Abstract
Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) is a rare, highly aggressive form of ovarian cancer primarily diagnosed in young women. We identified inactivating biallelic SMARCA4 mutations in 100% of the 12 SCCOHT tumors examined. Protein studies confirmed loss of SMARCA4 expression, suggesting a key role for the SWI/SNF chromatin-remodeling complex in SCCOHT.
Main
SCCOHT is a rare, aggressive form of ovarian cancer diagnosed in young women that is generally fatal when it spreads beyond the ovary. SCCOHT represents less than 1% of all ovarian cancer diagnoses, with fewer than 300 cases reported in the literature thus far1,2. The mean age at diagnosis is 23 years, and, unlike with the more common types of ovarian cancer, the majority of affected women present with early-stage disease3. Nonetheless, most patients relapse and die within 2 years of diagnosis, regardless of tumor stage, with a long-term survival rate of only 33%, even when disease is confined to the ovary at diagnosis4. There are no reliable adjuvant treatments that improve disease outcome, but multi-agent chemotherapy is thought to extend survival1,5.
The tissue of origin for SCCOHT remains speculative, and SCCOHT is still categorized as a miscellaneous tumor by the World Health Organization. Most tumors are unilateral, and tumor size of greater than 10 cm in diameter may be prognostically favorable1. Histological classification can be challenging, but commonly expressed immunohistochemical markers can be useful in excluding histological mimics6. Because there is an unmet need for better therapeutic options in SCCOHT, we performed target capture and massively parallel DNA sequencing to identify recurrent somatic mutations.
Sequencing of all protein-coding exons in 279 cancer-related genes for 12 paired tumor and normal SCCOHT samples identified inactivating biallelic SMARCA4 mutations in each case. The tumor and normal samples were sequenced to a mean depth of 442× across all genes. A minimum depth of 100× was achieved in 97% of targeted exons in tumors. The SMARCA4 somatic mutations identified in the tumor samples are shown in Supplementary Table 1. The probability of identifying SMARCA4 mutations in all 12 samples is less than 2.22 × 10−16. Only 4 additional non-recurrent somatic mutations were identified in any of the other 278 genes sequenced across all 12 samples (Supplementary Tables 2 and 3). In contrast, an analysis of 4,784 non-hypermutated tumors across The Cancer Genome Atlas (TCGA) identified somatic mutations in an average of 4.3 of these 279 genes for each tumor (s.d. of 4.4). TCGA samples with inactivating SMARCA4 mutations had more mutations in the other 278 genes sequenced (mean of 14) than the SCCOHT cases. Because the SMARCA2 and SMARCA4 proteins are mutually exclusive subunits within the BAF complex, we examined the co-occurrence of inactivating mutations in SMARCA2 and SMARCA4 in all non-hypermutated TCGA tumors. We found only one case with an inactivating mutation in SMARCA4 and a missense mutation in SMARCA2.
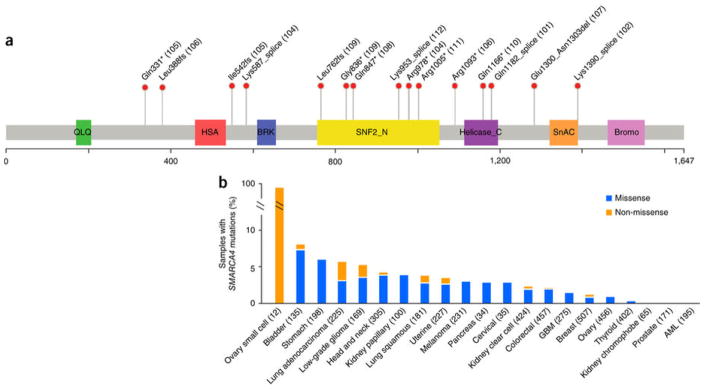
SMARCA4 mutations occurred throughout various exons and included nonsense, frameshift and splice-site mutations as well as a homozygous intragenic deletion of two exons (Fig. 1). No missense mutations of SMARCA4 were identified. We validated sequence variants from all 12 cases using Sanger sequencing. cDNA was sequenced in seven samples, and all were found to have mutations that were expressed in the mRNA transcripts (Supplementary Fig. 1). Four cases harbored two inactivating mutations each in SMARCA4. The remaining eight cases harbored single inactivating mutations accompanied by loss of heterozygosity at the SMARCA4 locus (supported by analysis of adjacent SNPs); in each case, the frequency of the mutant allele was 0.75 or greater. TCGA cases demonstrated a correlation between inactivating mutations in SMARCA4 and decreased SMARCA4 gene expression across various solid tumors, suggesting that truncating mutations induce nonsense-mediated decay (NMD) (Supplementary Fig. 2).
