Abstract
A fundamental question in developmental neuroscience is how hundreds of diverse cell types are generated to form specialized brain regions. The ganglionic eminences (GEs) are embryonic brain structures located in the ventral telencephalon that produce many inhibitory GABA (γ-Aminobutyric acid)-ergic cell types, including long-range projection neurons and local interneurons (INs), which disperse widely throughout the brain. While much has been discovered about the origin and wiring of these cells, a major question remains: how do neurons originating in the GEs become specified during development as one differentiated subtype versus another? This review will cover recent work that has advanced our knowledge of the mechanisms governing cortical interneuron subtype specification, particularly progenitors' spatial origin, birthdates, lineage, and mode of division.
Introduction
Beginning at the end of the 19th century, Orr and His proposed the Neuromeric model of brain development to explain how different brain regions are generated from the neural tube [1,2]. According to this model, the forebrain, midbrain, and hindbrain can be divided into a series of segmental compartments called prosomeres, mesomeres, and rhombomeres, respectively, from which different regions of the brain differentiate. Among the neuromeric divisions, rhombomeres have been the most extensively studied in vertebrates. Each rhombomere is a transverse section along the longitudinal axis of the neural tube with a bulge-like appearance, unique pattern of gene expression, and strict boundaries that prevent cells from migrating between adjacent compartments [3]. These demarcations have functional consequences. Precise combinatorial expression of transcription factors confers neuronal identity so that each rhombomere contributes in an exquisitely stereotyped way to the formation of individual nerves. For instance, in the chick embryo rhombomeres 2 and 3 contribute to the branchiomotor nucleus of the trigeminal nerve (CN V), rhombomeres 4 and 5 contribute to the facial nerve (CN VII), and rhombomeres 6 and 7 contribute to the glossopharyngeal nerve (CN IX) [4,5].
In the forebrain, neuronal diversity within the ventral telencephalon is associated with specific transient germinal zones, termed the medial, caudal and lateral ganglionic eminences (MGE, CGE, and LGE, respectively). While not forming longitudinal divisions in the neuraxis, the GEs otherwise satisfy many criteria of prosomeres: they are bulges in the embryonic brain, have spatial patterns of gene expression distinct from one another, and have been shown to contribute to specific cell types in the brain (Figure 1B). For example, the MGE gives rise to PV-expressing cortical interneurons, SST-expressing cortical interneurons, and the projection neurons of the globus pallidus (GP); the CGE gives rise to VIP-expressing cortical interneurons and Reelin-expressing cortical interneurons; and the LGE gives rise to olfactory bulb interneurons and the medium spiny projection neurons of the striatum. However, it has proven much harder to understand the logic governing cell fate decisions in the GEs compared to the rhombomeres of the hindbrain. While the projection neurons derived from the GEs populate subpallial structures including the striatum, globus pallidus and medial amygdala, cortical interneurons travel long and complex tangential migratory routes to their final settling positions throughout the forebrain [6] (Figure 1A). This has made fate-mapping and consistent classification of interneuron subtypes challenging. Complicating matters is the fact that most neurochemical markers that define interneuron subgroups are not expressed at early time points, posing a formidable barrier for understanding mechanisms of fate determination. Moreover, the embryonic preoptic area gives rise to a small yet diverse group of cortical interneurons [7] and the anatomical boundaries of each GE are not clearly demarcated (e.g. the caudal portions of both the MGE and LGE cannot easily be differentiated from the rostral CGE), calling into question the origin of some GABAergic cell types. Despite these challenges, recent work has advanced our knowledge of the mechanisms governing cortical interneuron subtype specification, which will be the main focus of this review.
Figure 1. Each ganglionic eminence produces GABAergic interneurons and projection neurons.
