Abstract
Background
Genomic sequence analyses have shown that horizontal gene transfer occurred during the origin of eukaryotes as a consequence of symbiosis. However, details of the timing and number of symbiotic events are unclear. A timescale for the early evolution of eukaryotes would help to better understand the relationship between these biological events and changes in Earth's environment, such as the rise in oxygen. We used refined methods of sequence alignment, site selection, and time estimation to address these questions with protein sequences from complete genomes of prokaryotes and eukaryotes.
Results
Eukaryotes were found to evolve faster than prokaryotes, with those eukaryotes derived from eubacteria evolving faster than those derived from archaebacteria. We found an early time of divergence (~4 billion years ago, Ga) for archaebacteria and the archaebacterial genes in eukaryotes. Our analyses support at least two horizontal gene transfer events in the origin of eukaryotes, at 2.7 Ga and 1.8 Ga. Time estimates for the origin of cyanobacteria (2.6 Ga) and the divergence of an early-branching eukaryote that lacks mitochondria (Giardia) (2.2 Ga) fall between those two events.
Conclusions
We find support for two symbiotic events in the origin of eukaryotes: one premitochondrial and a later mitochondrial event. The appearance of cyanobacteria immediately prior to the earliest undisputed evidence for the presence of oxygen (2.4–2.2 Ga) suggests that the innovation of oxygenic photosynthesis had a relatively rapid impact on the environment as it set the stage for further evolution of the eukaryotic cell.
Background
An emerging pattern found in gene and protein phylogenies that include prokaryotes (archaebacteria and eubacteria) and eukaryotes is the variable position of eukaryotes. In proteins involved in transcription and translation, eukaryotes often cluster with archaebacteria whereas in metabolic proteins they often cluster with eubacteria [1]. Among the latter proteins, eukaryotes sometimes group with α-proteobacteria, presumably reflecting the origin of mitochondria, and plants sometimes cluster with cyanobacteria, reflecting the origin of plastids. These patterns have been interpreted as a general signature of the symbiotic origin of eukaryotes [2,3] and horizontal gene transfer (HGT) of symbiont genes to the nucleus [4-9]. On the one hand, this complexity resulting from HGT can obscure some aspects of evolutionary history [8]. However, HGT also can provide the means to investigate otherwise difficult questions, such as inferring the number of symbiotic events and estimating the time of those events. This is the approach that we take in this study.
The goal of this study is to estimate the timing of evolutionary events involved in the origin of eukaryotes (Fig. 1), including the related origin of oxygenic photosynthesis. The latter is believed to have occurred only in cyanobacteria [10] and preceded the symbiotic event leading to the mitochondrion of eukaryotes. The earliest biomarker evidence of eukaryotes is at 2.7 Ga [11] and the earliest fossils appear 2.1 Ga [12]. The fossil record of cyanobacteria has been argued to extend to 3.5 Ga [13] but the biomarker evidence at 2.7–2.8 Ga [14,15] usually is considered to be the earliest record of cyanobacteria [10]. However, the 2-methylhopane biomarker of cyanobacteria has been detected in lower abundance in other prokaryotes, and many taxa (especially anaerobic species) have not been examined for the biomarker [15-17]. Also, the origin of oxygenic photosynthesis may have occurred at some time later than the origin of cyanobacteria. Geologic evidence bearing on the origin and rise in oxygen likewise has been debated [18,19]. Although the existence of banded iron formations prior to 3 Ga sometimes has been used as evidence for the early evolution of oxygenic photosynthesis, oxygen-independent mechanisms of iron deposition are known [20].
Figure 1.
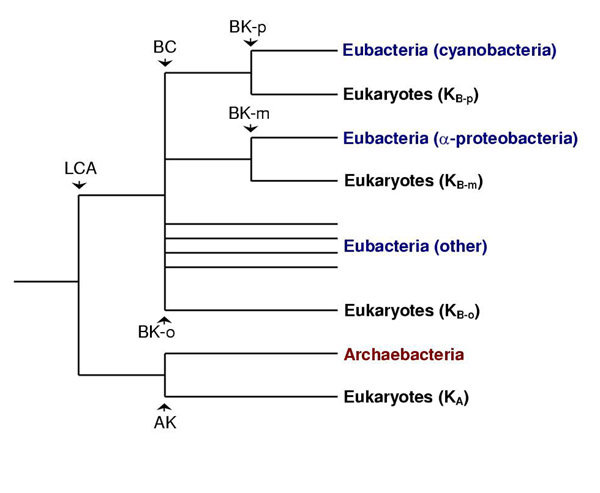
Working model of gene relationships used in this study. Eukaryotic proteins trace back to four different locations in the evolutionary tree of prokaryotes. The divergence between archaebacteria and eubacteria (last common ancestor, LCA), archaebacteria and eukaryotes (AK), and between cyanobacteria and other eubacteria (BC) are believed to represent speciation events between populations of prokaryotes. The remaining three divergence events are considered to reflect horizontal gene transfer following symbiosis: (1) between an archaebacterium and a eubacterium leading to the origin of eukaryotes (BK-o), (2) between an α-proteobacterium and a eukaryote leading to the origin of mitochondria (BK-m), and (3) between a cyanobacterium and a eukaryote leading to the origin of plastids (BK-p). In this study, divergence times are estimated for AK, BC, BK-o, and BK-m. The divergence time of a fifth event (not shown), the speciation event between a eukaryote (Giardia) and other eukaryotes (GK), also is estimated. Branch lengths are not proportional to time.