Figure 1. SMARCA4 mutations in SCCOHT and TCGA samples.
(a) Domain structure of the SMARCA4 protein (UniProt, SMCA4_HUMAN) overlaid with the alterations identified in 11 of the 12 SCCOHT cases in this study (case numbers in parentheses; case 103 with exon deletion is not shown). SNF2_N, SNF2 family N-terminal domain; helicase, helicase-conserved C-terminal domain; SnAC, Snf2-ATP coupling, chromatin- remodeling complex; bromo, bromodomain. Genomic coordinates for splice-site mutations can be found in Supplementary Table 5. (b) Percentages of samples with nonsynonymous SMARCA4 mutations in SCCOHT and TCGA non-hypermutated samples (numbers of samples per study in parentheses). Blue bars represent samples with missense-only mutations, and orange bars represent samples with non-missense (including nonsense, frameshift, splice-site and indel) mutations. GBM, glioblastoma multiforme; AML, acute myeloid leukemia.
We performed immunohistochemistry analysis of SMARCA4 (BRG1) in the nine cases with available tissue using commercially available antibodies. High-grade serous ovarian cancer with wild-type SMARCA4 sequence served as a positive control. In cases 104, 105, 106, 108, 109 and 110, nonsense mutations resulted in the introduction of a premature stop codon within the ORF of the mRNA transcript. In cases 104, 105, 106 and 109, nonsense mutations were heterozygous with frameshift or splice-site mutations. Immunohistochemistry demonstrated loss of SMARCA4 nuclear staining, compared to retention of staining in the internal positive control cells, in all four cases with nonsense mutations with available tissue (Supplementary Fig. 3). Case 111 had a heterozygous germline nonsense mutation with loss of the wild-type somatic allele and associated loss of protein expression in immunohistochemistry analysis. This case had a paucity of the pseudofollicular spaces that were more common in the remainder of the study cohort.
We confirmed a homozygous in-frame deletion of exons 25 and 26 detected in case 103 by Sanger sequencing of cDNA, with this alteration resulting in deletion of 102 amino acids of the helicase domain (Supplementary Fig. 4). Amplification with upstream and downstream primer pairs confirmed that transcription continued downstream of this deletion. Immunohistochemistry showed the retention of protein expression. However, sequencing data confirming an impaired C-terminal helicase domain suggest that this deletion results in the translation of a truncated, non-functional, catalytically dead product. A homozygous in-frame deletion within exon 27 resulting in the deletion of four amino acids (ETVN) was detected in case 107. No additional tissue was available to demonstrate the effect of this deletion on protein expression.
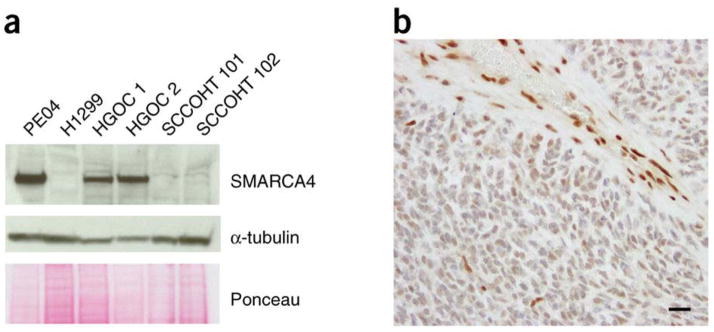
Because of the precise location of the biallelic splice-site mutations within intronic sequence in the highly conserved AG donor region, we tested whether introns were retained in cases 101, 102 and 112. We identified preferential intronic expression, as expected, in cDNA sequenced from a representative tumor sample with a splice-site mutation (Supplementary Fig. 5). One-step RT-PCR confirmed continuation of transcription downstream of the splice-site mutations, suggesting that the splice-site mutations do not cause mRNA truncation. However, immunoblots and immunohistochemistry showed clear loss of SMARCA4 protein in cases 101 and 102 (Fig. 2 and Supplementary Fig. 3). Case 112 had equivocal loss of protein expression in immunohistochemistry analysis, with tumor cells staining less intensely than normal tissue elements.