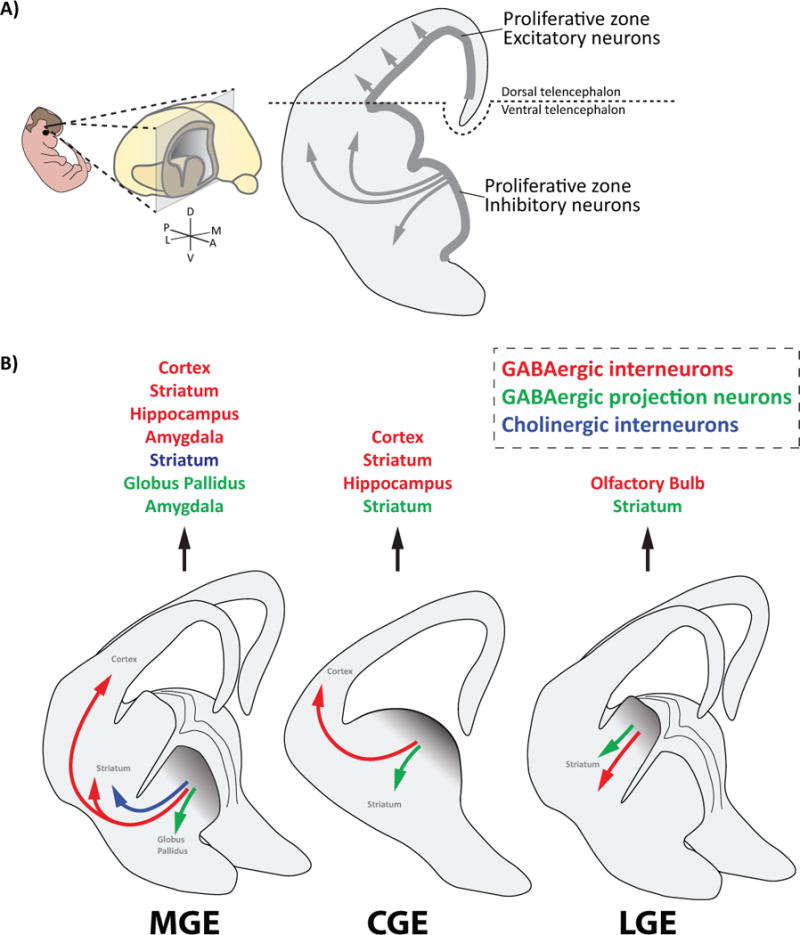
A) Image depicting the boundary between the dorsal telencephalon and the ventral telencephalon. The dorsal telencephalon produces excitatory pyramidal cells that migrate short distances radially into the overlying cortex. The ventral telencephalon produces interneurons that travel long and complex migratory routes to various regions throughout the brain.
B) Diagram depicting the cell types that each ganglionic eminence (GE) produces and the brain structures they occupy, which includes GABAergic interneurons (red), GABAergic projection neurons (green), and Cholinergic interneurons (dark blue).
Spatial domains
Nkx2.1 is a transcription factor expressed in the MGE that is critical for specifying interneurons towards an MGE rather than a CGE or LGE fate. Loss of function studies revealed that in the absence of Nkx2.1, MGE-derived interneurons undergo a fate switch to CGE and LGE subtypes [8,9]. Loss of Shh, which activates Nkx2.1, led to a similar result [10]. Thus, Nkx2.1 is an example of how a master regulator gene that is expressed in an anatomically defined region of the ventral telencephalon can be instrumental in cell fate decisions (e.g. MGE-type vs. CGE-type INs). However, the fact that multiple distinct cell types arise from the MGE (Figure 1B) suggests that smaller subdivisions of the GEs might also be involved in creating more refined subtypes of interneurons (such as PV or SST subtypes of MGE-type INs), similar to the spinal cord [11,12].
Indeed, there is evidence that spatially segregated progenitor domains exist within the MGE, CGE, and LGE that give rise to different neuronal subtypes [13–18]. When cells from the dorsal MGE (dMGE) of GFP+ donor animals were transplanted into the MGE of wild type host embryos, transplanted cells consistently produced a majority of SST+ cortical interneurons and only a minority of PV+ cortical interneurons. However, when GFP+ cells from the ventral MGE (vMGE) were transplanted into wild type hosts, a majority of GFP+ cells in the cortex were PV+ interneurons and only a minority were SST+ interneurons [13]. Thus, there was a spatial bias for the production of specific interneuron subtypes within subregions of the MGE. The same results were observed when dMGE and vMGE cells were transplanted into the postnatal cortex, excluding the possibility that the host MGE influenced transplanted cells’ differentiation [19–21]. In addition, there is evidence that the laminar fate of LGE/CGE-derived neocortical interneurons is also dependent on their progenitor domains [14]. A combination of extrinsic secreted factors acting through the induction of transcription factors creates these spatial domains within each eminence. For instance, while Shh is enriched in the dMGE, Wnt expression is enriched in the caudal portion of the MGE (unpublished data, G. Fishell). Similarly, transcription factors including Lhx8, Gbx2 and Otx all contribute to regional patterning within the MGE along the rostrocaudal and dorsoventral axis, and to varying degrees have been implicated in the specification of particular MGE-derived subtypes [22–24].
Temporal dynamics
Temporal dynamics within each GE also have a profound impact on the type of interneuron produced. For example, in the MGE SST+ interneurons have their peak of neurogenesis earlier than PV+ interneurons (Figure 2C). In particular Chandelier cells, which are a type of PV+ interneuron born late in the ventral germinal zone (VGZ), provide the best example of an interneuron population derived with both temporal and spatial precision [20]: When Nkx2.1+ cells were dissected late during embryogenesis (E16) from the VGZ and transplanted into the somatosensory cortex of P3 hosts, they reliably took on a Chandelier cell phenotype despite being placed in an ectopic heterochronic environment [25]. This implies that this population is committed to a Chandelier cell fate by an intrinsic genetic program late in embryogenesis. Birthdate also appears to be important for CGE-derived cortical interneuron subtype specification, as there is a shift in the subtypes produced across development [26,27].