The use of sequence changes to estimate the time of these early events also has its assumptions and limitations [21-23]. Nonetheless, many proteins contain conserved regions of amino acid sequence throughout prokaryotes and eukaryotes that permit alignment and analysis. The most extensive of these analyses have found that all major events related to the origin of eukaryotes occurred about 2.0–2.2 Ga [5,21]. This includes the divergence of archaebacteria and archaebacterial proteins in eukaryotes, the origin of cyanobacteria, and the divergence of eubacteria and eubacterial proteins in eukaryotes (the latter presumably reflecting symbiosis). However, these times were not adjusted for lineage-specific rate differences that have been discovered subsequently [23]. Here, we estimate the time of these events with protein sequences from complete genomes and consideration of lineage-specific rate variation.
Results
Rate differences
The shape parameter (α) of the gamma distribution used to account for rate variation among sites was found to differ consistently between calibration taxa and the overall data set for each gene (Fig. 2), requiring a dual-gamma approach (see Methods). Also, eukaryotic protein sequences were found to have an increased rate of evolution compared with prokaryotic sequences regardless of their archaebacterial or eubacterial origin (Fig. 3A). Average eukaryote rates were 1.37 (AK), 1.18 (BK-o), and 1.38 (BK-m) times the rate of the most closely related prokaryote in constant rate proteins (1.55, 1.24, and 1.56 in all proteins, respectively). Besides this general pattern, which may reflect fundamental differences between prokaryotes and eukaryotes (e.g., recombination), there are further differences among eukaryotes. In comparing rates of evolution in eukaryotic sequences derived independently from eubacteria and archaebacteria in the same protein, those derived from eubacteria (in all cases, BK-o) were found to be evolving at roughly twice the rate as their archaebacteria-derived counterparts (Fig. 3B). The slope was 2.01 and the correlation coefficient was 0.54 (n = 14 comparisons in seven proteins).
Figure 2.
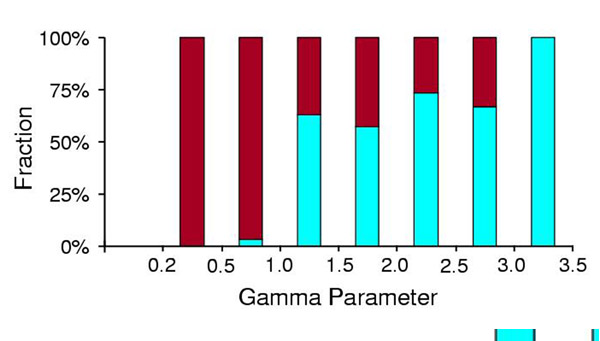
Differences in rate variation among sites (gamma parameter). Fraction of gamma parameters (64 proteins) measured from entire data sets for each protein (blue, prokaryotes and eukaryotes) and from subsets containing only calibration taxa (red, eukaryotes).
Figure 3.
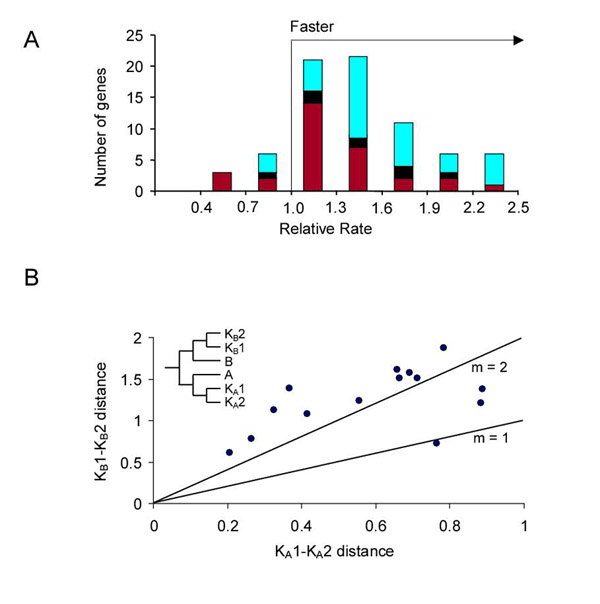
Differences in rates of protein evolution. (A) Prokaryotes versus eukaryotes. Histogram of ratios of eukaryote to prokaryote evolutionary rates. Eukaryotes are derived from three prokaryote lineages: BK-o (31 proteins, blue), BK-m (8 proteins, black), and AK (36 proteins, red). (B) Eukaryotes versus eukaryotes. Protein distances between two species of eukaryotes (KA1 and KA2 in inset) of archaebacterial origin compared with distances between the same two species of eubacterial origin (KB1 and KB2); slope (m) = 2.01. In each case, all sequences being compared are from the same protein. The mirrorlike phylogeny (inset) is the result of horizontal gene transfer and speciation rather than gene duplication.