Figure 2. Analyses of the splice-site mutation in case 102.
(a) Immunoblotting with antibody to the N terminus of SMARCA4. A high-grade serous ovarian cancer cell line (PEO4) and frozen tumor samples from two individuals with high-grade serous ovarian cancer (HGOC) were used as positive controls and retain protein expression. Protein extracted from H1299 non-small-cell lung cancer cells, deficient in SMARCA4, served as a negative control (Supplementary Fig. 6). Protein extracted from SCCOHT cases 101 and 102, both with donor-site splice-site mutations, show loss of SMARCA4 protein expression. (b) Loss of protein expression in archival tissue stained with a polyclonal antibody to SMARCA4. Note the intense staining of blood vessels and stromal cell nuclei as internal controls. Scale bar, 10 μm.
To determine the functional effects of SMARCA4 loss, we ectopically reintroduced SMARCA4 through electroporation in SMARCA4-null H1299 non –small-cell lung adenocarcinoma cells (Supplementary Fig. 6). Re-expression of SMARCA4 resulted in a dose-dependent suppression of cell growth. More p21 was also expressed, consistent with previous reports of the effect of SMARCA4 on cell cycle arrest7,8. We then stably depleted SMARCA4 in 293T cells using lentivirus-expressed short hairpin RNA (shRNA), which led to an increase in cell growth as measured by XTT proliferation assay (Supplementary Fig. 7).
Recurrent SMARCA4 mutations, found in 100% of the SCCOHT tumors sequenced as part of this study, were all inactivating and were associated with clear loss of protein expression in seven of nine cases, suggesting important oncogenic functions. SCCOHT tumors had few other mutations in the panel of 278 sequenced genes. Although the study was limited by a modest sample size due to the rarity of this disease, the identification of inactivating mutations in a single gene in all 12 tumors studied is consistent with the characteristics of a tumor suppressor and is supported by a pilot study of 2 SCCOHT cases with SMARCA4 mutations9. Most of the identified mutations affected the known helicase catalytic domains of SMARCA4, suggesting a potential role for these mutations in tumorigenesis. One case contained a germline mutation, which is consistent with previous reports suggesting a hereditary component to this disease10,11.
SMARCA4 mutations have previously been reported at low frequency in other solid tumors. Across all tumors characterized by TCGA thus far, SMARCA4 mutations have been detected in 3% of the 4,787 non-hypermutated samples. Mutation frequencies of 5–8% are present in bladder carcinoma, stomach adenocarcinoma, lung adenocarcinoma and lower grade glioma. Of the 128 somatic SMARCA4 mutations identified in the TCGA samples, 84% were missense variants of uncertain functional consequence (Fig. 1b). Inactivating mutations in SMARCA4 were most common in lung adenocarcinoma, although still infrequent, and were associated with poor outcome (Supplementary Fig. 8).
SWI/SNF complexes function as master regulators of gene expression by remodeling chromatin to alter nucleosome conformation, making it more accessible to transcriptional activation. The highly homologous SMARCA4 (BRG1) and SMARCA2 (BRM) proteins are mutually exclusive ATP-dependent catalytic subunits of the BAF complex and coexist with ARID1A (BAF250A) or ARID1B (BAF250B)12,13. SMARCA4 in the PBAF complex is associated with the PBRM1 (BAF180) subunit, which is absent in the BAF complex. Recent studies have demonstrated that silencing of SMARCA2 through RNA interference suppresses the growth of SMARCA4-deficient lung cancer cell lines and xenografts, suggesting a synthetic lethal relationship for these subunits14. ARID1A loss has recently been shown to be tumorigenic in an ovarian cancer lineage, suggesting that SMARCA4 loss might be sufficient for transformation mediated through BAF complex p53-dependent mechanisms15. Inhibitors of the SMARCA2 ATPase may be an effective approach for the treatment of SMARCA4-deficient tumors. Improved understanding of the function and characteristics of the SWI/SNF complex is now creating therapeutic opportunities for mutated tumors.