Figure 2. SST+ interneurons are born early in the VZ and PV+ interneurons are born late in the SVZ.
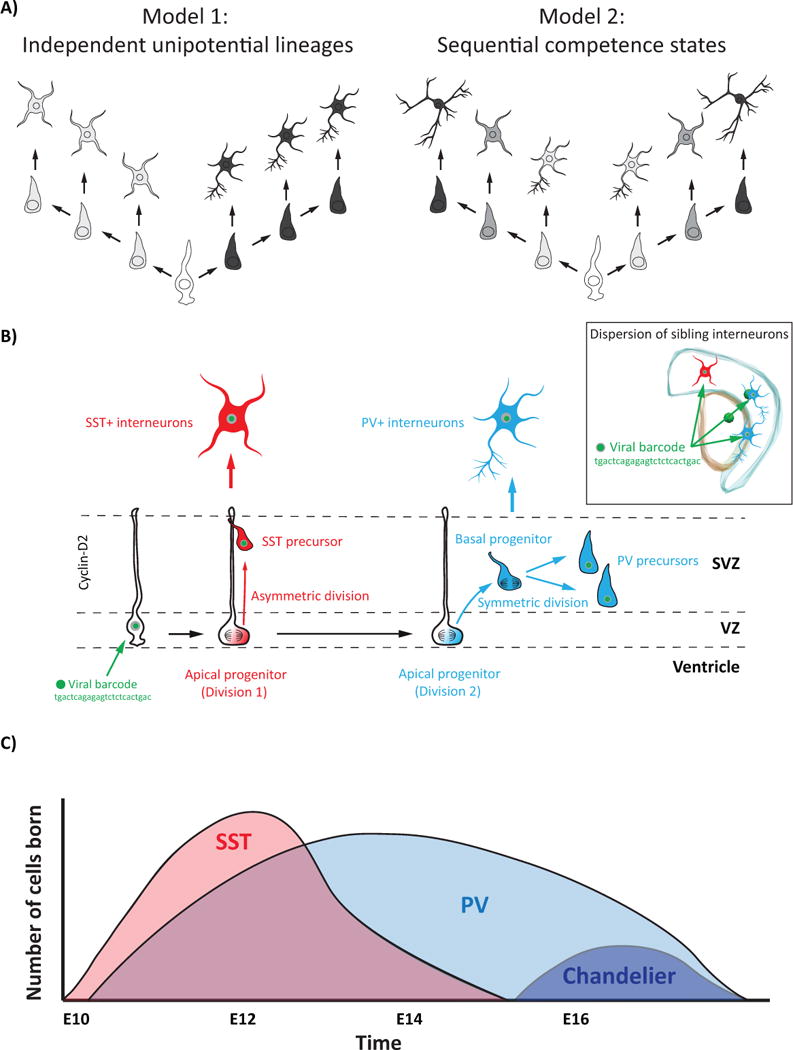
A) Two models of progenitor lineages in the GEs. Lineages might be restricted to producing particular interneuron subtypes (Model 1), or progenitors’ competencies might change over time to allow numerous lineages to be born in a precise sequence (Model 2).
B) Model depicting how clonally related SST+ and PV+ interneurons might be born in the MGE. First an Apical Progenitor (AP) in the VZ gives rise to an SST+ interneuron precursor, and then the AP transitions to a Basal Progenitor (BP) in the SVZ where it produces two PV+ interneuron precursors. Clonality is assessed by infecting progenitors with a unique DNA barcode (e.g. tgactcagagagtctctcactgac) that is passed on to that progenitor’s progeny.
C) Graph depicting the temporal order and number of select interneuron subtypes born in the MGE. SST+ interneurons (pink) are born early in the MGE, followed by PV+ interneurons (light blue), which includes Chandelier cells (dark blue).
Therefore there is a clear connection between birthdate and the type of interneuron generated. These temporal dynamics may be achieved by dedicated GE progenitors proliferating during restricted time windows to generate certain interneuron subtypes. Alternatively, GE progenitors might undergo progressive intrinsic changes in their competence to sequentially produce cell types in a defined order (Figure 2A). However, little evidence to date supports the idea that mitotic progenitors are fate restricted. Rather, there seems to be evidence for an inherent plasticity of progenitors, as demonstrated by the fate switch from MGE to CGE character upon removal of Nkx2.1. Furthermore, because the conditional removal of Nkx2.1 occurred at a time concurrent with terminal mitotic divisions [8], the correlation between subtype specification and birthdate may reflect fate commitment occurring at the end of a cell’s proliferative cycle (discussed further in Mode of Division section).
