Abstract
Effective metabolic engineering of microorganisms relies on balanced expression of both heterologous and endogenous genes to channel metabolic flux towards products of interest while achieving reasonable biomass buildup. To facilitate combinatorial pathway engineering and facile genetic operation, we engineered a set of modular cloning vectors compatible with BioBrick standards, called YaliBricks, to allow for rapid assembly of multigene pathways with customized genetic control elements (promoters, intronic sequences and terminators) in the oleaginous yeast Yarrowia lipolytica. We established a sensitive luciferase reporter and characterized a set of 12 native promoters to expand the oleaginous yeast genetic toolbox for transcriptional fine-tuning. We harnessed the intron alternative splicing mechanism and explored three unique gene configurations that allow us to encode genetic structural variations into metabolic function. We elucidated the role of how these genetic structural variations affect gene expression. To demonstrate the simplicity and effectiveness of streamlined genetic operations, we assembled the 12 kb five-gene violacein biosynthetic pathway in one week. We also expanded this set of vectors to accommodate self-cleavage ribozymes and efficiently deliver guide RNA (gRNA) for targeted genome-editing with a codon-optimized CRISPR-Cas9 nuclease. Taken together, the tools built in this study provide a standard procedure to streamline and accelerate metabolic pathway engineering and genetic circuits construction in Yarrowia lipolytica.
Keywords: Gene assembly, Intron alternative splicing, Luciferase reporter, Promoter activity, Yarrowia lipolytica, Gene configuration, Genome-editing
Highlights
-
•
Twelve modular cloning vectors YaliBricks developed for Y. lipolytica.
-
•
Luciferase reporter established and 12 native promoters characterized.
-
•
Intron alternative splicing harnessed to construct 3 gene configurations.
-
•
A 12-kb five-gene violacein biosynthetic pathway assembled in one week.
-
•
CRISPR-Cas9 genome-editing feature was incorporated into YaliBricks.
1. Introduction
The oleaginous yeast Yarrowia lipolytica has become the organism of choice for the production of oleochemicals (Abghari and Chen, 2014, Xu et al., 2016), biofuels (Ledesma-Amaro and Nicaud, 2016, Xu et al., 2017) and acetyl CoA-derived metabolites (Zhu and Jackson, 2015, Ledesma-Amaro et al., 2016). It has been extensively studied as a model organism for lipid accumulation (Beopoulos et al., 2009) and degradation (Fickers et al., 2005), dimorphism (Ruiz-Herrera and Sentandreu, 2002, Morales-Vargas et al., 2012) and protein secretory pathway (Beckerich et al., 1998). Y. lipolytica is also recognized as a “generally regarded as safe” (GRAS) organism (Groenewald et al., 2014) for the production of large quantities of citric acid (Papanikolaou et al., 2002, Papanikolaou et al., 2008), α-ketoglutarate (Zhou et al., 2010) and succinic acid (Cui et al., 2017) in the food industry. Coupled with its ability to natively utilize a wide range of substrates, such as glucose, fructose, glycerol and hydrocarbons, its low pH tolerance and strictly aerobic nature (Abghari and Chen, 2014, Ledesma-Amaro et al., 2016), these properties make this yeast a very attractive candidate for industrial biotechnology applications. Notable examples of its industrial applications include the production of eicosapentaenoic acid (EPA)-rich products (Xue et al., 2013, Xie et al., 2015), conjugated linoleic acid (Zhang et al., 2013), and cost-efficient production of lipid biofuels (Blazeck et al., 2014, Qiao et al., 2017, Xu et al., 2017).
Recent advances in understanding of Y. lipolytica's metabolic pathways, the sequencing of its genome (Liu and Alper, 2014) and further expansion of its genetic toolbox have resulted in several successful endeavors to metabolically engineer this unconventional yeast to produce high-value compounds, e.g. limonene (Cao et al., 2016) and γ-decalactone (Gomes et al., 2012, Braga and Belo, 2016). These achievements demonstrate the potential of Y. lipolytica as an industrial microbe to produce novel compounds and expand beyond its regular portfolio of fatty acids, fatty alcohols, biofuels, and protein production.
Although a number of gene over-expression and deletion tools have been established in Y. lipolytica (Hong et al., 2012), the library of available tools is not as developed as that of other yeasts such as Saccharomyces cerevisiae. This is due in part to Y. lipolytica having a high frequency of non-homologous end-joining (NHEJ), while S. cerevisiae mainly utilizes the highly efficient homologous recombination repair machinery to repair DNA double-strand breaks (Kretzschmar et al., 2013, Madzak, 2015). Fickers et al. developed a gene disruption method by combining the sticky-end polymerase chain reaction (SEP) method and Cre-lox recombination system (Fickers et al., 2003). However, acceptable rates of recombination only occur when long homology regions (0.5–1 kb) are used, making this method less efficient in the absence of the KU70 gene deletion (Madzak, 2015). Another hurdle in efficient genome-editing in Y. lipolytica lies in the fact that it has a less-defined non-coding RNA processing system. To solve this challenge, Schwartz et al. (2016) has used a hybrid tRNA promoter to express gRNA, allowing for efficient CRISPR-Cas9 targeting in Yarrowia. Alternatively, Gao et al. (2016) demonstrated that the Type II RNA polymerase, in combination with the highly efficient hammerhead ribozyme and HDV ribozyme, could efficiently deliver gRNA and achieved multiplex genome-editing in Y. lipolytica.
In addition to genome-editing tools, advances have been made to develop a multi-copy, plasmid-based gene expression system, which is suitable for gene-expression fine-tuning due to the simplicity of plasmid construction and high transformation efficiency compared to the laborious and time-consuming genome manipulation process (Liu et al., 2014, Madzak, 2015). For example, Liu et al. engineered the low-copy CEN plasmid by fusing different promoters upstream of the centromeric region and improved both the copy number and gene expression level (Liu et al., 2014).
In light of these advances, there is a pressing need for development of facile genetic tools that are tailored for modular and combinatorial pathway engineering in Y. lipolytica (Xu et al., 2013, Xu and Koffas, 2013). Indeed, metabolic engineering community has embraced the concepts of standardization and modularization to enable rapid construction of large pathway libraries covering a broad range of gene expression space (Xu et al., 2013, Xu et al., 2014, Xu et al., 2014). This is often achieved by creating interoperable genetic parts with well-defined gene expression metrics. Here we report the development of a set of BioBrick vectors and its application in metabolic engineering. The engineered 12 YaliBricks vectors comprise four compatible restriction enzyme sites to enable modular pathway engineering and facile genetic operation. We established a sensitive and reliable luciferase reporter platform and characterized 12 endogenous promoters that have broad range of transcriptional dynamics. We demonstrate the versatility of the engineered YaliBrick vectors to construct gene clusters with unique transcriptional configurations: operon, pseudo-operon, monocistronic. In addition, we constructed a 12 kb, 5-gene biosynthetic pathway for the antibiotic pigment violacein using YaliBrick as a proof of concept for quick pathway assembly and screening. We incorporated our YaliBrick vector with genome-editing feature and successfully introduced indel mutation and frame-deletion deletion of the CAN1 (arginine permease) genomic loci of Y. lipolytica. We envision that the YaliBrick vectors will provide a powerful platform to facilitate modular manipulation of multigene pathways and the construction of complex genetic circuits in oleaginous yeast. These pYaliBrick vectors, along with several other relevant plasmids used in this study, will be deposited to Addgene soon for public access.
2. Materials and methods
2.1. Strains and culture conditions
Escherichia coli NEB5α high efficiency competent cells were obtained from NEB for plasmid construction, preparation, propagation and storage. The Y. lipolytica wild type strain W29 was purchased from ATCC (ATCC20460). The auxotrophic Po1g (Leu−) was obtained from Yeastern Biotech Company (Taipei, Taiwan). All strains and plasmids are listed in Supplementary Table S1.
LB broth or agar plate with 100 µg/mL ampicillin was used to cultivate E. coli strains. Yeast rich medium (YPD) was prepared with 20 g/L Bacto peptone (Difco), 10 g/L yeast extract (Difco), and 20 g/L glucose (Sigma-Aldrich), and supplemented with 15 g/L Bacto agar (Difco) for solid plates. YNB medium was made with 1.7 g/L yeast nitrogen base (without amino acids and ammonium sulfate) (Difco), 5 g/L ammonium sulfate (Sigma-Aldrich), 0.69 g/L CSM-Leu (Sunrise Science Products, Inc.) or 0.67 g/L CSM-Leu-Ura (Sunrise Science Products, Inc.), and 20 g/L glucose. Selective YNB plates were made with YNB media supplemented with 15 g/L Bacto agar (Difco). For canavanine resistance selection, 0.70 g/L CSM-Arg (Sunrise Science Products, Inc.) and 15 g/L Bacto agar (Difco), supplemented with 50 μg/mL of canavanine, were used for YNB plates.