Methods
Online Methods
Subjects and samples
After obtaining institutional review board (IRB) approval, we identified all affected individuals at the Memorial Sloan-Kettering Cancer Center diagnosed with SCCOHT from 1998–2012. Because of the rarity of this disease, we identified only three cases with available tissue. We therefore contacted other academic centers and patient support groups to obtain nine additional cases. Informed consent was obtained from each patient or next of kin. A specialist in gynecologic pathology reviewed all cases to confirm diagnosis of SCCOHT. SCCOHT was diagnosed in the presence of a highly cellular and highly proliferative small cell malignancy with minimal stroma with follicle-like spaces (Supplementary Fig. 9). The presence of a large cell or rhabdoid component was accepted as part of the spectrum of this disease. The following entities were excluded from consideration with a combination of morphological examination and immunohistochemistry analysis: small cell neuroendocrine carcinoma, juvenile granulosa cell tumor, poorly differentiated Sertoli-Leydig cell tumor, desmoplastic small round cell tumor, metastatic melanoma, lymphoma and rhabdomyosarcoma (Supplementary Table 4). Clinical data collection was limited to only age and year of diagnosis owing to Health Insurance Portability and Accountability Act (HIPAA) regulations. No family history was available. DNA and RNA were extracted from paired formalin-fixed, paraffin-embedded tumors with at least 50% tumor cell nuclei and normal tissue samples according to standard protocols. Germline DNA was derived from either peripheral lymphocytes or formalin-fixed, paraffin-embedded blocks of anatomically distant tissues, such as benign lymph nodes, and used as the source of normal tissue.
IMPACT assays
Paired normal and tumor samples were sequenced to a depth of at least 100× using target capture and massively parallel sequencing. We profiled genomic alterations in 279 key cancer-associated genes using IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets), a custom hybrid capture-based deep sequencing assay16. The selected genes encompass all well-established oncogenes and tumor suppressor genes, including all druggable targets of therapies approved by the US Food and Drug Administration (FDA) and investigational agents in clinical trials at the Memorial Sloan-Kettering Cancer Center. Custom oligonucleotide probes were designed to capture all protein-coding exons and select introns from these 279 commonly implicated oncogenes, tumor suppressor genes and members of pathways deemed actionable by targeted therapies (Supplementary Table 3). We prepared barcoded sequence libraries (New England BioLabs, Kapa Biosystems) and performed exon capture on barcoded pools by hybridization (Nimblegen SeqCap) using an input of 97–250 ng of DNA, as previously described17. Captured pools were sequenced on an Illumina HiSeq 2000 (2 × 75-bp reads), and reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Alignment tool18. Filtering for duplicates, local multiple-sequence alignment and recalibration of base quality scores were performed using the Genome Analysis Toolkit (GATK) according to GATK best practices19. Sequence data were analyzed to identify three classes of somatic alterations: single-nucleotide variants, identified using MuTect20; small indels, identified using SomaticIndelDetector19; and copy number alterations, identified as previously described17. All candidate mutations and indels were manually reviewed using the Integrative Genomics Viewer (Supplementary Table 5)21.
All mutations were validated using Sanger sequencing of both genomic DNA and RNA transcripts (cDNA) to confirm the somatic nature of the alteration and transcript expression. Primers spanning exon-intron boundaries 24, 27 and 18 were generated for cases 101, 102 and 112, respectively, to detect the presence of retained introns. For cDNA synthesis, the SuperScript III One-Step RT-PCR System (Invitrogen) was used according to the manufacturer’s instructions.
Protein studies
Frozen tumor samples were available for two cases (cases 101 and 102).
Extracted protein was resolved by SDS-PAGE, transferred onto nitrocellulose and blotted with polyclonal antibody to Brg-1 (1:1,000 dilution; Santa Cruz Biotechnology, sc-10768) and with antibody to α-tubulin (1:1,000 dilution; Santa Cruz Biotechnology, sc-5546) as a loading control.