Taken together, it appears that while spatial domains and temporal dynamics strongly predict interneuron identity, their contribution may be more stochastic than absolute. Although the vMGE is biased for producing PV+ interneurons and the dMGE for SST+ interneurons, both PV+ and SST+ interneurons appear to originate from progenitors located throughout the entire MGE. And while the peak of neurogenesis for SST+ interneurons occurs earlier in the MGE than PV+ interneurons, SST+ interneurons are still being produced late at E15.5. Therefore, determining a cell’s fate is not as simple as determining its spatiotemporal origin. Rather, there is a stochastic mechanism underlying subtype specification that increases the likelihood that an interneuron will acquire a certain fate, but does not guarantee that outcome for every cell at a given time and location.
Lineage
How do the spatial domains and temporal dynamics within each GE refine the subtypes of interneurons generated? One hypothesis is that within each GE there are spatially segregated progenitor lineages with restricted fate potential dedicated to producing particular neuronal subtypes at a given time. Alternatively, a single lineage may give rise to multiple subtypes over time (Figure 2A). To address this question, four recent studies [28–31] sought to explore whether there are different progenitors dedicated to making either PV+ or SST+ interneurons, or if both PV+ and SST+ interneurons could be derived from the same progenitor. Using mouse genetics and retrovirus-mediated gene transfer to selectively label progenitors in the MGE, all four studies found mixed clones that contained both PV+ and SST+ interneurons, demonstrating that many progenitors were not restricted to making exclusively PV+ or SST+ interneurons (Figure 2B).
Two of these studies took advantage of a lineage fate mapping method devised by the Cepko laboratory, whereby dividing progenitor cells and their progeny can be tagged by a set of DNA barcodes to determine lineal relationships across individual cells in the adult. Both studies that utilized this unambiguous barcoding strategy to assign lineal relationships found that interneurons derived from a single progenitor (i.e. clonally related cells that share a lineage) within the forebrain can widely disperse across both functional and anatomical structures [29,30], in contrast to clonally related pyramidal cells that form functional radial units in the cortex (i.e. preferentially forming synapses with one another, as well as sharing functional relationships and physiological properties compared to nearby non-clonal cells [32–35]). This suggests that lineage plays no obvious role in the positioning of interneuron clones and does not prescribe the fate of ventral telencephalic progenitors in the same manner as in the dorsal telencephalon, although this is a topic of ongoing study and debate [36–38]. As such, more work is needed to determine if clonally related interneurons maintain any preferential spatial or functional relationship in adulthood. While it is possible that lineage plays no role in shaping interneuron fate, one could imagine that, even if they reside in different structures and domains, perhaps a subset of clonally related GABAergic populations share common properties. In such a model, specific lineages may give rise to progeny that share a common program that is contextually adapted after migration to different brain structures is complete. This would allow different regions of the telencephalon to acquire interneurons with particular properties, while also permitting them to fine tune their functional program in accordance with the requirements of specific brain structures [30,39].
Mode of division
Decades of work studying the development of excitatory pyramidal cells in the dorsal telencephalon has shown that different modes of cell division and cell cycle dynamics influence the ultimate number, subtype identity, and laminar distribution of postmitotic neurons [40–44]. Recent findings have begun to lend support to the importance of cell division modes within the ventral telencephalon [45,46], and more specifically, has provided insight into how PV+ and SST+ interneurons may originate from a single progenitor.
Similar to the dorsal telencephalon [47], each of the GEs contains three primary regions: the Ventricular Zone (VZ), Subventricular Zone (SVZ), and Mantle Zone (MZ) [45]. The VZ is the most apical portion of the GE that lines the ventricle and contains neural progenitors called Apical Progenitors (APs). APs have bipolar morphology with basal and apical processes visible during M-phase, undergo interkinetic nuclear migration, and divide at the VZ surface both symmetrically to expand the AP population, as well as asymmetrically, to produce another AP and a neurogenic Basal Progenitor (BP) [45]. The SVZ is located between the VZ and MZ, and contains BPs, which can further divide symmetrically to produce two neuronal precursor cells, although proliferative divisions of BPs have been detected in the SVZ of the LGE [45]. The MZ largely contains migratory postmitotic cells that are thought to be committed to particular cell fates (e.g. to PV- or SST-expressing interneurons) [31,45,48] (Figure 2B).