For promoter activity characterization, the media used for the shake flask fermentations contained 1.7 g/L yeast nitrogen base (without amino acids and ammonium sulfate), 5 g/L ammonium sulfate, 0.67 g/L CSM-Leu and 20 g/L glucose. Single Y. lipolytica colonies were picked from YNB selective plates and inoculated into YNB liquid media, which were grown at 30 °C overnight. Overnight cultures were inoculated into 50 mL of YNB media in 250 mL shake flasks with an initial cell density (OD600) of 0.01.
2.2. Plasmids construction
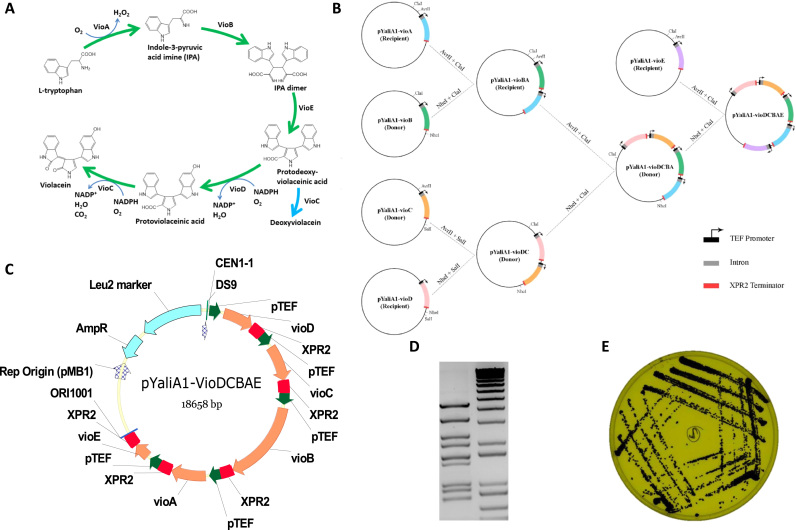
The 4 reporter genes hrGFP, mCherry, E. coli β-glucuronidase (EcUidA), and NanoLuc luciferase, and negative control E. coli biotin ligase (EcBirA) were PCR-amplified with the primers listed in Supplementary Table S2 and inserted into a pYaliA1 vector backbone at the SnaBI and KpnI site via Gibson assembly (Gibson et al., 2009). The coding sequences of vioA, vioB, vioC, vioD, and vioE were PCR-amplified from C. violaceum genomic DNA and inserted into a pYaliA1 vector backbone at the SnaBI and KpnI site via Gibson Assembly. Each PCR amplicon was cloned downstream of the Y. lipolytica TEF promoter in the vector pYaliA1 backbone containing TEF promoter and intron sequence. Upon sequence verification, vioA, vioB, vioC, vioD, and vioE were assembled into a single pYaliA1 vector backbone by restriction enzyme digestion subcloning using the restriction sites AvrII, NheI, and ClaI (Fermentas, Thermo Fisher Scientific). The detailed assembly procedure is listed in Fig. 4B.
Fig. 4.
Modular assembly of the 12 kb five-gene violacein biosynthetic pathway with pYaliBricks in Y. lipolytica. (A) Five biosynthetic steps lead to the synthesis of violacein from tryptophan. A detailed description of the biosynthetic reactions could be found in the main text. (B) In-parallel cloning procedure to assemble the five-gene pathway. (C) Plasmid map of the final five-gene construct. CEN1-1, yeast centromere containing the autonomous replication sequence; ORI1001, yeast replication origin; Leu2, leucine auxotrophic marker; TEF, promoter; XPR2, terminator; AmpR, ampicillin resistance maker. (D) Digestion pattern of the recombinant plasmid containing the five-gene violacein biosynthetic pathway. (E) Representative picture of violacein-producing Y. lipolytica grown on CSM-Leu plate.
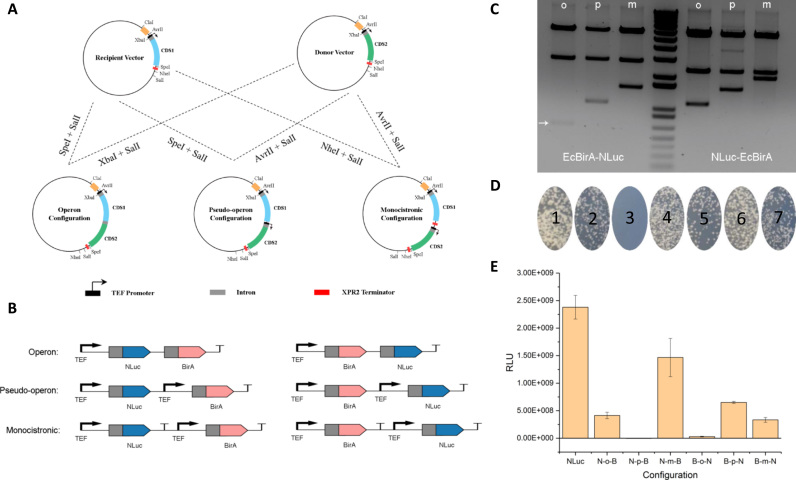
For construction of the operon configuration, the XbaI and SalI digested donor vector was gel purified and ligated to the SpeI and SalI digested destination vector. For pseudo-operon configuration, the AvrII and SalI digested donor vector was gel purified and ligated to the SpeI and SalI digested destination vector. For monocistronic configuration, the AvrII and SalI digested donor vector was gel purified and ligated to the NheI and SalI digested destination vector. Detailed cloning procedure is listed in Fig. 3A. This procedure was used to obtain all the six gene configurations N-o-B, N-p-B, N-m-B, B-o-N, B-p-N and B-m-N.
Fig. 3.
Intron alternative splicing allows the construction of three gene configurations in Y. lipolytica. (A) Modular cloning procedure to generate three gene configurations in Y. lipolytica. (B) A detailed layout of a two-gene pathway varying in gene configuration and positional order. (C) Gel digestion pattern of the six gene configurations: lane 1, B-o-N; lane 2, B-p-N, lane 3, B-m-N; lane 4, ladder; lane 5, N-o-B; lane 6, N-p-B and lane 7, N-m-B. (D) Yeast transformation assay to evaluate the efficiency of the Ura3 marker restoring Ura3 auxotrophic phenotype. Plate 1, Ura3 marker positive control; plate 2, U-o-M; plate 3, U-p-M; plate 4, U-m-M; plate 5, M-o-U; plate 6, M-p-U; plate 7, M-m-U. U is an abbreviation for Ura3 and M is an abbreviation for mCherry. (E) Positional effects of gene configuration on gene expression efficiency as evaluated by luminescence readout in the six genetic variations. RLU: relative luminescence unit. Error bars are the standard deviation calculated from triplicate readouts.
The 11 endogenous promoters were PCR-amplified using Y. lipolytica genomic DNA as the template. We typically amplify 1000 bp of gene sequence upstream of the structural DNA coding sequence. pYaliA1-NLuc was linearized with AvrII and XbaI and gel purified, then Gibson assembled with the 12 PCR amplicons, yielding a total of 12 pYaliBrick constructs including the original pYaliA1-NLuc plasmid.
Synthetic HHR-gRNA-Can1-tracrRNA-HDV (Supplementary Table S2) gene fragment was ordered from IDT and Gibson assembled into the XbaI and SpeI digested pYaliA1 vector. Humanized Cas9 attaching the nucleus localization signal was PCR amplified with p414-TEF1p-Cas9-CYC1t as template and Gibson assembled into the SnaBI and KpnI linearized pYaliA1 vector. All Gibson assembly cloned constructs were sequenced either through Genewiz or Quintara Biosciences. For assembling the gene construct containing both the Cas9 and CAN1 gRNA, AvII and NotI digested pYaliA1-gRNA-CAN1 was gel purified and ligated to the NheI and NotI digested pYaliA1-hCas9 to obtain the dual expression plasmid PYaliA1-hCas9-gRNA-ylCAN1.
All plasmids constructed were transformed into the Y. lipolytica host strain Po1g ΔLeu using the lithium acetate/single-strand carrier DNA/PEG method (Chen et al., 1997).
