Abstract
Carbon catabolite repression refers to the preference of microbes to metabolize certain growth substrates over others in response to a variety of regulatory mechanisms. Such preferences are important for the fitness of organisms in their natural environments, but may hinder their performance as domesticated microbial cell factories. In a Pseudomonas putida KT2440 strain engineered to convert lignin-derived aromatic monomers such as p-coumarate and ferulate to muconate, a precursor to bio-based nylon and other chemicals, metabolic intermediates including 4-hydroxybenzoate and vanillate accumulate and subsequently reduce productivity. We hypothesized that these metabolic bottlenecks may be, at least in part, the effect of carbon catabolite repression caused by glucose or acetate, more preferred substrates that must be provided to the strain for supplementary energy and cell growth. Using mass spectrometry-based proteomics, we have identified the 4-hydroxybenzoate hydroxylase, PobA, and the vanillate demethylase, VanAB, as targets of the Catabolite Repression Control (Crc) protein, a global regulator of carbon catabolite repression. By deleting the gene encoding Crc from this strain, the accumulation of 4-hydroxybenzoate and vanillate are reduced and, as a result, muconate production is enhanced. In cultures grown on glucose, the yield of muconate produced from p-coumarate after 36 h was increased nearly 70% with deletion of the gene encoding Crc (94.6 ± 0.6% vs. 56.0 ± 3.0% (mol/mol)) while the yield from ferulate after 72 h was more than doubled (28.3 ± 3.3% vs. 12.0 ± 2.3% (mol/mol)). The effect of eliminating Crc was similar in cultures grown on acetate, with the yield from p-coumarate just slightly higher in the Crc deletion strain after 24 h (47.7 ± 0.6% vs. 40.7 ± 3.6% (mol/mol)) and the yield from ferulate increased more than 60% after 72 h (16.9 ± 1.4% vs. 10.3 ± 0.1% (mol/mol)). These results are an example of the benefit that reducing carbon catabolite repression can have on conversion of complex feedstocks by microbial cell factories, a concept we posit could be broadly considered as a strategy in metabolic engineering for conversion of renewable feedstocks to value-added chemicals.
Keywords: Carbon catabolite repression, Catabolite repression control, Crc, Pseudomonas putida KT2440, ciscis-Muconate, Muconic acid, Lignin valorization
Highlights
-
•
Crc is a global regulator of carbon catabolite repression in pseudomonads.
-
•
The gene encoding Crc was deleted from muconate a producing P. putida strain.
-
•
Based on our proteomics data, expression of PobA and VanAB are regulated by Crc.
-
•
Deleting Crc improved conversion to muconate in the presence of glucose or acetate.
-
•
This may be a useful strategy toward developing pseudomonad cell factories.
1. Introduction
To successfully compete in the environmental niches they occupy, most microorganisms have developed innate preferences for certain growth substrates over others. This phenomenon has been singularly termed carbon catabolite repression (CCR), but the mechanisms that govern these preferences are as diverse as the organisms in which they have evolved (Reviewed in Görke and Stülke (2008)).
In pseudomonads, a preference for organic acids and amino acids over glucose, which is generally preferred over hydrocarbons and aromatic compounds, is imparted by a complex combination of global and operon-specific mechanisms (Reviewed in Rojo (2010)). Perhaps the most important of these mechanisms is the action of the Crc (catabolite repression control) protein, a global regulator that inhibits translation of targeted mRNAs by binding near ribosome binding sites. This binding occurs in association with another protein, Hfq, at catabolite activity (CA) sequence motifs that contain a AANAANAA core and the presence of Crc, Hfq, and the CA motif is essential for Crc regulation (Moreno et al., 2014, Moreno et al., 2009b, Sonnleitner et al., 2009, Sonnleitner et al., 2012). Crc has been shown to target catabolic pathways directly, by inhibiting translation of mRNAs encoding the enzymes themselves, and indirectly, by inhibiting translation of mRNAs encoding transcriptional regulators that drive expression of genes encoding catabolic enzymes as well as transporters required for substrates to enter the cell (Hernández-Arranz et al., 2013).