In the GEs, the cell cycle regulator cyclin-D2 is enriched in BPs within the SVZ. Interestingly, cyclin-D2-null mice exhibit a 30–40% reduction of PV+ interneuron numbers in the neocortex with no change in SST+ interneuron numbers [49,50]. In vivo fate mapping revealed that cortical interneurons originating from MGE-derived APs were biased toward generating SST+ cortical interneurons. When MGE progenitors were genetically driven toward dividing as APs or BPs, their fates were biased toward SST+ and PV+ interneurons, respectively [51], indicating that interneurons’ fate is in part malleable by the mode of division. Taken together, this data suggests that early in development asymmetric divisions in the VZ of the MGE preferentially generate SST+ interneurons, whereas PV+ interneurons are predominantly produced later by BPs in the SVZ of the MGE. Therefore, it is possible that in the MGE a single progenitor first divides as an SST-producing AP and then transitions to a PV-producing BP, which neatly explains both, how mixed clones can contain SST+ and PV+ interneurons, as well as why SST+ interneurons tend to be born earlier in the MGE than PV+ interneurons (Figure 2B).
As in the cortex, new types of progenitors are still being discovered in the proliferative zones of the GEs. A new type of progenitor called ‘subapical progenitor,’ which undergo nonsurface mitoses within the VZ, was recently found in the MGE and LGE [45]. Extended live-cell imaging revealed LGE progenitors frequently progressed through a stereotyped sequence, advancing from AP to subapical progenitor to BP. Subapical progenitors predominantly contributed to larger clones, although clones of all sizes were distributed equally along the D-V axis of the LGE. In contrast to the cerebral cortex [52], in the LGE cell cycle length became progressively shorter with subsequent divisions and a majority of divisions were basal as opposed to apical [45]. It is possible that additional modes of division exist within the VZ and SVZ of the GEs, and the mechanisms regulating how these modes of division influence cell fate are not fully understood. Indeed, given the greater diversity of interneurons versus pyramidal cells, GE progenitor diversity may ultimately prove to have an important role in generating the incredible breadth of interneuron subtypes.
Conclusion
Neurogenic proliferative zones become increasingly complex in higher organisms, reflecting a need for increased neuronal diversity and cell numbers [53,54]. Many questions remain that require a deeper understanding about transcriptional profiles of individual GE progenitor cells: Are temporal changes in gene expression global or only present within a particular subset of progenitors within the VZ or SVZ? Are different types of progenitors dedicated to producing specific neuron subtypes? How does gene expression change over development and across evolution, and how do these changes affect progenitor competencies? How many further genes are critical for directing neurons towards MGE vs. CGE vs. LGE fates?
It is clear that numerous mechanisms are simultaneously regulating cell fate decisions in the GEs. For example, evidence is emerging that epigenetic regulation adds an additional dimension to the spatial-temporal determinants of interneuron fate determination [55]. When neural stem cells were differentiated into immature GABAergic interneurons in vitro, methylation changes occured at many promoters, restricting cell fate potential [56]. In vivo, histone deacetylase was shown to inhibit tyrosine hydroxylase transcription to suppress dopaminergic neurotransmitter expression in migrating olfactory bulb interneurons [57], and the chromatin organizer Satb1 is important for SST+ interneuron specification in an activity dependent manner [58,59]. Therefore, while differences in gene expression may not be detected between progenitors, this does not mean that their chromatin landscapes are identical. For instance, loss of Satb1 results in distinct deficits in SST+ versus PV+ interneurons [58], and Prox1 removal differentially affects distinct CGE-derived interneuron subtypes [60]. Furthermore, although Nkx2.1 is found throughout the entire MGE from very early in development, a recent epigenetic study found that Nkx2.1 differentially regulates gene expression in progenitors. While Nkx2.1 binds to distal regulatory elements to promote transcriptional repression in VZ progenitors, it promotes a permissive chromatin state and transcriptional activation in SVZ and MZ progenitors [61]. Thus, it seems inevitable that further investigation will provide additional examples of transcription factors that have different affects depending on what portion of the genome is accessible in a given cell. Epigenetic changes in interneurons might also contribute to psychiatric diseases, as epigenetic dysregulation at GABAergic promotors in PV cells is associated with schizophrenia [62]. More work needs to be done to understand the molecular mechanisms regulating GE development, particularly: how cell cycle dynamics and mode of division influence cell fate; what contribution, if any, does lineage have on subtype specification; what are the epigenetic mechanisms regulating cell fate decisions; and how does early network activity modify intrinsic genetic programs in various interneuron subtypes? Elucidating these processes are crucial for fundamentally understanding brain development, and for ultimately treating the numerous disorders associated with interneuron dysfunction [63–65].
Highlights.