Two other rate comparisons were limited by a small number of proteins: eubacteria versus eukaryotes (KA) and eubacteria versus archaebacteria. Only three proteins were available in the first comparison and all three showed a faster rate in eukaryotes (1.43, 1.12, 1.23; x = 1.26). This result differs from that reported elsewhere [23], in which the two rates were not significantly different. In the second case, we found that archaebacteria are evolving at a slower rate than eubacteria, as was noted elsewhere [23]. In our case, regression of archaebacterial branch length versus eubacterial branch length, fixed through the origin, resulted in a slope of 0.93 and correlation coefficient of 0.65 (n = 9 proteins). However, in both of these comparisons, rate tests did not yield significant rate differences probably because of the short length of most proteins. Sample size (eight protein sets) also was limited in the Kollman and Doolittle study [23]. Taken together these data suggest the following relative order of rate differences: archaebacteria < eubacteria < eukaryotes (archaebacterial origin) < eukaryotes (eubacterial origin). As additional genomic data become available, more proteins will be useful and greater precision in these rates and rate differences will be possible.
Phylogeny and time estimation
It has been suggested that eukaryotic genes and proteins of archaebacterial origin are more closely related to one lineage of archaebacteria (Crenarchaeota; "eocytes") than the other major lineage (Euryarchaeota) [24]. If true, this would bear on our time estimate for the divergence of archaebacteria and eukaryotes. Thus, we conducted a phylogenetic analysis of 72 proteins containing representatives of the two major groups of archaebacteria, eukaryotes, and eubacteria. At the 95% bootstrap significance level, 19 proteins supported archaebacterial monophyly whereas none supported the eocyte hypothesis (Crenarchaeota + Eukaryota). This indicates that the lineage of archaebacteria leading to the eukaryote nuclear genome diverged prior to the split between the Crenarchaeota and Euryarchaeota. As noted previously [1], most (in this case, 21 out of 36) eukaryotic proteins with archaebacterial affinity are informational (involved in transcription, translation, and related processes).
Among 41 eukaryotic proteins with eubacterial affinities, Rickettsia is most closely related to eukaryotes in phylogenetic analyses of nine individual proteins. This agrees with the genetic and cell biological evidence implicating an α-proteobacterium as progenitor of the mitochondrion [25] and supports the hypothesis that these nine eukaryotic proteins owe their origin to that symbiotic event [2]. However, the remaining 32 proteins do not show this pattern but instead identify other species or groups of eubacteria as closest relative. Unlike Rickettsia, no other single species appears as closest relative in more than three proteins, but rather most (19/32 proteins) identify groups of species as closest relative (e.g, Fig. 4A). To further explore this question we combined sequences of all 11 proteins with a full representation of eubacterial taxa (11 species). In the combined analysis, eukaryotes fall significantly outside of the well-defined clade containing α- and γ-proteobacteria (Fig. 4B). The relatively basal and unresolved position of eukaryotes is consistent with the preponderance of single proteins showing different groups of species as closest relative. Three individual proteins showed significant bootstrap support for a Rickettsia-eukaryote cluster in four-taxon analyses (rooted with an archaebacterium) whereas four proteins significantly supported a Rickettsia-Escherichia cluster that excluded the eukaryote.
Figure 4.
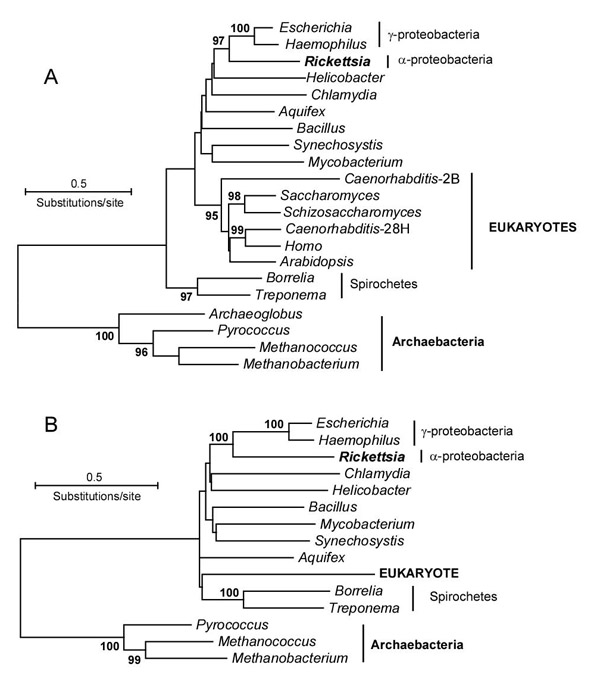
Phylogenetic relationships of eubacteria and eukaryotes rooted with archaebacteria. Neighbor-joining bootstrap consensus trees showing significant (≥ 95%) bootstrap values; maximum-likelihood and maximum parsimony produced identical topologies for significant nodes. (A) Cytoplasmic alanyl tRNA synthetase, showing BK-o pattern: eukaryotes most closely related to eubacteria but not closely related to the α-proteobacterium (Rickettsia). (B) Combined analysis of all proteins with full complement (11 species) of eubacterial taxa and showing eubacterial-eukaryote relationship (11 proteins, 1596 amino acids); significant groups remain after removal of cytoplasmic alanyl tRNA synthetase.