2.3. hrGFP and mCherry fluorescence assay
The strains bearing the fluorescent protein reporters GFP (excitation = 488 nm, emission = 509 nm) and mCherry (excitation = 587 nm, emission = 610 nm) were assayed with a Biotek H1 Synergy microtiter plate reader using whole cell cultures. Cell cultures were cultivated for 12 h in minimal YNB medium supplemented with 2% glucose at 30 °C and normalized to an OD600 of 1.
2.4. Lysate preparation for GUS and MUG assay
Cells expressing EcUidA were picked from single colonies and grown in 3 mL minimal medium overnight at 30 °C. 50 mL shake flask cultures were inoculated with the overnight culture at an initial OD600 ≈ 0.01 and the cultures were harvested the following day at late-exponential phase. The cells harvested were normalized to an OD600 of 1 and pelleted via centrifugation. Crude protein extracts were obtained by a combination of incubation in cell lysis buffer supplemented with 1 mg/L Zymolyase and 1 min of mechanical lysing. Total protein concentration of lysates was quantified using Bradford reagent (Thermo Fisher Scientific). These lysates were then used for both the MUG assay and histochemical GUS staining assay as outlined in the methodology by Hong et al. (2012). For each MUG assay reaction, 100 μL of lysate was added to 700 μL of GUS assay buffer (1 g/L 4-methylumbelliferyl β-D-glucuronide) and incubated at 37 °C for one hour. Aliquots of 100 μL were collected at 0, 30, 60 min time points and the quenched in 900 μL stop buffer (1 M Na2CO3). Fluorescence intensity measurements were taken with a Biotek H1 Synergy microtiter plate reader, at an excitation wavelength of 360 nm and emission wavelength of 455 nm. For the colorimetric GUS staining assay, 100 μL crude lysate was added to 500 μL staining buffer (2 mM 5-bromo-4-chloro-3-indolyl glucuronide) and incubated overnight at 37 °C.
2.5. Luciferase assay for promoter library characterization
All promoters were PCR-amplified from Y. lipolytica strain Po1g genomic DNA and cloned upstream of the luciferase reporter gene NLuc with the intronic sequence in the pYaliBrick vector bearing a leucine auxotrophic marker. Constructs were sequence-verified and transformed into the Y. lipolytica host strain Po1g ΔLeu. Single colonies were picked for culturing and subsequently harvested for protein extraction to perform the luciferase assay using the Nano-Glo Luciferase Assay System kit from Promega. Luminescence was measured with a Biotek H1 Synergy microplate reader.
2.6. Genome editing with CRISPR-Cas9
pYaliA1-hCas9 along with CAN1 sgRNA PCR product were transformed into Y. lipolytica Po1g ΔLeu strain to transiently disrupt the CAN1 gene. pYaliA1-hCas9-gRNA-ylCAN1 was transformed into Y. lipolytica Po1g ΔLeu for plasmid-based CAN1 disruption. All transformation cultures were plated onto CSM-Leu plates and incubated at 30 °C for two days. Colonies obtained were then spotted on CSM-Arg (yeast complete synthetic media without arginine) supplemented with 50 μg/mL canavanine and incubated at 30 °C incubator for three days to select for positive mutants. Canavanine-resistant colonies were inoculated into 2 mL YPD media and grown at 30 °C with shaking at 250 rpm overnight. Y. lipolytica genomic DNA was purified with the Promega Wizard genomic DNA purification kits. CAN1 gene was PCR amplified with the primer pairs Can1_F and Can1_R and analyzed by DNA gel electrophoresis. PCR products were cleaned-up by Zymo PCR concentrator clean-up kits and sent to Genewiz for DNA sequencing with Can1_seq as the sequencing primer.
3. Results and discussions
3.1. Developing a bioluminescence reporter system
Effective metabolic engineering of microorganisms relies on assembling and rewiring biosynthetic pathways to channel carbon flux towards producing compounds of interest. One of the most important aspects is to modulate gene expression and ideally achieve a balanced pathway expression with no metabolic bottlenecks. As such, it is crucial for metabolic engineers to be able to monitor gene expression levels via a stable, consistent, and reliable reporter system. In the case of genetic parts characterization, there are several reporter systems developed for this application; most assays utilize fluorescence or luminescence to detect and quantify gene expression. Fluorescence reporter activity can be measured either by the fluorescence intensity of the protein itself (as in the case of hrGFP and mCherry) or the specific enzymatic activity of the reporter using a fluorescent substrate (e.g. 4-methylumbelliferyl-β-D-galactopyranoside, MUG assay) (Haugwitz et al., 2008). On the other hand, bioluminescent assays utilize luciferase and, depending on the intrinsic nature of the enzyme, they require either luciferin and ATP or only coelenterazine as the substrate (Haugwitz et al., 2008).
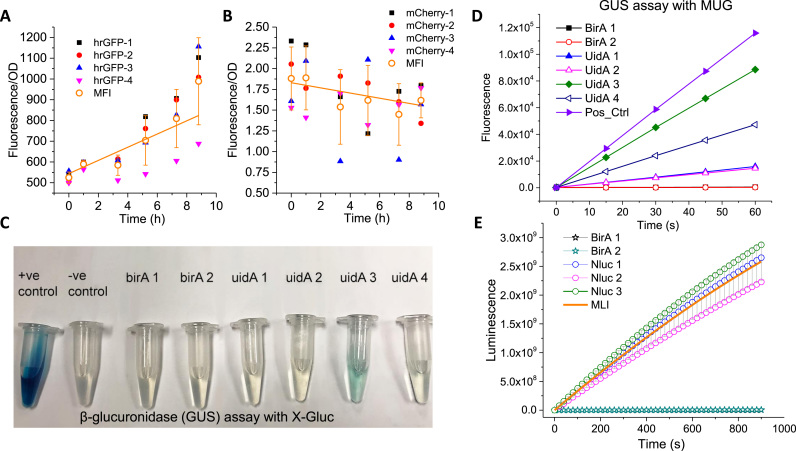
To determine the best reporter system, four reporter genes hrGFP, mCherry, E. coli β-glucuronidase (EcUidA), and luciferase gene NanoLuc were constructed under the control of a constitutive TEF promoter carried by a self-replicating plasmid. We observed that the fluorescence readout was choppy and there was no consistent fluorescence expression pattern in both hrGFP or mCherry (Fig. 1A and B). Humanized hrGFP exhibited increased mean fluorescence intensity (MFI) with time across the four parallel biological replicates (Fig. 1A), albeit the sample-to-sample variation was large. Interestingly, mean fluorescence intensity (MFI) of mCherry was observed leveling off with time, possibly due to yeast cell autofluorescence (wavelength of mCherry emission is 610 nm). This indicates that either the expression of hrGFP and mCherry under the constitutive TEF promoter is very weak, or at least the yeast autofluorescence interferes with the detection. Humanized GFP (hrGFP) probably demonstrated better stability and functionality than mCherry under the tested condition; this is further corroborated by the fact that hrGFP reporter could be detected in Y. lipolytica using flow cytometry (Blazeck et al., 2011, Hussain et al., 2016), albeit with low sensitivity and detection limit.
Fig. 1.
Evaluating the reliability and consistency of different reporter systems in Y. lipolytica. (A) hrGFP fluorescence protein as the reporter. MFI, mean fluorescence intensity. Four biological replicates were tested. (B) mCherry fluorescence protein as the reporter. Four biological replicates were tested. (C) β-glucuronidase (GUS) reporter with X-gluc (5-Bromo-4-chloro-3-indolyl-β-D-glucuronide) as substrate. Blue product could be quantified based on a spectrophotometric absorbance assay. (D) β-glucuronidase (GUS) reporter with MUG (4-methylumbelliferyl-beta-D-glucuronide) as substrate. Fluorescence product could be quantified with a fluorometric assay. Four biological replicates were tested. (E) Luciferase Nanoluc as the reporter. MLI, mean luminescence intensity. Three biological replicates were tested.
Next, we tested whether we could use the E. coli β-glucuronidase (encoded by uidA) as a reporter. β-glucuronidase (GUS) enzymatic activity in Y. lipolytica was measured using both fluorescent substrate (MUG, 4-methylumbelliferyl-β-D-galactopyranoside) and histochemical staining substrate (X-gluc, 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid). The colorimetric GUS staining assay took about 24 h to develop the color and did not demonstrate satisfactory detection of GUS activity (Fig. 1C). On the other hand, the fluorescent MUG assay was able to detect varying levels of enzymatic activity across all biological replicates of EcUidA-expressing strains with minimal background noise (Fig. 1D). Also, intron characterization was performed with the MUG assay by comparing enzymatic activity in stains with and without the intronic sequence fused downstream of the TEF promoter, revealing a 238-fold difference in fluorescence intensity in the strain with the intron-fused EcUidA over the intron-less EcUidA. This demonstrates the MUG assay's broad dynamic range spanning across about 2 orders of magnitude.