The GEs produce many types of GABAergic projection neurons and a majority of GABAergic interneurons
Spatial origin, birthdate, and mode of division influence interneuron fate
Subdomains exist within each GE, producing different neuronal subtypes over time
PV and SST-expressing interneurons can originate from the same progenitor
Different modes of divisions are biased for producing different interneuron subtypes
Acknowledgments
We are grateful to Dr. Timothy Petros for comments on the manuscript. Research in the Fishell laboratory is supported by NIH (NS 081297, MH095147, P01NS074972) and the Simons Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of outstanding interest
- 1.Puelles L, Rubenstein JLR. Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends in Neurosciences. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 2.Rubenstein J, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- 3.Fraser S, Keynes R, Lumsden A. Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature. 1990;344:431–435. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- 4.Guthrie S, Lumsden A. Motor neuron pathfinding following rhombomere reversals in the chick embryo hindbrain. Development. 1992;114:663–673. doi: 10.1242/dev.114.3.663. [DOI] [PubMed] [Google Scholar]
- 5.Jacob J, Guthrie S. Facial Visceral Motor Neurons Display Specific Rhombomere Origin and Axon Pathfinding Behavior in the Chick. The Journal of Neuroscience. 2000;20:7664–7671. doi: 10.1523/JNEUROSCI.20-20-07664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marin O, Rubenstein JLR. A long, remarkable journey: Tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 7.Gelman D, Griveau A, Dehorter N, Teissier A, Varela C, Pla R, Pierani A, Marín O. A Wide Diversity of Cortical GABAergic Interneurons Derives from the Embryonic Preoptic Area. The Journal of Neuroscience. 2011;31:16570. doi: 10.1523/JNEUROSCI.4068-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butt SJ, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008;59:722–732. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 10.Xu Q, Guo L, Moore H, Waclaw RR, Campbell K, Anderson SA. Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron. 2010;65:328–340. doi: 10.1016/j.neuron.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Briscoe J, Pierani A, Jessell TM, Ericson J. A Homeodomain Protein Code Specifies Progenitor Cell Identity and Neuronal Fate in the Ventral Neural Tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 13.Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Torigoe M, Yamauchi K, Kimura T, Uemura Y, Murakami F. Evidence That the Laminar Fate of LGE/CGE-Derived Neocortical Interneurons Is Dependent on Their Progenitor Domains. J Neurosci. 2016;36:2044–2056. doi: 10.1523/JNEUROSCI.3550-15.2016. Study used in utero electroporation to label progenitors from different spatial domains within the LGE/CGE, and found they gave rise to interneurons in distinct cortical lamina. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waclaw RR, Ehrman LA, Pierani A, Campbell K. Developmental origin of the neuronal subtypes that comprise the amygdalar fear circuit in the mouse. J Neurosci. 2010;30:6944–6953. doi: 10.1523/JNEUROSCI.5772-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun K, Garel S, Fischman S, Rubenstein JL. Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J Comp Neurol. 2003;461:151–165. doi: 10.1002/cne.10685. [DOI] [PubMed] [Google Scholar]
- 17*.Zechel S, Zajac P, Lönnerberg P, Ibáñez CF, Linnarsson S. Topographical transcriptome mapping of the mouse medial ganglionic eminence by spatially resolved RNA-seq. Genome Biology. 2014;15:486. doi: 10.1186/s13059-014-0486-z. Laser-capture microdissection was used in combination with RNA-sequencing to systematically sample MGE tissue in a regular grid and make a topographical map of the MGE transcriptome, identifying a ventro-lateral gradient of increasing migratory capacity within the MGE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenman JM, Wang B, Campbell K. Tlx Controls Proliferation and Patterning of Lateral Telencephalic Progenitor Domains. The Journal of Neuroscience. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inan M, Welagen J, Anderson SA. Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb Cortex. 2012;22:820–827. doi: 10.1093/cercor/bhr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wonders CP, Taylor L, Welagen J, Mbata IC, Xiang JZ, Anderson SA. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev Biol. 2008;314:127–136. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Chatterjee M, Li JY. The mouse homeobox gene Gbx2 is required for the development of cholinergic interneurons in the striatum. J Neurosci. 2010;30:14824–14834. doi: 10.1523/JNEUROSCI.3742-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development. 2009;136:3841–3851. doi: 10.1242/dev.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Hoch RV, Lindtner S, Price JD, Rubenstein JL. OTX2 Transcription Factor Controls Regional Patterning within the Medial Ganglionic Eminence and Regional Identity of the Septum. Cell Rep. 2015;12:482–494. doi: 10.1016/j.celrep.2015.06.043. An Otx2 conditional knock out mouse was used in combination with Otx2 ChIP-seq to study the transcription factor's role in patterning the MGE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi H, Lu J, Huang ZJ. The Spatial and Temporal Origin of Chandelier Cells in Mouse Neocortex. Science. 2013;339:70–74. doi: 10.1126/science.1227622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28**.Ciceri G, Dehorter N, Sols I, Huang ZJ, Maravall M, Marin O. Lineage-specific laminar organization of cortical GABAergic interneurons. Nat Neurosci. 2013;16:1199–1210. doi: 10.1038/nn.3485. [30] [DOI] [PubMed] [Google Scholar]
