3B). Steady state kinetic parameters for the reaction of PaNAS with L-histidine were determined by fitting the Michaelis–Menton equation to a secondary plot of initial rates measured by stopped flow spectrometry at varying L-histidine concentrations (Figure S4A). Using this fit, we determined a kcat of 1.07 ± 0.02 min−1, a Km of 5.4 ± 0.4 μM, and a kcat/Km of 3400 ± 200 M−1 s−1 (Figure 3D). These results demonstrate that PaNAS is specific for L-histidine and that the P. aeruginosa biosynthetic pathway incorporates L-histidine, aminobutyrate and α-ketoglutarate to form an opine metallophore product (Figure 3E).
To confirm the mass of this product, we incubated the P. aeruginosa enzymes and substrates for 2 h at room temperature and performed mass spectrometry, predicting that an opine product composed of one L-histidine, one aminobutyrate, and one α-ketoglutarate would have a neutral mass of 386.14 Da. We considered the possibility that NAS could incorporate two aminobutyrate moieties as in M. thermautotrophicus15 or three as in plant NAS enzymes.18 This would result in neutral product masses of 487.20 and 588.25 Da. The product m/z species for the P. aeruginosa reaction had an [M + H]+ HRMS m/z of 387.1526 (Figure S5A–D). Also observed was an ion at m/z 258.04 matching the [M + H]+ for the NAS product. No m/z matching products with zero, two, or three aminobutyrates were observed, leading us to propose an overall reaction scheme for the biosynthesis of a novel opine metallophore we name pseudopaline (Figure 3E).
As opine dehydrogenases use diverse amino acid and α-keto acid substrates, the substrate specificity of the S. aureus enzymes was examined. All 42 amino acids were tested in conjunction with pyruvate in the plate reader screening assay. Only in the presence of D-histidine were we able to measure significant oxidation of NADPH (Figure 3C and Figure S3A). A secondary plot of SaNAS initial reaction rates measured by stopped flow spectrometry at varying concentrations of D-histidine allows us to determine a kcat of 1.79 ± 0.02 min−1, a Km of 13.0 ± 0.7 μM, and a kcat/Km of 2300 ± 100 M −1 s−1 (Figure S4B). As key intermediates in the citric acid cycle, we reasoned that oxaloacetate and α-ketoglutarate would be available for opine biosynthesis. Reactions with pyruvate and D-histidine resulted in a NADPH oxidation rate of 3.2 ± 0.3 μmol L−1 min−1. In the presence of oxaloacetate, the rate was 1.1 ± 0.2 μmol L−1 min−1, and for α-ketoglutarate, the rate was within error of zero (Figure 4). Catalysis with oxaloacetate suggests that S. aureus could be producing two staphylopine variants, one with pyruvate as in Figure 2A and an analogue with oxaloacetate. However, the significantly lower initial rate with oxaloacetate may suggest that its incorporation is not physiologically relevant. While we can readily identify staphylopine with pyruvate incorporated by mass spectrometry (Figure S6A–C), we have been unable to confirm the production of the oxaloacetate variant.
Figure 4.
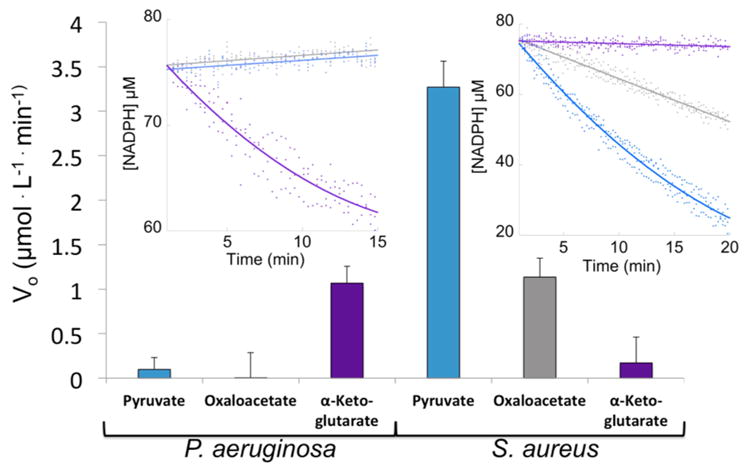
Opine dehydrogenase substrate specificity. Initial reaction rates of PaODH (left) and SaODH (right) with pyruvate, oxaloacetate, or α-ketoglutarate. Rates were derived from the inset data (left, PaODH; right, SaODH). P. aeruginosa reactions used L-His, while S. aureus reactions used D-His. The enzyme (2 μM), 75 μM NADPH, 500 μM SAM, 2 mM histidine, and 2 mM α-keto acid were combined in 50 mM KPi (Pa) or Tris (Sa) pH 8 buffer.