Divergence time estimates from the multigene (MG) and average distance (AD) approaches are similar, but rate-adjusted times are older than unadjusted times (Table 1). The time estimate for the AK divergence averages 4.0 Ga and the remaining times range from 1.8 to 2.7 Ga. The time estimate for BK-o (2.7 ± 0.20 Ga) was older than the estimate for BK-m (1.8 ± 0.20 Ga) whereas the time estimate for the origin of Giardia (2.2 ± 0.12 Ga) was intermediate. The BC time estimate was 2.6 ± 0.26 Ga.
Table 1.
Divergence time estimates (billion years ago)
| Multigene | Average-distance | |||||
| Comparison | All | constant | all | constant | Mean ± SE* | |
| Archaebacteria-eukaryotes (AK) | 3.18 | 3.42 | 3.05 | 3.58 | 3.50 ± 0.25 | |
| Rate adjusted | 4.11 | 3.86 | 3.69 | 4.09 | 3.97 ± 0.32 | |
| Eubacteria-eukaryotes (BK-o) | 2.31 | 2.45 | 2.27 | 2.48 | 2.46 ± 0.14 | |
| Rate adjusted | 2.54 | 2.76 | 2.51 | 2.70 | 2.73 ± 0.20 | |
| Eubacteria-cyanobacteria (BC) | 1.68 | 1.73 | 1.92 | 1.85 | 1.79 ± 0.29 | |
| Rate adjusted | 2.56 | 2.52 | 2.66 | 2.60 | 2.56 ± 0.26 | |
| Giardia-eukaryotes (GK) | 2.82 | 2.54 | 3.32 | 2.46 | 2.50 ± 0.22 | |
| Rate adjusted | 2.72 | 2.31 | 2.04 | 2.16 | 2.23 ± 0.12 | |
| Eubacteria-eukaryotes (BK-m) | 1.70 | 1.72 | 1.47 | 1.39 | 1.56 ± 0.29 | |
| Rate adjusted | 2.02 | 2.07 | 1.72 | 1.61 | 1.84 ± 0.20 | |
*Mean (Multigene, constant rate + Average-distance, constant rate) ± standard error (Multigene, constant rate).
Discussion
The purpose of this study was to examine the temporal relationship between the origin of eukaryotes and events in Earth history. However, some unexpected results required refinement in methodology. These included finding greater among-site rate variation in the calibration group and different rates of sequence change between prokaryotes and eukaryotes, and between eukaryotes derived from different groups of prokaryotes. By taking into account these variables, the resulting time estimates are more robust and have fewer assumptions. For example, the time estimate for the origin of eukaryotes (BK-o) is not based on a general assumption of rate constancy between prokaryotes (or even eubacteria) and eukaryotes because rates are adjusted for each protein and each comparison. Also, the calibration used for BK-o is not a general eukaryotic calibration but one based exclusively on eukaryote sequences derived from eubacteria. A tradeoff in these improved methods was a reduction in the number of proteins that could be used, which increased the variance of the time estimates. Nonetheless, the phylogenies and time estimates obtained in this study have a bearing on current models for the evolution of eukaryotes.
Until about five years ago, it was generally accepted that there was a prior period (before mitochondria) in the history of eukaryotes [2,26]. The basal position of eukaryotes lacking mitochondria (amitochondriate) in phylogenetic trees [27] was consistent with this supposition as was evidence from sequence signatures [6]. However, molecular phylogenetic studies of several proteins in recent years have suggested that some or all amitochondriate eukaryotes once possessed mitochondria in the past [9]. Based on this new evidence, most current models for the origin of eukaryotes assume only a single symbiotic or fusion event between an archaebacterium and an α-proteobacterium [8,28,29].
Under the single-symbiosis model, eukaryotes should cluster exclusively with an α-proteobacterium (e.g., Rickettsia), among eubacteria. However, our phylogenetic analyses (Fig. 4) instead indicate, significantly, that many eukaryotic proteins originated from one (or more) eubacterial lineages other than α-proteobacteria. The reduced genome of Rickettsia[25] would not explain this result because Rickettsia possesses all of the proteins used in the combined analysis (Fig. 4B). Protein function and location also are consistent with a premitochondrial origin. Only one of the 32 BK-o proteins is restricted to the mitochondrion whereas eight of the nine BK-m proteins are restricted to that organelle. Also, all six of the proteins involved in cellular respiration are in the BK-m group. Based on the serial endosymbiosis theory, the first symbiotic event involved a spirochete [3]. On the other hand, sequence signatures of the heat shock molecular chaperone protein HSP-70 and other evidence have indicated that the first symbiotic event involved a gram-negative eubacterium [6]. Our data are unable to distinguish between these two alternatives but agree with both in implicating an earlier, premitochondrial event. Predation by prokaryotes on early eukaryotes also may have led to HGT.
If two or more symbiotic events were involved, this does not necessarily confirm that any of the living lineages of amitochondriate eukaryotes arose prior to the second (mitochondrial) event. All may have once possessed mitochondria. However, because Giardia arose at an early time (Table 1) and branches near the base of the eukaryote phylogeny, the simplest explanation is that it never possessed mitochondria and is a primary (not secondary) amitochondriate. Although the position of Giardia in some protein phylogenies [30] has been proposed as evidence that it is a secondary amitochondriate, others have urged caution until additional, more conclusive, data become available [6].