- 29**.Harwell CC, Fuentealba LC, Gonzalez-Cerrillo A, Parker PR, Gertz CC, Mazzola E, Garcia MT, Alvarez-Buylla A, Cepko CL, Kriegstein AR. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron. 2015;87:999–1007. doi: 10.1016/j.neuron.2015.07.030. [30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Mayer C, Jaglin XH, Cobbs LV, Bandler RC, Streicher C, Cepko CL, Hippenmeyer S, Fishell G. Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries. Neuron. 2015;87:989–998. doi: 10.1016/j.neuron.2015.07.011. Four studies characterized clonally related MGE-derived interneurons [28–31]. All four studies found that SST+ and PV+ interneurons can be clonally related, meaning they originated from the same progenitor cell in the MGE. Two of these studies [28,31] used spatial criteria to define interneuron clusters. The other two studies [29,30] used DNA barcodes to identify clonally related interneurons independent of their spatial relationship and found that interneuron clones can disperse widely in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Brown KN, Chen S, Han Z, Lu C-H, Tan X, Zhang X-J, Ding L, Lopez-Cruz A, Saur D, Anderson SA, et al. Clonal Production and Organization of Inhibitory Interneurons in the Neocortex. Science. 2011;334:480–486. doi: 10.1126/science.1208884. [30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Lu H, Cheng PL, Ge S, Xu H, Shi SH, Dan Y. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. 2012;486:118–121. doi: 10.1038/nature11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu YC, He S, Chen S, Fu Y, Brown KN, Yao XH, Ma J, Gao KP, Sosinsky GE, Huang K, et al. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. 2012;486:113–117. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 36*.Turrero García M, Mazzola E, Harwell Corey C. Lineage Relationships Do Not Drive MGE/PoA-Derived Interneuron Clustering in the Brain. Neuron. 92:52–58. Showed that the spatial parameters used in Brown et al. 2011 to cluster interneurons fails to identify lineal boundaries in the datasets of either Mayer et al. 2015 or Harwell et al. 2015, and found no evidence that lineage influences interneuron clustering in the forebrain. [Google Scholar]
- 37*.Mayer C, Bandler Rachel C, Fishell G. Lineage Is a Poor Predictor of Interneuron Positioning within the Forebrain. Neuron. 92:45–51. Demonstrate that cortical interneurons are similarly distributed whether or not they share a lineal relationship, and therefore suggest that there is no compelling evidence that clonality influences the position or function of interneurons. Argue that a random computer simulation cannot be used to assess the influence of clonality on interneuron development because the entire population of cortical interneurons is not randomly dispersed in vivo. [Google Scholar]
- 38*.Sultan Khadeejah T, Han Z, Zhang X-J, Xianyu A, Li Z, Huang K, Shi S-H. Clonally Related GABAergic Interneurons Do Not Randomly Disperse but Frequently Form Local Clusters in the Forebrain. Neuron. 92:31–44. doi: 10.1016/j.neuron.2016.09.033. Compared the distribution of interneurons published in Mayer et al. 2015 and Harwell et al. 2015 to a random computer simulation, and based on this analysis found that clonally related interneurons do not randomly disperse but frequently form local clusters. Based on this finding, they argue that lineage influences the spatial distribution of forebrain interneurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci U S A. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of notchl immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhong W, Feder JN, Jiang M-M, Jan LY, Jan YN. Asymmetric Localization of a Mammalian Numb Homolog during Mouse Cortical Neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 44.Kornack DR, Rakic P. Radial and horizontal deployment of clonally related cells in the primate neocortex: Relationship to distinct mitotic lineages. Neuron. 1995;15:311–321. doi: 10.1016/0896-6273(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 45.Pilz GA, Shitamukai A, Reillo I, Pacary E, Schwausch J, Stahl R, Ninkovic J, Snippert HJ, Clevers H, Godinho L, et al. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat Commun. 2013;4:2125. doi: 10.1038/ncomms3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheth AN, Bhide PG. Concurrent cellular output from two proliferative populations in the early embryonic mouse corpus striatum. The Journal of Comparative Neurology. 1997;383:220–230. [PubMed] [Google Scholar]