CCR is undoubtedly important for the fitness of saprophytic soil bacteria like Pseudomonas putida KT2440, which is well-suited for its native environment because of its ability to judiciously degrade a wide range of natural and xenobiotic substrates, including those derived from the three major fractions of plant biomass, namely cellulose, hemicellulose, and lignin. Lignin is a heterogeneous polymer of aromatic monomers that is an important component of the plant cell wall and can account for up to 40% of the carbon in terrestrial biomass (Ragauskas et al., 2014, Zakzeski et al., 2010). We have recently reported the development of P. putida KT2440-based biocatalysts for conversion of lignin monomers such as p-coumarate and ferulate to muconate (Fig. 1) (Johnson et al., 2016; Vardon et al., 2016, 2015), which can be converted to adipic acid (Vardon et al., 2015, Vardon et al., 2016) and diethyl terephthalate (Lu et al., 2015), precursors to the commodity plastics nylon and polyethylene terephthalate, respectively. Muconate can also be utilized directly or partially hydrogenated to produce novel materials (Rorrer et al., 2016, Rorrer et al., 2017). p-Coumarate and ferulate are common, ester-linked hydroxycinnamic acids in lignin (Vanholme et al., 2010) and represent the predominant monomeric, lignin-derived species generated by alkaline treatment of herbaceous feedstocks, such as corn stover and switchgrass, accounting for up to 40%, on a mass basis, of the solubilized lignin (Karp et al., 2014, Karp et al., 2016, Karp et al., 2015, Munson et al., 2016). The resulting liquor also contains organic acids including sugar degradation products and acetate from acetyl groups in hemicellulose. In the conversion of lignin to a product such as muconate, we envision that lignin monomers would be converted to the targeted product while other substrates, such as residual organic acids or carbohydrate-derived products, could be utilized to generate energy and carbon required for growth (Beckham et al., 2016). However, such a strategy could be problematic in light of the preference of P. putida KT2440 for organic acids and sugars over aromatic molecules such as those derived from lignin.
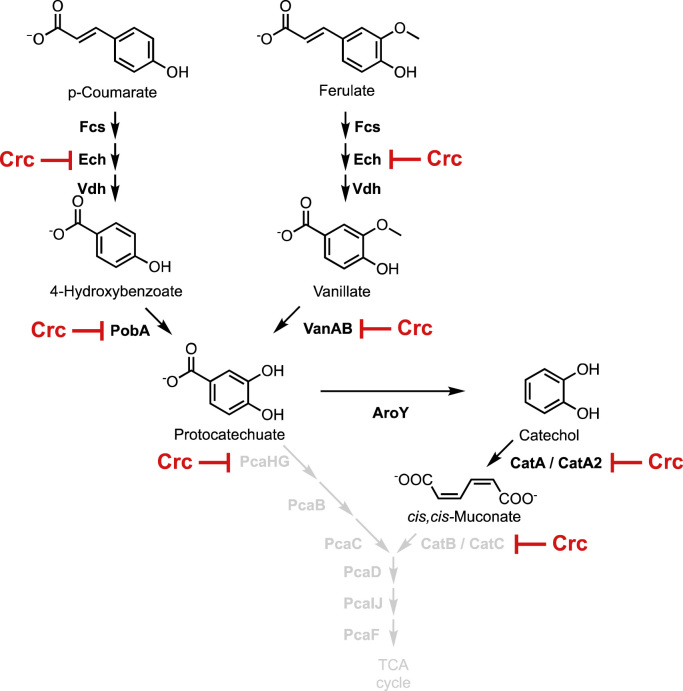
Fig. 1.
Crc regulation of metabolic pathways for production of muconate in the engineered strain, P. putida KT2440-CJ102 (Vardon et al., 2016). The lignin monomers p-coumarate and ferulate are metabolized via protocatechuate, which is redirected to catechol by deletion of pcaHG and integration of aroY, which encodes a protocatechuate decarboxylase from Enterobacter cloacae (Vardon et al., 2016, Vardon et al., 2015). Catechol then undergoes intradiol ring cleavage by action of the CatA and CatA2 dioxygenases to form cis,cis-muconate, which accumulates due to deletion of catB and catC. Putative targets of Crc regulation are shown (Browne et al., 2010, Hernández-Arranz et al., 2013, Morales et al., 2004).