Our results demonstrate that opine metallophores are not exclusive in the genus Staphylococcus as they are also found in the widely diverged species P. aeruginosa. Pseudopaline represents a third identified metallophore produced by P. aeruginosa in addition to the well-characterized pyoverdin and pyochelin molecules. Pseudopaline and staphylopine incorporate an imidazole functional group from histidine with opposite stereochemistry. Additionally, pseudopaline incorporates α-ketoglutarate into the metallophore scaffold, which is bulkier and contains an extra carboxyl group compared to the pyruvate-derived metallophore found in staphylopine, providing additional flexibility and potentially increased metal affinity. The S. aureus opine dehydrogenase catalytic activity with α-ketoglutarate is within error of zero, but SaODH accepts oxaloacetate, also containing an additional carboxylate group, as a substrate alternative at 35% the steady state rate of pyruvate (Figure 4). Conversely, P. aeruginosa shows no catalytic activity in the presence of pyruvate or oxaloacetate (within error of zero), but full activity with α-ketoglutarate (Figure 4).
The functional role of opine metallophores is not well-established but is associated with virulence in S. aureus and P. aeruginosa. Homologous operons are found in Serratia marcescens (WW4) and Yersenia pestis (CO92), suggesting that opine metallophores are produced by these pathogens. Gi et al.12 predicted the presence of NAS in the P. aeruginosa operon. Their work demonstrates that the operon is involved in iron uptake during growth in culture derived airway mucus. This is significant as previous work has addressed the uptake of iron by pyoverdin and pyochelin, but the production of pseudopaline by P. aeruginosa must now also be considered in model systems. Staphylopine is involved in nickel and cobalt acquisition but also takes up iron in metal-limited media.6 The conservation of the histidine moiety in pseudopaline suggests a potential role in nickel and cobalt uptake in P. aeruginosa given the established role of histidine-dependent nickel transport by S. aureus NikA and staphylopine-dependent nickel and cobalt transport by CntA.19 The role of trace elements in pathogenesis, such as nickel and cobalt, has received less scrutiny than that of iron. However, urease, a nickel-dependent enzyme, is an established virulence factor in S. aureus where it is expressed in 90% of methicillin resistant strains.20 Urease is also produced by P. aeruginosa.21 Cobalt is required for the P. aeruginosa cobalamin-dependent ribonucleotide reductase (NrdJab), which is necessary for biofilm development under oxygen-limited conditions.22 While the presence of pseudopaline in P. aeruginosa cultures will still need to be established to confirm production in vivo, these findings point to the importance of nickel and cobalt uptake in pathogenesis, suggesting a potential role for pseudopaline in the transport of these trace metals.
This work establishes the existence of an opine metallophore biosynthetic pathway in a second major pathogen, P. aeruginosa, and proposes the structure of the small molecule pseudopaline. The characterization of the enzymes, nicotianamine synthase and opine dehydrogenase, reveals the recognition of amino acid substrates with opposite stereochemistry by SaNAS and PaNAS, and the incorporation of different α-keto acids, pyruvate, and oxaloacetate by SaODH and α-ketoglutarate by PaODH.
Supplementary Material
Acknowledgments
Funding
This publication was made possible by funds from National Science Foundation Grant CHE-1403293. J.S.M. was supported by the National Institutes of Health Graduate Training Program in the Dynamic Aspects of Chemical Biology via Grant T32 GM008545.
Special thanks to Annemarie Chilton and Kathy Meneely for their advice regarding the design of experiments, Ben Neuenswander and Matthew Hoag for mass spectrometry analysis, and Joe Kappock and Graham Moran for critically reading the manuscript.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.7b00804.