The number of symbiotic events was important for our primary concern of estimating a timescale for the early evolution of eukaryotes. We find that the divergence between archaebacteria and the lineage leading to eukaryotes (KA) was quite early (~4 Ga), which is about the time of the earliest biomarker evidence of life (3.9–3.8 Ga) [31]. We interpret that divergence to be a speciation event between two lineages of archaebacteria, with KA not becoming "eukaryotic" until the first symbiotic event at 2.7 Ga. The remaining time estimates cluster around the mid-life of Earth (1.8–2.7 Ga). The order of those events falls in a logical sequence: BK-o, BC, and BK-m. For example, the origin of mitochondria appears as the second (not first) symbiotic event, and the origin of cyanobacteria comes before the oxygen-utilizing organelles, mitochondria. Moreover, the timing of these biological events is consistent with the timing of events in geologic and atmospheric history (Fig. 5). Cyanobacteria appear before the major (undisputed) evidence of the rise in oxygen (2.4–2.2 Ga) and mitochondria appear after the rise in oxygen. Also, the estimates for the origin of cyanobacteria and eukaryotes are consistent (within one SE) with the earliest biomarker evidence for those two groups (~2.7 Ga.) [11,15]. Phylogenetic analyses of photosynthetic genes and sequence signatures also support a relatively late order of appearance of cyanobacteria among photosynthetic prokaryotes [32,33].
Figure 5.
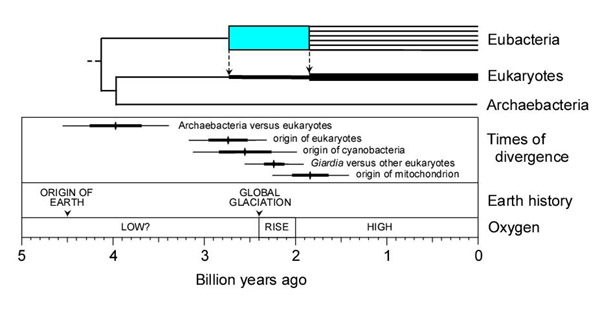
Summary diagram showing relationship between timing of evolutionary events (Table 2) and that of Earth and atmospheric histories. Time estimates are shown with ± 1 standard error (thick line) and 95% confidence interval (narrow line). The phylogenetic tree illustrates the radiation of extant eubacterial lineages (blue), and dashed lines with arrows indicate the origin of eukaryotes (BK-o) and origin of mitochondria (BK-m). The earliest divergence (last common ancestor) was not estimated but is placed (arbitrarily) just prior to the AK divergence. The increasing thickness of the eukaryote lineage represents eubacterial genes added to the eukaryote genome through two major episodes of horizontal gene transfer. The rise in oxygen represents a change from <1% to >15% present atmospheric level [34,52], although the time of the transition period and levels have been disputed [19,53].
Extensive glaciations occurred in the Paleoproterozoic (~2.4 Ga), and may have been global in extent [34]. It has been proposed that a major rise in oxygen at this time lowered global temperatures and may have triggered the glaciations [35]. If this is true, and given the time estimates here, the evolutionary innovation of oxygenic photosynthesis may have had a relatively rapid impact on the environment. Moreover, this innovation may have caused a mass extinction of prokaryotes at that time, as a result of the toxic effects of oxygen, as suggested by the virtual absence of lineages prior to ~2.5 Ga and subsequent rapid radiation of lineages (Figs. 4,5).
Conclusions
Our analyses of prokaryotic and eukaryotic genomic sequence data support two symbiotic events in the origin of eukaryotes: one premitochondrial (2.7 billion years ago, Ga) and a later mitochondrial event (1.8 Ga). Our time estimate for the divergence of an early-branching eukaryote (Giardia) that lacks mitochondria, 2.2 Ga, suggests that it is a primary and not secondary amitochondriate organism. Our time estimate for the origin of cyanobacteria (2.6 Ga) is more recent than expected and suggests that earlier fossils claimed to be of cyanobacteria are of other organisms (or artifacts). Moreover, the appearance of cyanobacteria immediately prior to the earliest undisputed evidence for the presence of oxygen (2.4–2.2 Ga) suggests that the innovation of oxygenic photosynthesis had a relatively rapid impact on the environment as it set the stage for further evolution of the eukaryotic cell.
Materials and Methods
Sequence data and alignment
We assembled and aligned protein sequences of all 467 potentially orthologous groups from complete genome databases and these were supplemented with additional eukaryote taxa from the sequence database of the National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/entrez/. Data from the following species were assembled into presumptive orthology groups (hereafter, proteins) and aligned [36]: Aquifex aeolicus, Bacillus subtilis, Borrelia burgdorferi, Chlamydia trachomatis, Escherichia coli, Haemophilus influenzae, Helicobacter pylori, Mycobacterium tuberculosis, Mycoplasma genitalium, Mycoplasma pneumoniae, Rickettsia prowazekii, Synechocystis sp., and Treponema pallidum (Eubacteria), Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum, Methanococcus jannaschii, Pyrococcus abyssi, and P. horikoshii (Archaebacteria: Euryarchaeota), and Arabidopsis thaliana, Caenorhabditis elegans, Dictyostelium discoideum, Drosophila melanogaster, Gallus gallus, Homo sapiens, Mus musculus, Oryza saticva, Plasmodium falciparum, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Trypanosoma sp., and Xenopus laevis (Eukaryota). The genome sequence of Aeropyrum pernix (Archaebacteria: Crenarchaeota) became available during completion of study and was included in phylogenetic analyses only.