The main disadvantage of the MUG assay is the requirement to individually quench each sample in stop buffer before measuring fluorescence intensity, making it a rather inefficient and laborious process for multiplex assays. Another disadvantage is that the MUG assay is not amenable to time-course analysis of gene expression studies due to the fast kinetics of the MUG fluorophore emission.
To address these concerns, we turned to test the NanoLuc luciferase reporter by measuring the rate of luminescence emission from lysates of NLuc-expressing Y. lipolytica cells. Integration of the rate of luminescence emission exhibited a linear response curve with relatively small deviation from the mean luminescence intensity (mL) (Fig. 1E). Against the negative control BirA, the luminescence reporter in the NLuc-expressing strain yielded 104-fold higher signal than the baseline (BirA strain). The sensitivity of both the luciferase assay and MUG assay were similar when the strong, constitutive TEF promoter was used.
Bioluminescent luciferase assays have become the preferred reporter system for gene expression studies due to their high sensitivity and broad dynamic range (Masser et al., 2016). The biggest advantage of using NanoLuc luciferase is the minimal background noise due to its independence of ATP. In contrast to the larger GUS reporter (EcUidA is encoded by 603 amino acid residues) and hrGFP reporter (240 amino acid residues), the NanoLuc luciferase only contains 171 amino acid residues, giving NanoLuc the advantage of quicker translation and folding and better expressibility. This makes NanoLuc luciferase an ideal reporter for monitoring rapid gene expression changes with minimal protein expression burden. Its smaller size is also beneficial in the construction of fusion proteins or split-proteins due to less probability of interfering with the functionality of the tagged protein.
3.2. Characterizing transcriptional activity of 12 promoters
The highly complex transcriptional machinery in eukaryotes has resulted in many engineering efforts to identify and characterize core promoters and their proximal sequences. Given that endogenous gene expression elements are often used in pathway engineering in eukaryotes, and their transcriptional machinery is poorly understood and hard to predict, Hussain et al. systematically examined several common promoter components in yeast and engineered hybrid promoters by using in-tandem repeats of upstream activating sequences in conjunction with downstream truncated promoters to allow for a degree of predictability in gene expression (Hussain et al., 2016). For example, they have identified several promoters that are sensitive to oleic acid, and created an oleic acid-responsive hybrid acyl-CoA oxidase (POX2) promoter with modified core promoter and proximal sequences (Hussain et al., 2016). Blanchin-Roland et al. (1994) reported that Y. lipolytica extracellular protease (XPR2) promoter was regulated by a multitude of factors including pH, carbon, nitrogen and peptones. In addition, upstream activating sequence (UAS) regions have been suggested as the major player in mediating the response to those factors (Wagner and Alper, 2016). Dulermo et al. engineered variants of the TEF and LEU promoters by coupling different UAS1 elements in tandem and found that there is a positive correlation between the number of tandem UAS elements and expression levels (Dulermo et al., 2017). In all, promoter engineering efforts in Y. lipolytica are highly restricted to a number of well-characterized promoters. Expanding the number of available endogenous promoters will provide alternative targets for further transcriptional engineering. It will also lead to the discovery of orthogonal promoters with minimal interdependence on native cell transcriptional machinery when there are considerations to decouple heterologous gene expression from the biomass precursor pathway. This allows for a more customizable approach in combinatorial gene expression optimization to selectively target different metabolic pathways.
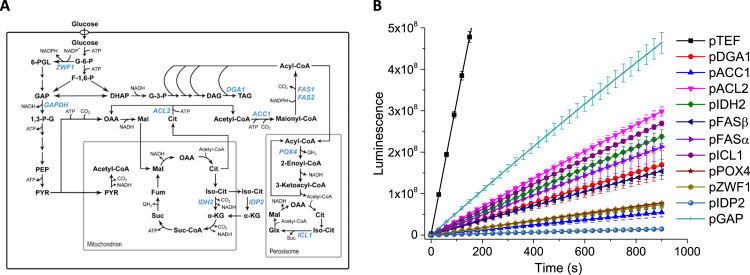
We selected 11 endogenous promoters in addition to the well-characterized TEF promoter for genetic parts characterization (Table 1). Given Y. lipolytica's ability to accumulate up to 50% of its dry cell weight as lipids (Shi et al., 2016), the promoters chosen are primarily centering around the lipogenic pathways: the supply of acetyl-CoA and malonyl-CoA, the TCA metabolic pathway, glycolysis, the supply of NADPH from pentose phosphate pathway, and the lipid oxidation pathway (Fig. 2A). Unsurprisingly, the TEF promoter exhibited strongest expression levels, since the native TEF gene codes for the abundant translational elongation factor 1-α, which is essential for gene translation in all cells. Furthermore, in the case of Y. lipolytica, the TEF gene only exists as a single copy in the chromosome, implying that its promoter is very strong. The remaining 11 promoters derived from TCA cycle, glycolysis, pentose phosphate pathway or lipid oxidation pathway, demonstrated relative transcriptional activity ranging from 0.7% to 29.7% of TEF promoter activity (Fig. 2B). The top three candidates in this group are promoters found in glycolysis (pGAP promoter driving the expression of glyceraldehyde-3-phosphate dehydrogenase), glyoxylate cycle (pICL1 driving the expression of the isocitrate lyase), and a key precursor step of fatty acid biosynthesis (pACL2 driving the expression of the ATP: citrate lyase). The bottom two candidates in this promoter panel are involved in the supply of malonyl-CoA (pACC1driving the expression of acetyl-CoA carboxylase) and the supply of NADPH (pZWF1 driving the expression of the PP pathway glucose-6-phosphate dehydrogenase), which have been identified as the rate-limiting steps for efficient lipid biosynthesis (Tai and Stephanopoulos, 2013, Wasylenko et al., 2015).
Table 1.
List of promoters and their corresponding vector names.
| Transcription unit | Annotation | Promoter name | Vector name |
|---|---|---|---|
| Translational elongation factor EF−1 alpha | YALI0C09141g | ylTEF promoter | pYaliA1 |
| Acyl-CoA: diacylglycerol acyltransferase | YALI0E32769g | ylDGA1 promoter | pYaliB1 |
| Acetyl-CoA-carboxylase 1 | YALI0C11407g | ylACC promoter | pYaliC1 |
| ATP citrate lyase 2 | YALI0D24431g | ylACL2 promoter | pYaliD1 |
| Isocitrate dehydrogenase NAD+ subunit 2 mitochondrial | YALI0D06303g | ylIDH2 promoter | pYaliE1 |
| Fatty acid synthase subunit beta | YALI0B15059g | ylFAS2 promoter | pYaliF1 |
| Fatty acid synthase subunit alpha | YALI0B19382g | ylFAS1 promoter | pYaliG1 |
| Isocitrate lyase 1 | YALI0C16885g | ylICL1 promoter | pYaliH1 |
| POX4 Fatty-acyl coenzyme A oxidase | YALI0E27654g | ylPOX4 promoter | pYlaliJ1 |
| ZWF1 Glucose-6-phosphate dehydrogenase | YALI0E22649g | YlZWF1 promoter | pYaliK1 |
| Cytosolic NADP-specific isocitrate dehydrogenase | YALI0F04095g | ylIDP2 promoter | pYaliL1 |
| Glyceraldehyde 3-phosphate dehydrogenase | YALI0C06369g | ylGAPHD promoter | pYaliM1 |
Fig. 2.
Determining the transcriptional activity of 12 native promoters in Y. lipolytica. (A) Metabolic map of the 12 chosen structural genes that encode critical enzymes catalyze lipid biosynthesis in Y. lipolytica. (B) Characterizing the transcriptional activity of the 12 chosen promoters with luminescence as the readout. The details of the 12 chosen promoters can be found in Table 1.