The immunohistochemistry staining method for SMARCA4 was optimized using several antibodies under a variety of different conditions until one was chosen on the basis of its ability to demonstrate consistent nuclear staining patterns in a small group of high-grade serous ovarian cancers serving as positive controls. One whole formalin-fixed, paraffin-embedded section from each of the available SCCOHT cases was evaluated with a commercially available polyclonal antibody against SMARCA4 (Upstate Cell Signaling Solutions, 07-478). Whole sections underwent epitope retrieval using heat by steaming with EDTA at pH 8 for 30 min. Antigen retrieval was followed by overnight incubation with the primary antibody at 4 °C (1:4,000 dilution). Detection of bound antibody was accomplished with biotinylated anti-rabbit IgG (1:500 dilution; Vector Laboratories, BA-1000) and ABC (Vector Laboratories, PK-6100). Diaminobenzidine (DAB) was used as the chromogen. For tumors with mutant SMARCA4 and loss of SMARCA4 protein expression, positive staining of blood vessels and stromal cells was used as an internal positive control. Absence of nuclear staining in tumor cells in the presence of internal positive control staining was scored as ‘loss of expression’.
Before protein extraction, cells were washed with ice-cold PBS. Cells were lysed in RIPA buffer, and extracted protein was resolved by SDS-PAGE, transferred onto nitrocellulose and blotted with antibodies to Brg-1 (1:1,000 dilution; Santa Cruz Biotechnology, sc-10768), p21 (1:1,000 dilution; Cell Signaling Technology, 2947) and β-actin (1:10,000 dilution; Santa Cruz Biotechnology, sc-69879) as a loading control.
Computational and biostatical analyses
Mutation frequencies across TCGA tumor types were collated from data contained within the cBioPortal for Cancer Genomics22,23. Background mutation frequencies for the 279 genes sequenced as part of this study were also obtained for TCGA tumor types, excluding hypermutated cases that carried more than 1,000 nonsynonymous mutations.
The probability of finding 1 gene mutated in all 12 samples when 279 genes are sequenced is given by 1 – (1 – p12 )279. This equation assumes that mutation of a given gene in a case is a Bernoulli trial with probability p, that the gene mutations are exchangeable and that these mutations are also independently and identically distributed across cases. We used P = 0.015, derived from TCGA samples as explained above.
Functional studies
Cell lines
The H1299 human non–small-cell lung carcinoma cell line was a kind gift from the Powell Laboratory (Memorial Sloan-Kettering Cancer Center). The cell line was authenticated in June 2013 by the STR DNA profiling method (Genetica DNA Laboratories) using the DSMZ database. H1299 cells were cultured in RPMI medium supplemented with 10% FCS. 293T cells were obtained from the American Type Culture Collection (ATCC) in September 2011. Cells were cultured in DME-HG medium supplemented with 10% FCS. All cells tested negative for mycoplasma.
SMARCA4 overexpression in H1299 cells
Plasmid containing SMARCA4 cDNA (pCMV6-XL5; Origene, SC323288) was transfected into H1299 cells by electroporation using Nucleofector (Amaxa). After 24 h, cells were counted using a TC10 Automated Cell Counter (Bio-Rad), and protein was extracted and analyzed by immunoblotting.
SMARCA4 knockdown and XTT proliferation assays in 293T cells
To knock down SMARCA4, the GIPZ SMARCA4 shRNA Viral Particle Starter kit (Thermo Scientific) was used. Transduction was performed according to the manufacturer’s suggestions. Stable knockdown was achieved by selection with 2 μg/ml puromycin. XTT proliferation assays (ATCC, 30-1011K) were performed according to the manufacturer’s suggestions. Cells (40,000) were seeded in a 24-well dish, and absorbance was measured at different time points after seeding (24–96 h later) using a Synergy HT Plate Reader (BioTek) and used to report relative cell proliferation.
Supplementary Material
1. Sequence analyses for SMARCA4 in SCCOHT cases.
2. SMARCA4 gene expression across TCGA tumors for cases with available mutation and RNA-seq data (RSEM).
3. Immunohistochemistry for SMARCA4 in SCCOHT cases.
4. Analysis of homozygous deletion in case 103.
5. Analysis of splice-site variant in case 102.
6. Sequence analyses for SMARCA4 in the H1299 cell line.
7. SMARCA4 effect on cell proliferation.
8. Overall survival among lung adenocarcinoma TCGA cases based on inactivating SMARCA4 mutations.
9. Histopathological features of an SCCOHT.
Acknowledgments
We appreciate the assistance of the Small Cell Ovarian Cancer Foundation with recruitment. We are grateful to all the patients and family members who generously donated biospecimens so that we could conduct this study. We thank G. Monemvasitis for editorial assistance. This work was supported by the Katie Oppo Research Fund, the Chia Family Foundation, the Geoffrey Beene Cancer Research Center, the Farmer Family Foundation and a Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209).