References
- 1.Skaar EP. The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts. PLoS Pathog. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer J, Neely A, Stintzi A, Georges C, Holder IA. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale SE, Doherty-Kirby A, Lajoie G, Heinrichs DE. Role of Siderophore Biosynthesis in Virulence of Staphylococcus aureus: Identification and Characterization of Genes Involved in Production of a Siderophore. Infect Immun. 2004;72:29–37. doi: 10.1128/IAI.72.1.29-37.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindsay JA, Riley TV, Mee BJ. Production of siderophore by coagulase-negative staphylococci and its relation to virulence. Eur J Clin Microbiol Infect Dis. 1994;13:1063–1066. doi: 10.1007/BF02111829. [DOI] [PubMed] [Google Scholar]
- 6.Ghssein G, Brutesco C, Ouerdane L, Fojcik C, Izaute A, Wang SL, Hajjar C, Lobinski R, Lemaire D, Richaud P, Voulhoux R, Espaillat A, Cava F, Pignol D, Borezee-Durant E, Arnoux P. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science. 2016;352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 7.Remy L, Carriere M, Derre-Bobillot A, Martini C, Sanguinetti M, Borezee-Durant E. The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Mol Microbiol. 2013;87:730–743. doi: 10.1111/mmi.12126. [DOI] [PubMed] [Google Scholar]
- 8.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H, Min J, Ikeguchi Y, Zeng H, Dong A, Loppnau P, Pegg AE, Plotnikov AN. Structure and mechanim of spermidine synthases. Biochemistry. 2007;46:8331–8339. doi: 10.1021/bi602498k. [DOI] [PubMed] [Google Scholar]
- 10.Moore LW, Chilton WS, Canfield ML. Diversity of Opines and Opine-Catabolizing Bacteria Isolated from Naturally Occurring Crown Gall Tumors. Appl Environ Microbiol. 1997;63:201–207. doi: 10.1128/aem.63.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strahl J, Dringen R, Schmidt MM, Hardenberg S, Abele D. Metabolic and physiological responses in tissues of the long-lived bivalve Arctica islandica to oxygen deficiency. Comp Biochem Physiol, Part A: Mol Integr Physiol. 2011;158:513–519. doi: 10.1016/j.cbpa.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Gi M, Lee KM, Kim SC, Yoon JH, Yoon SS, Choi JY. A novel siderophore system is essential for the growth of Pseudomonas aeruginosa in airway mucus. Sci Rep. 2015;5:14644. doi: 10.1038/srep14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreyfus C, Lemaire D, Mari S, Pignol D, Arnoux P. Crystallographic snapshots of iterative substrate translocations during nicotianamine synthesis in Archaea. Proc Natl Acad Sci U S A. 2009;106:16180–16184. doi: 10.1073/pnas.0904439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bielecki P, Komor U, Bielecka A, Musken M, Puchalka J, Pletz MW, Ballmann M, Martins dos Santos VA, Weiss S, Haussler S. Ex vivo transcriptional profiling reveals a common set of genes important for the adaptation of Pseudomonas aeruginosa to chronically infected host sites. Environ Microbiol. 2013;15:570–587. doi: 10.1111/1462-2920.12024. [DOI] [PubMed] [Google Scholar]
- 17.Bielecki P, Puchalka J, Wos-Oxley ML, Loessner H, Glik J, Kawecki M, Nowak M, Tummler B, Weiss S, dos Santos VA. In-vivo expression profiling of Pseudomonas aeruginosa infections reveals niche-specific and strain-independent transcriptional programs. PLoS One. 2011;6:e24235. doi: 10.1371/journal.pone.0024235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shojima S, Nishizawa N, Fushiya S, Nozoe S, Irifune T, Mori S. Biosynthesis of Phytosiderophores. Plant Physiol. 1990;93:1497–1503. doi: 10.1104/pp.93.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebrette H, Borezee-Durant E, Martin L, Richaud P, Boeri Erba E, Cavazza C. Novel insights into nickel import in Staphylococcus aureus: the positive role of free histidine and structural characterization of a new thiazolidine-type nickel chelator. Metallomics. 2015;7:613–621. doi: 10.1039/c4mt00295d. [DOI] [PubMed] [Google Scholar]
- 20.Konieczna I, Zarnowiec P, Kwinkowski M, Kolesinska B, Fraczyk J, Kaminski Z, Kaca W. Bacterial Urease and its Role in Long-Lasting Human Disease. Curr Protein Pept Sci. 2012;13:789–806. doi: 10.2174/138920312804871094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics. 2009;10:78. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crespo A, Pedraz L, Astola J, Torrents E. Pseudomonas aeruginosa Exhibits Deficient Biofilm Formation in the Absence of Class II and III Ribonucleotide Reductases Due to Hindered Anaerobic Growth. Front Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.