Christen et al. performed metabolic flux analysis on several aerobic species of yeast including Y. lipolytica and found that the flux through the TCA cycle and pentose phosphate pathway equals the glycolytic flux (Christen and Sauer, 2011). In addition, Liu et al. demonstrated that the glyoxylate shunt is always active throughout different stages of growth, thus ensuring that the metabolite pools for the TCA cycle are replenished (Liu et al., 2016). Taken together, the flux through the glyoxylate shunt and the reactions leading to acetyl-CoA formation would be high in correlation to measured metabolite pools. However, there is no clear correlation between high flux and promoter strength. Additional studies would be needed to determine optimal activation conditions for the promoters.
It is generally accepted that promoter architecture in eukaryotic organisms is extremely sophisticated in their regulatory mechanisms and the transcriptional responses are not as predictable compared to prokaryotic systems (Hussain et al., 2016, Portela et al., 2017). A notable example is the Snf1-mediated acetyl-CoA carboxylase regulation (Seip et al., 2013, Shi et al., 2014). The 11 promoters identified in this study have not been fully characterized under different carbon, nitrogen and oxygen conditions. It is also noted that the sampling was performed at the late-exponential phase; it is known that gene activation and repression are correlated to the stages of growth. As such, the expression levels determined using the reporter assay may not reflect their optimal potential. Promoter activation and regulation can be mediated by different factors such as carbon source, nitrogen availability, pH, and metabolite levels (Madzak, 2015, Hussain et al., 2016). Further investigation is needed to fully elucidate these new promoters to better understand each promoter element and determine its function in regulating gene expression and lipogenesis. In addition, characterizing promoters responsible for gene expression in central carbon metabolism and lipogenesis would better inform future metabolic engineering efforts in fine-tuning native pathways without the need for replacement of the native promoter, which avoids potential disruption of regulatory mechanisms.
3.3. Gene configuration engineering
Comparative genomic studies revealed that Y. lipolytica is phylogenetically distant from other hemiascomycetous yeasts. It is considered a non-conventional, intron-poor species; however, among other hemiascomycetes, it has the highest number of introns in its genome to date. Yarrowia's relatively high intron density of 0.17 indicates that alternative splicing (AS) may play a major role in increasing the variance and complexity of its transcriptome (Mekouar et al., 2010). The outcome of AS of mRNA generally falls into two categories: mRNA that can be translated into functional proteins or nonsense-containing mRNA that may generate truncated, potentially non-functional proteins. For example, alternative splicing has led to generation of two unique MDH transcripts encoding malate dehydrogenases with different N-terminal signaling peptides; upon translation, the expressed malate dehydrogenase is either localized in the mitochondria, cytosol or peroxisome (Kabran et al., 2012). Eukaryotes have a quality control mechanism, nonsense-mediated mRNA decay (NMD), that targets mRNA with premature termination codons for degradation, thus preventing their translation. Mekouar et al. investigated several instances of AS in Y. lipolytica gene models and found that it exhibits all modes of alternative splicing, predominantly generated by intron retention (Mekouar et al., 2010). However, the resulting transcripts usually incorporated an early termination codon and led to nonsense-containing mRNAs.
Regardless, this concept of transcriptional interference provides a potential strategy to regulate gene expression levels without the need for in-depth promoter engineering and understanding of the complicated regulatory mechanisms underlying each promoter. In our pYaliBrick vectors, four compatible restriction enzyme sites are strategically placed in between the gene regulatory elements to allow for easy genetic operation when performing DNA assembly: operon, pseudo-operon, and monocistronic gene configurations (Fig. 3A). By taking advantage of isocaudomers with compatible ends, we can reuse the restriction sites infinitely in subsequent cloning steps, enabling rapid assembly of gene fragments without the need to design unique primers to incorporate new sites. This method is particularly useful for assembling pathways with multiple similar homologous regions or repeated parts, which is technically challenging for homologous recombination-based methods such as Gibson assembly or yeast transformation-based gene assembly (Xu et al., 2013a).
The three gene configuration mimics the prokaryotic gene expression system. An operon configuration encodes two structural gene sharing the same promoter and terminator, with each of the gene containing an intron derived from the TEF gene at their 5′ end. The intron is built upstream of the structural gene but downstream of the ATG exon to allow spliceosome processing of multiple intron sequences (Blumenthal, 1998) and potentially generate functional mRNA transcripts encoding different enzyme functionality (Fig. 3B). Similarly, a pseudo-operon configuration encodes two structural gene sharing the same terminator but driven by separate promoters. And a monocistronic configuration encodes two structural gene driven by separate promoters and terminators (Fig. 3B). Our standard cloning procedure allows us to streamline the genetic operation and rapidly create the three unique gene configurations (Fig. 3A). For a 2-gene pathway, six unique genetic organization could be easily obtained (Fig. 3B) and later screened by double digestion analysis (Fig. 3C). A ladder pattern of gel electrophoresis image indicates the three unique gene configurations, as demonstrated in Fig. 3C.
To test how these three gene configurations affect expression levels, we assembled six distinct genetic constructs varying in gene order and configuration. The two genes encode the yeast auxotrophic marker Ura3 and a fluorescence reporter (Supplementary Fig. S4). Upon transformation into the Ura3-deficient (Ura3-) strain, we evaluated which gene configuration could better complement the Ura3-deficient phenotype. Colony based screening on CSM-Ura media allowed us to compare the relative expression level of the Ura3 gene and inform us how genetic structural variation correlates with mRNA transcription (Fig. 3D). This complementation experiment demonstrates that six out of the seven constructs could functionally express the Ura3 gene, with the exception of plate No. 3 which corresponds to Ura3 and mCherry arranged in pseudo-operon form (Fig. 3D). To further validate this result, we then constructed six additional plasmids bearing two genes: BirA and NLuc. BirA is used as a control to investigate positional effects. All constructs displayed weaker expression of luminescence relative to the single NLuc control (Fig. 3E), which is not unusual as gene expression becomes weaker in tandem genetic constructs driven by similar promoters due to transcriptional inhibition (Shearwin et al. 2005). Constructs harboring NLuc as the first gene exhibited strongest light emission in the monocistronic format (N-m-B, Fig. 3E) followed by operon (N-o-B, Fig. 3E). However, the pseudo-operon configuration (N-p-B, Fig. 3E) showed baseline signal, indicating the possibility of non-functional NLuc. The pseudo-operon configuration N-p-B contains an additional promoter within the transcript (Fig. 3B). Given that NLuc is functionally expressed when it is configured as an operon (Fig. 3D and E), we can infer that the non-functionality observed in the pseudo-operon configuration may be due to the lack of the transcriptional termination signal, thus preventing the functional maturation of the first messenger RNA which encodes the NLuc luciferase. Thus, the transcript generated cannot be translated into functional NLuc, although BirA is functionally expressed.
When NLuc is located downstream of BirA, the intron in front of NLuc moves further down along the genetic construct. The weak luminescence signal detected in the operon construct (B-o-N, Fig. 3E) indicates that the spliceosome prefers to splice the intron which is in close proximity to the exon. That is, splicing the intron on the first gene BirA may inhibit splicing of the intron on the second gene (NLuc). This alternative splicing inhibition provides a novel approach to adjust the transcripts ratio of two proteins. On the other hand, BirA-NLuc in both pseudo-operon (B-p-N) and monocistronic (B-m-N) configurations retained NLuc functionality due to the production of functional NLuc mRNA and thus functional proteins, as indicated by the relatively strong luminescence signal (Fig. 3E). In addition, positional effects within the monocistronic configuration exhibited a 4.4-fold decrease in NLuc gene expression when NLuc is placed downstream of BirA compared to the N-m-B construct where NLuc is placed upstream of BirA (Fig. 3E). Taking this into consideration, designing metabolic pathways in a tandem configuration will require stronger promoters to drive the transcription of the downstream genes due to this positional effect. These results establish a foundation to study how introns affect the processing of mRNA. It also provides an alternative approach to encode complex metabolic activity into a well-defined genetic configuration.
3.4. Functional expression of multigene pathway
Violacein is a pigmented, indolocarbazole compound with interesting bioactivities such as antibacterial, anticancer, antiviral, trypanocidal, and antioxidant properties (Leal et al., 2015, Durán et al., 2016). These properties make violacein an attractive antibiotic target. The gene cluster vioABCDE isolated from Chromobacterium violaceum is responsible for converting the precursor tryptophan to violacein via a combination of enzymatic and non-enzymatic steps. VioA, VioC, and VioD are flavin-dependent oxygenases, with VioA catalyzing the first key step in converting L-tryptophan to 2-iminoindole pyruvate (Fig. 4A). VioB is a heme protein that yields the dimeric structure due to the C-C bond formation between the β-carbons in the benzylic rings of the two tryptophan molecules. The unstable intermediate formed by VioAB then goes through an indole shift catalyzed by VioE. At this point, the VioABE intermediate protodeoxyviolaceinic acid (Hirano et al., 2008) can directly be catalyzed either by VioD followed by VioC, or directly by VioC, to form violaceinic acid and deoxyviolaceinic acid (Fig. 4A), respectively (Hoshino, 2011).