Footnotes
Competing interests
The authors declare no competing financial interests.
Contributions
J.J.M. and D.A.L. conceived the study. N.S., M.F.B. and D.A.L. supervised analyses. P.J., J.J.M. and F.D. designed and performed analyses. R.A.S. and D.A.L. provided patient materials. N.O. processed tissue samples. R.A.S. performed pathology review. M.G. performed statistical analyses. S.N.S. and R.S. performed massively parallel DNA sequencing. J.G. performed computational analyses. P.J., J.J.M. and D.A.L. wrote the manuscript with contributions from all other authors.
URLs.
cBioPortal for Cancer Genomics, http://cbioportal.org/; DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) database, http://www.dsmz.de/.
Contributor Information
Petar Jelinic, Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Jennifer J Mueller, Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Narciso Olvera, Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Fanny Dao, Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Sasinya N Scott, Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Ronak Shah, Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Jian Jiong Gao, Computational Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Nikolaus Schultz, Computational Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Mithat Gonen, Department of Epidemiology and Biostatistics, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Robert A Soslow, Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Michael F Berger, Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
Douglas A Levine, Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, New York, USA.
References
- 1.Estel R, Hackethal A, Kalder M, Munstedt K. Arch Gynecol Obstet. 2011;284:1277–1282. doi: 10.1007/s00404-011-1846-5. [DOI] [PubMed] [Google Scholar]
- 2.Young RH, Oliva E, Scully RE. Am J Surg Pathol. 1994;18:1102–1116. doi: 10.1097/00000478-199411000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Harrison ML, et al. Gynecol Oncol. 2006;100:233–238. doi: 10.1016/j.ygyno.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Seidman JD. Gynecol Oncol. 1995;59:283–287. doi: 10.1006/gyno.1995.0023. [DOI] [PubMed] [Google Scholar]
- 5.Pautier P, et al. Ann Oncol. 2007;18:1985–1989. doi: 10.1093/annonc/mdm376. [DOI] [PubMed] [Google Scholar]
- 6.McCluggage WG. Adv Anat Pathol. 2004;11:288–296. doi: 10.1097/01.pap.0000138146.357376.1e. [DOI] [PubMed] [Google Scholar]
- 7.Hendricks KB, Shanahan F, Lees E. Mol Cell Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napolitano MA, et al. J Cell Sci. 2007;120:2904–2911. doi: 10.1242/jcs.004002. [DOI] [PubMed] [Google Scholar]
- 9.Kupryjańczyk J, et al. Pol J Pathol. 2013;64:238–246. doi: 10.5114/pjp.2013.39331. [DOI] [PubMed] [Google Scholar]
- 10.Longy M, Toulouse C, Mage P, Chauvergne J, Trojani M. J Med Genet. 1996;33:333–335. doi: 10.1136/jmg.33.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald JM, et al. J Pediatr Surg. 2012;47:588–592. doi: 10.1016/j.jpedsurg.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Reisman D, Glaros S, Thompson EA. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 13.Wilson BG, Roberts CW. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 14.Oike T, et al. Cancer Res. 2013;73:5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 15.Guan B, Wang TL, Shih IM. Cancer Res. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. J Vis Exp. 2013;80:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagle N, et al. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA, et al. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cibulskis K, et al. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson JT, et al. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, et al. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, et al. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Sequence analyses for SMARCA4 in SCCOHT cases.
2. SMARCA4 gene expression across TCGA tumors for cases with available mutation and RNA-seq data (RSEM).
3. Immunohistochemistry for SMARCA4 in SCCOHT cases.
4. Analysis of homozygous deletion in case 103.
5. Analysis of splice-site variant in case 102.
6. Sequence analyses for SMARCA4 in the H1299 cell line.
7. SMARCA4 effect on cell proliferation.
8. Overall survival among lung adenocarcinoma TCGA cases based on inactivating SMARCA4 mutations.
9. Histopathological features of an SCCOHT.