- 47.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 48.Halliday AL, Cepko CL. Generation and migration of cells in the developing striatum. Neuron. 1992;9:15–26. doi: 10.1016/0896-6273(92)90216-z. [DOI] [PubMed] [Google Scholar]
- 49.Glickstein SB, Monaghan JA, Koeller HB, Jones TK, Ross ME. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J Neurosci. 2009;29:9614–9624. doi: 10.1523/JNEUROSCI.2284-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glickstein SB, Moore H, Slowinska B, Racchumi J, Suh M, Chuhma N, Ross ME. Selective cortical interneuron and GABA deficits in cyclin D2-null mice. Development. 2007;134:4083–4093. doi: 10.1242/dev.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.Petros TJ, Bultje RS, Ross ME, Fishell G, Anderson SA. Apical versus Basal Neurogenesis Directs Cortical Interneuron Subclass Fate. Cell Rep. 2015;13:1090–1095. doi: 10.1016/j.celrep.2015.09.079. Study showed that different modes of progenitor division were biased towards producing different interneuron subtypes in the MGE. Apical Progenitors in the VZ were biased towards producing SST+ interneurons, whereas Basal Progenitors in the SVZ were biased towards producing PV+ interneurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caviness VS, Goto T, Tarui T, Takahashi T, Bhide PG, Nowakowski RS. Cell Output, Cell Cycle Duration and Neuronal Specification: a Model of Integrated Mechanisms of the Neocortical Proliferative Process. Cerebral Cortex. 2003;13:592–598. doi: 10.1093/cercor/13.6.592. [DOI] [PubMed] [Google Scholar]
- 53.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 54.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 55.Nord Alex S, Pattabiraman K, Visel A, Rubenstein John LR. Genomic Perspectives of Transcriptional Regulation in Forebrain Development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burney MJ, Johnston C, Wong KY, Teng SW, Beglopoulos V, Stanton LW, Williams BP, Bithell A, Buckley NJ. An epigenetic signature of developmental potential in neural stem cells and early neurons. Stem Cells. 2013;31:1868–1880. doi: 10.1002/stem.1431. [DOI] [PubMed] [Google Scholar]
- 57.Banerjee K, Akiba Y, Baker H, Cave JW. Epigenetic control of neurotransmitter expression in olfactory bulb interneurons. Int J Dev Neurosci. 2013;31:415–423. doi: 10.1016/j.ijdevneu.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Close J, Xu H, De Marco Garcia N, Batista-Brito R, Rossignol E, Rudy B, Fishell G. Satb1 is an activity-modulated transcription factor required for the terminal differentiation and connectivity of medial ganglionic eminence-derived cortical interneurons. J Neurosci. 2012;32:17690–17705. doi: 10.1523/JNEUROSCI.3583-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denaxa M, Kalaitzidou M, Garefalaki A, Achimastou A, Lasrado R, Maes T, Pachnis V. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Rep. 2012;2:1351–1362. doi: 10.1016/j.celrep.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60*.Miyoshi G, Young A, Petros T, Karayannis T, McKenzie Chang M, Lavado A, Iwano T, Nakajima M, Taniguchi H, Huang ZJ, et al. Prox1 Regulates the Subtype-Specific Development of Caudal Ganglionic Eminence-Derived GABAergic Cortical Interneurons. J Neurosci. 2015;35:12869–12889. doi: 10.1523/JNEUROSCI.1164-15.2015. Study shows that Prox1 differentially regulates the development of particular CGE-derived interneuron subtypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Sandberg M, Flandin P, Silberberg S, Su-Feher L, Price James D, Hu Jia S, Kim C, Visel A, Nord Alex S, Rubenstein John LR. Transcriptional Networks Controlled by NKX2-1 in the Development of Forebrain GABAergic Neurons. Neuron. 2016;91:1260–1275. doi: 10.1016/j.neuron.2016.08.020. Used a variety of techniques including ChIP-seq and histone ChIP-seq from basal ganglia at embryonic day E13.5 to study the transcriptional networks regulated by Nkx2.1. The study found that Nkx2.1 preferentially binds to distal regulatory elements (as opposed to transcription start sites) and played a differential role in the VZ versus the SVZ and MZ. More specifically, Nkx2.1 led to a repressed epigenetic state in the VZ and a permissive chromatin state in the SVZ and MZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morishita H, Kundakovic M, Bicks L, Mitchell A, Akbarian S. Interneuron epigenomes during the critical period of cortical plasticity: Implications for schizophrenia. Neurobiol Learn Mem. 2015;124:104–110. doi: 10.1016/j.nlm.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inan M, Petros TJ, Anderson SA. Losing your inhibition: linking cortical GABAergic interneurons to schizophrenia. Neurobiol Dis. 2013;53:36–48. doi: 10.1016/j.nbd.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marin O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 65.Volk DW, Edelson JR, Lewis DA. Altered expression of developmental regulators of parvalbumin and somatostatin neurons in the prefrontal cortex in schizophrenia. Schizophr Res. 2016 doi: 10.1016/j.schres.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