Previous studies have taken advantage of its strong violet color to develop quick visual screening tools. The purple pigment has allowed for evaluating DNA assembly strategies and balancing expression for various biotechnological applications (Lee et al., 2013, Mitchell et al., 2015). To test our design strategies in a quick and effective manner, we constructed the 12 kb five-gene pathway for violacein production in our standardized pYaliBrick vectors. Our engineered pYaliBricks support the modular and in-parallel assembly of multiple gene constructs via streamlined genetic manipulation (Fig. 4B). To construct the five-gene pathway along with the regulatory control elements (promoters, introns and terminators), we performed three rounds of cloning in one week and assembled the five-gene pathway in a monocistronic configuration as illustrated in Fig. 4C. Sequence verification was quickly confirmed by the unique fragmentation pattern via restriction digestion (Fig. 4D). Upon transformation into the Po1g Y. lipolytica host, colonies exhibited various color ranging from white to deep purple (Supplementary Fig. S5). The phenotypic variance seen indicates the possibility of genetic instability of our Y. lipolytica host in retaining all functional genes in the violacein biosynthetic pathway. A dark purple colony was subsequently chosen (Fig. 4E) and cultivated in CSM-Leu minimal media, HPLC quantification of the chosen strain yielded about 31.5 mg/L violacein. This is the first demonstration of the functional expression of the five-gene violacein pathway in oleaginous yeast, albeit further strain optimization is required to improve the cost-competitiveness.
The pYaliBrick assembly vectors provide a rapid method of assembling large pathways containing multiple reused parts (promoter, intron, terminator), which otherwise would not be attainable through Gibson assembly cloning. Given that the process involves no PCR amplification and only relies on subcloning using highly specific restriction enzymes, it is not necessary to sequence the construct for each assembly. The high fidelity and specificity of restriction enzyme ensures the accuracy of the pYaliBrick assembly platform. The simplicity and ease of manipulation to combine and configure multiple genes in one vector makes the pYaliBrick DNA assembly method highly predictable and efficient. In addition to the 12 promoters that are built into our pYaliBrick cloning system, the constructed vectors provide a unique approach to balancing gene expression. Collectively, we were able to construct a 12 kb natural product biosynthetic pathway containing vioABCDE and all necessary gene regulatory elements into the pYaliBrick vector and demonstrated violacein production in Y. lipolytica.
3.5. Accommodating genome-editing feature into pYaliBricks
CRISPR-Cas9 mediated genome editing technique has revolutionized the field of metabolic engineering and synthetic biology (Qi et al., 2013; Wang et al., 2016). The RNA-guided DNA nuclease (Cas9) specifically targets the genomic loci that are complementary to the designed spacer (or guide) sequence adjacent to a PAM motif (protospacer adjacent motif). A crRNA (CRISPR RNA) containing the 20 bp designed protospacer sequence joins with the tracrRNA (trans-activating CRISPR RNA) to form an active scaffold and recruit Cas9 endonuclease to target the genome and induce a site-specific double-strand break (Jiang et al., 2013, Wagner and Alper, 2016). The double-strand break can be repaired either through non-homologous end joining (NHEJ) or homology-directed repair (HDR). A synthetic guide RNA (sgRNA) can be designed to contain both the crRNA and the tracrRNA for the facile delivery of guide RNA. The processing of tracrRNA and crRNA largely relies on the type III RNA polymerase and the activity of the ribonuclease P to deliver non-coding RNA. The less-characterized non-coding RNA processing system poses a potential challenge to efficiently deliver functional gRNA in Y. lipolytica (Dujon et al., 2004, Acker et al., 2008).
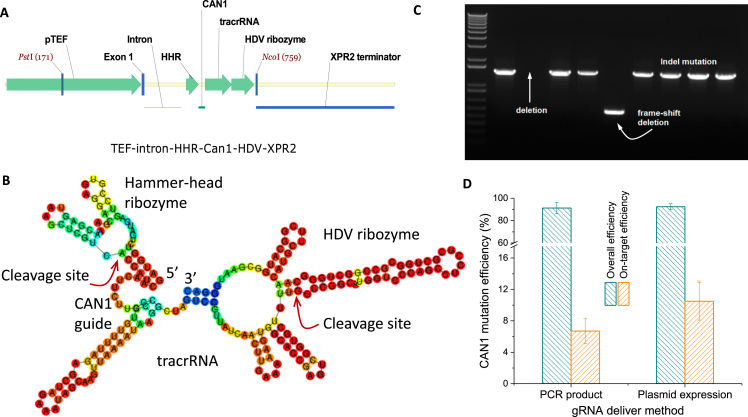
To encode gRNA into pYaliBrick vectors, we adopted the highly efficient self-cleavage ribozyme strategy (Bayer and Smolke, 2005, Gao and Zhao, 2014, Gao et al., 2016) and modified the type II RNA promoter to drive the expression of the single guide RNA (sgRNA). We chose to edit the CAN1 (arginine permease) genomic locus, as this genetic marker allows us to easily evaluate the mutation efficiency based on canavanine resistance. The sgRNA along with the tracrRNA are flanked by the hammer-head ribozyme at the 5′ end and the HDV ribozyme at the 3′ end (Fig. 5A). Functional expression of the designed gene cassette leads to the generation of hybrid ribozyme and gRNA transcripts. The two ribozymes catalyze a selective hydrolysis reaction that cleaves the linking phosphodiester to give rise to a functional sgRNA transcript (Fig. 5B).
Fig. 5.
CRISPR-Cas9 mediated genome-editing of CAN1 gene in Y. lipolytica. (A) Design of single guide RNA (sgRNA) with type II promoter (TEF) driving the expression of CAN1 single guide RNA. CAN1: gRNA that target the arginine permease; HHR: hammer-head ribozyme; tracrRNA: trans-activating CRISPR RNA. (B) Secondary structure of the transcribed sgRNA flanked by upstream hammer-head ribozyme and downstream HDV ribozyme. Red arrow indicates the self-cleavage site. (C) PCR product fragments of the mutant CAN1 analyzed by gel electrophoresis. (D) CAN1 mutation efficiency in both transient sgRNA delivery and plasmid-based sgRNA delivery. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Selective screening of yeast colonies grown on CSM-Arg plate supplemented with canavanine allowed us to easily identify canavanine resistance. PCR amplification of the mutant CAN1 led to the production of distinct gene fragment sizes, with some of the colonies completely lacking the CAN1, while others possessed a truncated form of CAN1 (Fig. 5C). Transient expression of sgRNA in linear cassette form (the exact PCR product is shown in Fig. 5A) along with a NLS-tagged Cas9 results in about 92% of colonies that were resistant to canavanine. Plasmid-based delivery of sgRNA was also tested and revealed no significant difference in the mutation rate compared to the transient expression of sgRNA (Fig. 5D). PCR product of the mutant CAN1 was later sequenced to verify the exact mutation across the CAN1 genomic locus. Our sequencing results indicate plasmid-based gRNA delivery yields about 12.5% on-target indel mutation, compared to the 7.2% on-target indel mutation in the transient expression (Fig. 5D). We believe that the low on-target indel mutation arose from the NHEJ (non-homologous end joining) repair mechanism, which is dominant in Y. lipolytica. Interestingly, we also observed complete deletion and partial deletion of CAN1 in the mutants (Fig. 5C), indicating the complexity of the DSB repair mechanism in this yeast.
4. Conclusion
The NanoLuc luciferase assay is a simple and efficient reporting system for gene expression analysis. It also has the potential of being integrated into an automated workflow to vastly improve its throughput, and with its rapid optical detection method, this assay makes an indispensable tool for characterizing genetic parts in large-scale studies. In the context of genetic toolbox development in model organisms, we have established a reliable reporter system and opened the doors for a biosensor-based, top-down approach to pathway engineering and genetic circuits evaluation in Y. lipolytica. Given the complexity of eukaryotic gene expression, this reporter system will facilitate the characterization of novel genetic elements, as well as the elucidation of unique regulatory mechanisms.