Global alignment algorithms differ from local alignment algorithms in that they sometimes align unrelated (non-homologous) sites together with homologous sites. Using a computational tool, xcons [36], such unrelated sites were removed from these CLUSTALW [37] alignments to increase probability of site homology. During construction of protein alignments using the WAT system [36], short fragmented sequences were manually removed. Of the 204 proteins that could be calibrated for time estimation, the orthology of roughly half (116 proteins) was ambiguous for unknown reasons (e.g., lateral gene transfer, gene loss, or poor phylogenetic resolution) leaving 87 proteins for phylogeny and time estimation. The seven shortest (<75 amino acids) of those were used only in phylogenetic analyses; the remaining proteins averaged 196 amino acids each. Where possible, proteins were rooted by duplicate proteins (duplicate genes); otherwise, they were midpoint-rooted.
Separately, for timing the origin of Giardia, sequences of 17 proteins were obtained from the public databases and aligned [37] in which the following taxa were available: Giardia and other eukaryotes (including calibration taxa; see below), archaebacteria, and eubacteria. Correspondence and requests for materials should be addressed to S.B.H. (e-mail: sbh1@psu.edu) or see http://www.evogenomics.org/Publications/data/Eukaryotes/ for alignments and other information.
Time estimation
Methods are described elsewhere [38] except as follows. Our initial goal was to estimate divergence times for the last common ancestor (LCA), the divergence between archaebacteria and eukaryotes (AK), cyanobacteria and closest eubacterial relatives (origin of cyanobacteria, BC), eubacteria and mitochondrial eukaryotes (origin of mitochondria, BK-m), and Giardia and other eukaryotes (GK) (Fig. 1). The importance of Giardia is its lack of mitochondria and basal location in many phylogenies of eukaryotes [27,39].
However, our initial phylogenetic analyses revealed that many eukaryotes did not cluster with Rickettsia, the α-proteobacterium, as predicted by current genomic models [8,25]. Instead, they typically formed a basal lineage among eubacteria in the tree. This result was consistent with the serial endosymbiosis theory [3] and with other findings [6] and therefore we designated this divergence as BK-o (origin of eukaryotes). Estimation of the divergence time of the origin of plastids (BK-p) was not a goal of this study, and the LCA was not estimated because of an insufficient number of duplicate proteins needed for reciprocal rooting [23]. Thus, five divergence times were studied: AK, BC, BK-o, BK-m, and GK. Eukaryotes derived from different prokaryotes are referred to herein as KA (from AK), KB-o (from BK-o), and KB-m (from BK-m).
Because of the large amount of sequence conservation in these proteins, it was not possible to calibrate directly by extrapolation from vertebrates [40], for which an extensive fossil record exists. For example, sequences often were identical among rodents, primates, and birds. Instead, multiple calibrations were used from older divergences among kingdoms (plants, animals, fungi) and animal phyla, derived from analysis of 75 nuclear proteins calibrated with the vertebrate fossil record [38]. This two-step calibration reduced the error involved in extrapolation. Two classes of time estimation methods were used and compared. The multigene (MG) approach uses the mean or mode of many single-gene time estimates [40,41] whereas the average-distance (AD) approach [42-44] involves the combining of distances and rates among genes or proteins to yield a single time estimate. For the AD approach, we weight each single-gene distance, before combining, by the length of the protein (aligned amino acids).
Protein-specific rates were estimated by regression, fixed through the origin, of these calibration points within eukaryotes: arthropod-chordate (0.993 Ga), chordate-nematode (1.177 Ga), and plant-animal-fungi (1.576 Ga) [38]. During the course of the study, it was discovered that the shape parameter (α) of the gamma distribution used to account for rate variation among sites, estimated by a likelihood method [45], differed consistently between calibration taxa (average, 1.99) and the overall data set (1.44) for each gene (Fig. 2). Therefore, a dual-gamma approach was taken whereby the eukaryote rate was estimated using the eukaryote gamma parameter and the time estimate (involving prokaryotes) was made using the overall gamma parameter. There is insufficient evidence at present to determine whether or not this difference is biologically based, related to the covarion model [46], or follows a simple scaling relationship with time or total protein distance. If the relationship is scaled, additional modification in methods may be necessary in the future.