In addition, the 12 endogenous promoters characterized here have expanded our library of genetic parts for fine-tuning gene expression. Positional effects and gene configurations were also examined in the context of the prokaryotic operon, pseudo-operon, and monocistronic gene arrangements to elucidate how genetic structural variation could lead to distinct gene expression patterns. Using these three genetic configurations, we proposed a novel strategy to generate functional operon-type gene expression through intron alternative-splicing mechanism. We studied the positional effects and discovered the mutual transcriptional inhibition among the adjacent gene cassette. This invention facilitates the control and optimization of gene expression at the proper level while avoiding the need to modify the complex transcriptional machinery. The five-gene pathway for the pigmented compound violacein was rapidly assembled in Y. lipolytica using the self-replicating YaliBrick vectors. Applying the gene expression-tuning strategies outlined in this paper, a high-producing violacein-producing strain was quickly identified via visual screening, and titers were subsequently quantified with HPLC. Incorporating the highly efficient self-cleavage ribozyme allows us to generate functional single guide RNA (sgRNA) and introduce site-specific indel mutations and frame-shift mutations, paving the way for further genome-editing optimization in Y. lipolytica. Taken together, the tools and strategies developed in this work can streamline the genetic manipulation of Y. lipolytica and accelerate the construction of multiple gene pathways for diverse industrial applications. Our work provides an efficient and versatile toolkit to upgrade Y. lipolytica as a powerful workhorse for natural product and oleochemical production.
Funding
The authors would like to acknowledge the Department of Chemical, Biochemical and Environmental Engineering, College of Engineering and Information Technology, Office of the Vice President for Research at University of Maryland Baltimore County for funding support.
Acknowledgments
Conflict of interest
None declared.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2017.09.001.
Appendix A. Supplementary material
Supplementary material
References
- Abghari A., Chen S. Yarrowia lipolytica as an oleaginous cell factory platform for the production of fatty acid-based biofuel and bioproducts. Front. Energy Res. 2014:2. [Google Scholar]
- Acker J., Ozanne C., Kachouri-Lafond R., Gaillardin C., Neuvéglise C., Marck C. Dicistronic tRNA–5S rRNA genes in Yarrowia lipolytica: an alternative TFIIIA-independent way for expression of 5S rRNA genes. Nucleic Acids Res. 2008;36(18):5832–5844. doi: 10.1093/nar/gkn549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T.S., Smolke C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotech. 2005;23(3):337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- Beckerich J.M., Boisramé-Baudevin A., Gaillardin C. Yarrowia lipolytica: a model organism for protein secretion studies. Int. Microbiol. 1998;1 [PubMed] [Google Scholar]
- Beopoulos A., Chardot T., Nicaud J.M. Yarrowia lipolytica: a model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie. 2009;91(6):692–696. doi: 10.1016/j.biochi.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Blanchin-Roland S., Cordero Otero R.R., Gaillardin C. Two upstream activation sequences control the expression of the XPR2 gene in the yeast Yarrowia lipolytica. Mol. Cell. Biol. 1994;14(1):327–338. doi: 10.1128/mcb.14.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazeck J., Hill A., Liu L., Knight R., Miller J., Pan A., Otoupal P., Alper H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014;5:3131. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- Blazeck J., Liu L., Redden H., Alper H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl. Environ. Microbiol. 2011;77(22):7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T. Gene clusters and polycistronic transcription in eukaryotes. Bioessays. 1998;20(6):480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Braga A., Belo I. Biotechnological production of γ-decalactone, a peach like aroma, by Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2016;32(10):169. doi: 10.1007/s11274-016-2116-2. [DOI] [PubMed] [Google Scholar]
- Cao X., Lv Y.-B., Chen J., Imanaka T., Wei L.-J., Hua Q. Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction. Biotechnol. Biofuels. 2016;9(1):214. doi: 10.1186/s13068-016-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.C., Beckerich J.M., Gaillardin C. One-step transformation of the dimorphic yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 1997;48(2):232–235. doi: 10.1007/s002530051043. [DOI] [PubMed] [Google Scholar]
- Christen S., Sauer U. Intracellular characterization of aerobic glucose metabolism in seven yeast species by 13C flux analysis and metabolomics. FEMS Yeast Res. 2011;11(3):263–272. doi: 10.1111/j.1567-1364.2010.00713.x. [DOI] [PubMed] [Google Scholar]
- Cui Z., Gao C., Li J., Hou J., Lin C.S.K., Qi Q. Engineering of unconventional yeast Yarrowia lipolytica for efficient succinic acid production from glycerol at low pH. Metab. Eng. 2017;42:126–133. doi: 10.1016/j.ymben.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., Montigny J., Marck C., Neuveglise C., Talla E. Genome evolution in yeasts. Nature. 2004;430 [Google Scholar]
- Dulermo R., Brunel F., Dulermo T., Ledesma-Amaro R., Vion J., Trassaert M., Thomas S., Nicaud J.-M., Leplat C. Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microbial Cell Fac. 2017;16:31. doi: 10.1186/s12934-017-0647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán N., Justo G.Z., Durán M., Brocchi M., Cordi L., Tasic L., Castro G.R., Nakazato G. Advances in Chromobacterium violaceum and properties of violacein-Its main secondary metabolite: a review. Biotechnol. Adv. 2016;34(5):1030–1045. doi: 10.1016/j.biotechadv.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Fickers P., Benetti P.H., Waché Y., Marty A., Mauersberger S., Smit M.S., Nicaud J.M. Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005;5(6-7):527–543. doi: 10.1016/j.femsyr.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Fickers P., Le Dall M.T., Gaillardin C., Thonart P., Nicaud J.M. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods. 2003;55(3):727–737. doi: 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Gao S., Tong Y., Wen Z., Zhu L., Ge M., Chen D., Jiang Y., Yang S. Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J. Ind. Microbiol. Biotechnol. 2016;43(8):1085–1093. doi: 10.1007/s10295-016-1789-8. [DOI] [PubMed] [Google Scholar]
- Gao Y., Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integ. Plant Biol. 2014;56(4):343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6 doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gomes N., Teixeira J.A., Belo I. Fed-batch versus batch cultures of Yarrowia lipolytica for γ-decalactone production from methyl ricinoleate. Biotechnol. Lett. 2012;34(4):649–654. doi: 10.1007/s10529-011-0824-0. [DOI] [PubMed] [Google Scholar]
- Groenewald M., Boekhout T., Neuvéglise C., Gaillardin C., van Dijck P.W.M., Wyss M. Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 2014;40(3):187–206. doi: 10.3109/1040841X.2013.770386. [DOI] [PubMed] [Google Scholar]
- Haugwitz M., Nourzaie O., Garachtchenko T., Hu L., Gandlur S., Olsen C., Farmer A., Chaga G., Sagawa H. Multiplexing bioluminescent and fluorescent reporters to monitor live cells. Curr. Chem. Genom. 2008;1:11–19. doi: 10.2174/1875397300801010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S., Asamizu S., Onaka H., Shiro Y., Nagano S. Crystal structure of VioE, a key player in the construction of the molecular skeleton of violacein. J. Biol. Chem. 2008 doi: 10.1074/jbc.M708109200. [DOI] [PubMed] [Google Scholar]
- Hong S.-P., Seip J., Walters-Pollak D., Rupert R., Jackson R., Xue Z., Zhu Q. Engineering Yarrowia lipolytica to express secretory invertase with strong FBA1IN promoter. Yeast. 2012;29(2):59–72. doi: 10.1002/yea.1917. [DOI] [PubMed] [Google Scholar]
- Hoshino T. Violacein and related tryptophan metabolites produced by Chromobacterium violaceum: biosynthetic mechanism and pathway for construction of violacein core. Appl. Microbiol. Biotechnol. 2011;91(6):1463. doi: 10.1007/s00253-011-3468-z. [DOI] [PubMed] [Google Scholar]
- Hussain M.S., Gambill L., Smith S., Blenner M.A. Engineering promoter architecture in oleaginous yeast Yarrowia lipolytica. ACS Synth. Biol. 2016:5. doi: 10.1021/acssynbio.5b00100. [DOI] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabran P., Rossignol T., Gaillardin C., Nicaud J.M., Neuvéglise C. Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res. 2012;19(3):231–244. doi: 10.1093/dnares/dss007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar A., Otto C., Holz M., Werner S., Hübner L., Barth G. Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr. Genet. 2013;59(1):63–72. doi: 10.1007/s00294-013-0389-7. [DOI] [PubMed] [Google Scholar]