We compared rates of change in archaebacterial versus eubacterial sequences using paralogous sequences (those related by gene duplication) as a root for relative rate tests [47,48]. To examine rate differences between eukaryotic sequences and their closest prokaryote orthologs (those representing the same gene), we used the more distant prokaryote (archaebacteria or eubacteria) as root. For examining rates in eukaryotic sequences derived from either archaebacteria or eubacteria, we compared pairwise distances of the same taxa present in both locations (e.g., one pair clustering with archaebacteria and the other with eubacteria) in the same protein. The discovery of rate differences among prokaryotes and eukaryotes required rate adjustments for all proteins and comparisons, including those accepted in rate tests. These adjustments were made by estimating time only with the eukaryote lineage, or in the case of BC, using a cyanobacterial rate adjusted by direct comparison of the cyanobacteria branch and eukaryote branch in rate tests. For example, the AK divergence time was estimated only with the KA calibration and the BC, BK-o, and BK-m divergence times were estimated only with the KB-o calibration. These restrictions further reduced the number of proteins available for time estimation to the following: AK (36 total, 21 constant-rate), BK-o (25, 16), BK-m (7, 5), BC (20, 16), and Giardia-eukaryotes (17, 11).
Modes were used in the MG approach, as described previously [40], except with the BK-m comparison where the median was used because of the small number of proteins. The mode is preferred over the mean or median because it eliminates or reduces the effect of outliers (e.g., unusually high estimates resulting from paralogous comparisons). In this study, a large number of overlapping bins was used initially to better define the distribution of time estimates, followed by use of a smaller number of non-overlapping bins and standard estimation of mode by interpolation. This two-step procedure was found to reduce the influence of bin size on mode estimation. For the AD approach, single-gene distances and rates were weighted by sequence length and then combined distances were divided by combined rates.
Phylogeny estimation
Phylogenetic trees [49] were constructed for each gene from amino acid data to assist in gene selection and interpretation. A gamma distance was used for all trees, with α estimated from the entire data set [45]. An analysis involving combined protein alignments was performed with maximum likelihood [50], neighbor joining [49,51], and maximum parsimony [51], using bootstrapping. Bootstrap consensus trees show branch-lengths estimated by ordinary least-squares method [51]. Bootstrap support ≥ 95% was considered significant.
Acknowledgments
Acknowledgements
We thank L. E. Brahmakulam, B. R. Eidell, D. S. Heckman, A. R. Pfaff, J. L. Shoe, and R. L. Stauffer for assistance with analyses, and D. J. Des Marais, J. Kasting, L. Margulis, W. Martin, M. Nei, and M. Rohmer for comments or discussion. S.B.H. was supported by NASA and NSF, and S.K. by NIH and NSF.
Contributor Information
S Blair Hedges, Email: sbh1@psu.edu.
Hsiong Chen, Email: hxc166@psu.edu.
Sudhir Kumar, Email: s.kumar@asu.edu.
Daniel YC Wang, Email: dyw1@hotmail.com.
Amanda S Thompson, Email: ast116@psu.edu.
Hidemi Watanabe, Email: watanabe@gsc.riken.go.jp.
References
- Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Origin of Eukaryotic Cells. New Haven, Connecticut, Yale University Press. 1970.
- Margulis L. Archael-eubacterial mergers in the origin of Eukarya: Phylogenetic classification of life. Proc Natl Acad Sci USA. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding GB, Gupta R. Protein-based phylogenies support a chimeric origin of the eukaryotic genome. Mol Biol Evol. 1995;12:1–6. doi: 10.1093/oxfordjournals.molbev.a040178. [DOI] [PubMed] [Google Scholar]
- Feng D-F, Cho G, Doolittle RF. Determining divergence times with a protein clock: Update and reevaluation. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Micro Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. A paradigm gets shifty. Nature. 1998;392:15–16. doi: 10.1038/32033. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Lang DF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- DesMarais DJ. When did photosynthesis emerge on earth? Science. 2000;289:1703–1705. [PubMed] [Google Scholar]
- Brocks JJ, Logan GA, Buick R, Summons RE. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- Han T-M, Runnegar B. Megascopic eukaryotic algae from the 2.1 billion-year-old Negaunee iron-formation, Michigan. Science. 1992;257:232–235. doi: 10.1126/science.1631544. [DOI] [PubMed] [Google Scholar]
- Schopf JW. Microfossils of the early Archean apex chert: New evidence of the antiquity of life. Science. 1993;260:640–646. doi: 10.1126/science.260.5108.640. [DOI] [PubMed] [Google Scholar]
- Knoll AH. A new molecular window on early life. Science. 1999;285:1025–1027. doi: 10.1126/science.285.5430.1025. [DOI] [PubMed] [Google Scholar]
- Summons RE, Jahnke LL, Hope JM, Logan GA. 2-methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature. 1999;400:554–557. doi: 10.1038/23005. [DOI] [PubMed] [Google Scholar]
- Rohmer M, Bisseret P, Neunlist S. The hopanoids, prokaryotic triterpenoids and precursors of ubiquitous molecular fossils. Biological Markers in Sediments and Petroleum : A Tribute to Wolfgang K Seifert (Edited by Moldowan JM, Albrecht P, Philp RP) Englewood Cliffs, New Jersey, Prentice Hall. 1992. pp. 1–17.