- Leal A.M. d S., de Queiroz J.D.F., de Medeiros S.R.B., Lima T.K. d S., Agnez-Lima L.F. Violacein induces cell death by triggering mitochondrial membrane hyperpolarization in vitro. BMC Microbiol. 2015;15:115. doi: 10.1186/s12866-015-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma-Amaro R., Lazar Z., Rakicka M., Guo Z., Fouchard F., Coq A.-M.C.-L., Nicaud J.-M. Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab. Eng. 2016;38:115–124. doi: 10.1016/j.ymben.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Ledesma-Amaro R., Nicaud J.-M. Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog. Lipid Res. 2016;61:40–50. doi: 10.1016/j.plipres.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Lee M.E., Aswani A., Han A.S., Tomlin C.J., Dueber J.E. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Alper H.S. Draft genome sequence of the oleaginous yeast yarrowia lipolytica PO1f, a commonly used metabolic engineering host. Genome Announc. 2014;2:4. doi: 10.1128/genomeA.00652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Otoupal P., Pan A., Alper H.S. Increasing expression level and copy number of a Yarrowia lipolytica plasmid through regulated centromere function. FEMS Yeast Res. 2014:14. doi: 10.1111/1567-1364.12201. [DOI] [PubMed] [Google Scholar]
- Liu N., Qiao K., Stephanopoulos G. 13C metabolic flux analysis of acetate conversion to lipids by yarrowia lipolytica. Metab. Eng. 2016;38:86–97. doi: 10.1016/j.ymben.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Madzak C. Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl. Microbiol. Biotechnol. 2015;99(11):4559–4577. doi: 10.1007/s00253-015-6624-z. [DOI] [PubMed] [Google Scholar]
- Masser A.E., Kandasamy G., Kaimal J.M., Andréasson C. Luciferase NanoLuc as a reporter for gene expression and protein levels in Saccharomyces cerevisiae. Yeast. 2016;33(5):191–200. doi: 10.1002/yea.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekouar M., Blanc-Lenfle I., Ozanne C., Da Silva C., Cruaud C., Wincker P., Gaillardin C., Neuvéglise C. Detection and analysis of alternative splicing in Yarrowia lipolytica reveal structural constraints facilitating nonsense-mediated decay of intron-retaining transcripts. Genome Biol. 2010;11(6) doi: 10.1186/gb-2010-11-6-r65. R65-R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L.A., Chuang J., Agmon N., Khunsriraksakul C., Phillips N.A., Cai Y., Truong D.M., Veerakumar A., Wang Y., Mayorga M., Blomquist P., Sadda P., Trueheart J., Boeke J.D. Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res. 2015;43(13):6620–6630. doi: 10.1093/nar/gkv466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Vargas A.T., Domínguez A., Ruiz-Herrera J. Identification of dimorphism-involved genes of Yarrowia lipolytica by means of microarray analysis. Res. Microbiol. 2012;163(5):378–387. doi: 10.1016/j.resmic.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Papanikolaou S., Galiotou-Panayotou M., Fakas S., Komaitis M., Aggelis G. Citric acid production by Yarrowia lipolytica cultivated on olive-mill wastewater-based media. Bioresour. Technol. 2008;99(7):2419–2428. doi: 10.1016/j.biortech.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Papanikolaou S., Muniglia L., Chevalot I., Aggelis G., Marc I. Yarrowia lipolytica as a potential producer of citric acid from raw glycerol. J. Appl. Microbiol. 2002;92(4):737–744. doi: 10.1046/j.1365-2672.2002.01577.x. [DOI] [PubMed] [Google Scholar]
- Portela R.M.C., Vogl T., Kniely C., Fischer J.E., Oliveira R., Glieder A. Synthetic core promoters as universal parts for fine-tuning expression in different yeast species. ACS Synth. Biol. 2017;6(3):471–484. doi: 10.1021/acssynbio.6b00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Lei S., Larson Matthew H., Gilbert Luke A., Doudna Jennifer A., Weissman Jonathan S., Arkin Adam P., Lim Wendell A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao K., Wasylenko T.M., Zhou K., Xu P., Stephanopoulos G. "Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism.". Nat Biotechnol. 2017;35(2):173–177. doi: 10.1038/nbt.3763. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Sentandreu R. Different effectors of dimorphism in Yarrowia lipolytica. Arch. Microbiol. 2002;178 doi: 10.1007/s00203-002-0478-3. [DOI] [PubMed] [Google Scholar]
- Schwartz C.M., Hussain M.S., Blenner M., Wheeldon I. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowialipolytica. ACS Synth. Biol. 2016;5 doi: 10.1021/acssynbio.5b00162. [DOI] [PubMed] [Google Scholar]
- Seip J., Jackson R., He H., Zhu Q., Hong S.P. Snf1 is a regulator of lipid accumulation in Yarrowia lipolytica. Appl. Environ. Microbiol. 2013;79(23):7360–7370. doi: 10.1128/AEM.02079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin K.B., Callen B.P., Egan J.B. Transcriptional interference – a crash course. Trends Genet. 2005;21(6):339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Chen Y., Siewers V., Nielsen J. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. mBio. 2014;5:3. doi: 10.1128/mBio.01130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Ji H., Siewers V., Nielsen J. Improved production of fatty acids by Saccharomyces cerevisiae through screening a cDNA library from the oleaginous yeast Yarrowia lipolytica. FEMS Yeast Res. 2016;16(1) doi: 10.1093/femsyr/fov108. fov108-fov108. [DOI] [PubMed] [Google Scholar]
- Tai M., Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Wagner J.M., Alper H.S. "Synthetic biology and molecular genetics in non-conventional yeasts: Current tools and future advances.". Fungal Genetics and Biology. 2016;89:126–136. doi: 10.1016/j.fgb.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Ann. Rev. Biochem. 2016;85(1):227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Wasylenko T.M., Ahn W.S., Stephanopoulos G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab. Eng. 2015;30:27–39. doi: 10.1016/j.ymben.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Xie D., Jackson E.N., Zhu Q. Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: from fundamental research to commercial production. Appl. Microbiol. Biotechnol. 2015;99(4):1599–1610. doi: 10.1007/s00253-014-6318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Bhan N., Koffas M.A.G. Engineering plant metabolism into microbes: from systems biology to synthetic biology. Curr. Opin. Biotechnol. 2013;24(2):291–299. doi: 10.1016/j.copbio.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Xu P., Gu Q., Wang W., Wong L., Bower A.G.W., Collins C.H., Koffas M.A.G. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 2013;4:1409. doi: 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- Xu P., Koffas M.G. Vol. 1073. Humana Press; 2013. Assembly of multi-gene pathways and combinatorial pathway libraries through ePathBrick Vectors; pp. 107–129. (Synthetic Biology). [DOI] [PubMed] [Google Scholar]
- Xu P., Li L., Zhang F., Stephanopoulos G., Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. USA. 2014;111(31):11299–11304. doi: 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Wang W., Li L., Bhan N., Zhang F., Koffas M.A.G. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli. ACS Chem. Biol. 2014;9(2):451–458. doi: 10.1021/cb400623m. [DOI] [PubMed] [Google Scholar]
- Xu P., Qiao W.S., Ahn W.S. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. USA. 2016;113(39):10848–10853. doi: 10.1073/pnas.1607295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Qiao K., Stephanopoulos G. Engineering oxidative stress defense pathways to build a robust lipid production platform in Yarrowia lipolytica. Biotechnol. Bioeng. 2017;114(7):1521–1530. doi: 10.1002/bit.26285. [DOI] [PubMed] [Google Scholar]
- Xue Z., Sharpe P.L., Hong S.P., Yadav N.S., Xie D., Short D.R., Damude H.G., Rupert R.A., Seip J.E., Wang J. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 2013:31. doi: 10.1038/nbt.2622. [DOI] [PubMed] [Google Scholar]
- Zhang B., Chen H., Li M., Gu Z., Song Y., Ratledge C., Chen Y.Q., Zhang H., Chen W. Genetic engineering of Yarrowia lipolytica for enhanced production of trans-10, cis-12 conjugated linoleic acid. Microb. Cell Fact. 2013;12(1):70. doi: 10.1186/1475-2859-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhou H., Du G., Liu L., Chen J. Screening of a thiamine-auxotrophic yeast for α-ketoglutaric acid overproduction. Lett. Appl. Microbiol. 2010;51(3):264–271. doi: 10.1111/j.1472-765X.2010.02889.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Jackson E.N. Metabolic engineering of Yarrowia lipolytica for industrial applications. Curr. Opin. Biotechnol. 2015;36 doi: 10.1016/j.copbio.2015.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material