- Rohmer M. The biosynthesis of triterpenoids of the hopane series in the Eubacteria: A mine of new enzyme reactions. Pure Appl Chem. 1993;65:1293–1298. [Google Scholar]
- Ohmoto H. When did the earth's atmosphere become oxic? The Geochemical News. 1997;93:12–26. [Google Scholar]
- Watanabe Y, Martini JEJ, Ohmoto H. Geochemical evidence for terrestrial ecosystems 2.6 billion years ago. Nature. 2000;408:574–578. doi: 10.1038/35046052. [DOI] [PubMed] [Google Scholar]
- Widdel F, Schnell S, Heising S, Ehrenreich A, Assmus B. Ferrous iron oxidation by anoxygenic phototrophic bacteria. Nature. 1993;362:834–838. doi: 10.1038/362834a0. [DOI] [Google Scholar]
- Doolittle RF, Feng D-F, Tsang S, Cho G, Little E. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fitch WM. Dating the cenancestor of organisms. Science. 1996;274:1750. doi: 10.1126/science.274.5293.1750. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Doolittle RF. Determining the relative rates of change for prokaryotic and eukaryotic proteins with anciently duplicated paralogs. J Mol Evol. 2000;51:173–181. doi: 10.1007/s002390010078. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Lake JA. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992;257:74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UCM, Podowski RM, Naslund AK, Eriksson A-S, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Gunderson JH, Elwood HJ, Alonso RA, Peattie DA. Phylogenetic meaning of the kingdom concept: An unusual ribosomal RNA from G iardia lamblia. Science. 1989;243:75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- Embley TM, Hirt RP. Early branching eukaryotes? Curr Opin Genet Dev. 1998;8:624–629. doi: 10.1016/S0959-437X(98)80029-4. [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Svard SG, Tovar J, Clark CG, Smith MW, Gillin FD, Sogin ML. A mitochondrial-like chaperonin 60 gene in G iardia lamblia : Evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojzsis SJ, Arrhenius G, McKeegan KD, Harrison TM, Nutman AP, Friend CRL. Evidence for life on Earth before 3,800 million years ago. Nature. 1996;384:55–59. doi: 10.1038/384055a0. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Mukhtar T, Singh B. Evolutionary relationships among photosynthetic prokaryotes (Heliobacterium chlorum, Chloroflexus aurantiacus, cyanobacteria, Chlorobium tepidum and proteobacteria): Implications regarding the origin of photosynthesis. Mol Microbiol. 1999;32:893–906. doi: 10.1046/j.1365-2958.1999.01417.x. [DOI] [PubMed] [Google Scholar]
- Xiong J, Fischer WM, Inoue K, Nakahara M, Bauer CE. Molecular evidence for the early evolution of photosynthesis. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- Kirschvink JL, Gaidos EJ, Bertani LE, Beukes NJ, Gutzmer J, Maepa LN, Steinberger RE. Paleoproterozoic snowball Earth: Extreme climatic and geochemical global change and its biological consequences. Proc Natl Acad Sci USA. 2000;97:1400–1405. doi: 10.1073/pnas.97.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov AA, Kasting JF, Brown LL. Greenhouse warming by CH4 in the atmosphere of early Earth. J Geophys Res. 2000;105:11981–11990. doi: 10.1029/1999JE001134. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. The genome sequence of Buchnera sp. APS, an endocellular bacterial symbiont of aphids. Nature. 2000;407:81–86. doi: 10.1016/S0168-9002(97)01373-9. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DY-C, Kumar S, Hedges SB. Divergence time estimates for the early history of animal phyla and the origin of plants, animals, and fungi. Proc R Soc Lond B. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML. History assignment: When was the mitochondrion founded? Curr Opin Genet Dev. 1997;7:792–799. doi: 10.1016/S0959-437X(97)80042-1. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Parker PH, Sibley CG, Kumar S. Continental breakup and the ordinal diversification of birds and mammals. Nature. 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Molecular evolutionary clocks. Molecular Evolution (Edited by Ayala FJ) Sunderland, MA, Sinauer Associates. 1976. pp. 160–178.
- Gu X. Early metazoan divergence was about 830 million years ago. J Mol Evol. 1998;47:369–371. doi: 10.1007/pl00013150. [DOI] [PubMed] [Google Scholar]
- Lynch M. The age and relationships of the major animal phyla. Evolution. 1999;53:319–325. doi: 10.1111/j.1558-5646.1999.tb03768.x. [DOI] [PubMed] [Google Scholar]
- Yang Z. Paml: A program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 1970;4:579–593. doi: 10.1007/BF00486096. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- Kumar S. Phyltest: A program for testing phylogenetic hypotheses. University Park, Pennsylvania, Institute of Molecular Evolutionary Genetics, Pennsylvania State University. 1996.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. Molphy version 2.3: Programs for molecular phylogenetics based on maximum likelihood. Tokyo, Institute of Statistical Mathematics. 1996.
- Kumar S, Tamura K, Nei M. Mega: Molecular evolutionary genetic analysis. University Park, Pennsylvania State University. 1993.
- Holland HD. Early proterozoic atmosphere change. Early life on Earth (Edited by Bengston S) New York, Columbia University Press. 1994. pp. 237–244.
- Ohmoto H. Evidence in pre – 2.2 Ga paleosols for the early evolution of atmospheric oxygen and terrestrial biota. Geology. 1996;24:1135–1135. doi: 10.1130/0091-7613(1996)024<1135:EIPGPF>2.3.CO;2. [DOI] [PubMed] [Google Scholar]