During production of muconate from p-coumarate or ferulate, cultures of our engineered P. putida KT2440 strain, KT2440-CJ102, growing on glucose or acetate exhibit a marked accumulation, respectively, of 4-hydroxybenzoate (4-HBA) and vanillate. These intermediates are converted to protocatechuate (PCA), which is further metabolized, in this engineered strain, to produce muconate via catechol (Fig. 1) (Vardon et al., 2015, Vardon et al., 2016). Interestingly, both the 4-HBA hydroxylase, PobA, and the vanillate demethylase, VanAB, that convert these molecules to PCA have been identified as putative targets of Crc regulation (Browne et al., 2010, Hernández-Arranz et al., 2013, Morales et al., 2004). Here we have confirmed these predictions using mass spectrometry (MS)-based proteomics and demonstrated that deletion of the gene encoding Crc enhances metabolism of both 4-HBA and vanillate, leading to enhanced muconate production from p-coumarate or ferulate when either glucose or acetate are supplied as a source of carbon and energy.
2. Materials and methods
2.1. Deletion of crc
Construction of P. putida KT2440-CJ102 (ΔcatRBCA::Ptac:catA ΔpcaHG::Ptac:aroY) has been described previously (Vardon et al., 2016). The crc gene was deleted from P. putida KT2440-CJ102 using the antibiotic selection/sucrose counter-selection method (Blomfield et al., 1991). To construct the suicide vector for this deletion, 1000 bp targeting regions 5’ and 3’ of the crc gene (PP_5292, GenBank: AE015451.1:6039987..6040766) were amplified from P. putida KT2440 gDNA using Q5® Hot Start High-Fidelity 2X Master Mix (New England Biolabs) and primer pairs oCJ255 (5’-AACAGCTATGACATGATTACGAATTCGTAGCGTAGTGTGACTTGAAGGGCACAC-3’) & oCJ256 (5’-TTGCGGCCGCAAATGGCCCCATAAATCTCGTGCGTGTATG-3’) and oCJ257 (5’-GGGGCCATTTGCGGCCGCAAGGCCATTGGGGCTGCATTG-3’) & oCJ258 (5’-GCCTGCAGGTCGACTCTAGAGGATCCGCTTCCTCAAAGGCCAGGGC-3’), respectively, synthesized by Integrated DNA Technologies (IDT). These fragments were then assembled into pK18mobsacB (ATCC 87097) (Schäfer et al., 1994) digested with EcoRI and BamHI using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs) and transformed into NEB® 5-alpha F'Iq competent Escherichia coli (New England Biolabs) according to the manufacturer's instructions. Transformants were selected on LB (Lennox) plates containing 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, and 15 g/L agar, supplemented with 50 μg/mL kanamycin and grown at 37 °C. Clones were screened by diagnostic restriction digest and confirmed by Sanger sequencing performed by GENEWIZ, Inc.
The resulting plasmid, pCJ033, was transformed into KT2440-CJ102 by electroporation (Choi et al., 2006). Isolates in which the plasmid had recombined into the genome were selected on LB (Lennox) plates supplemented with 50 μg/mL kanamycin. Subsequent counter-selection for recombination of the plasmid out of the genome was performed on YT+25% sucrose plates containing 10 g/L yeast extract, 20 g/L tryptone, 250 g/L sucrose, and 18 g/L agar, as described previously (Johnson and Beckham, 2015). Isolates in which the crc gene was deleted were identified by amplification of a 2046 bp product (rather than the 2826 WT product) using primers oCJ261 (5’-GCCATGAATAGCTGCTCC-3’) & oCJ262 (5’-GGTCAGCGTTGAAAACGG-3’) in diagnostic colony PCR with MyTaq™ HS Red Mix (Bioline). It was also noted that isolates in which crc had been deleted formed smaller colonies relative to those expressing Crc, consistent with a lower growth rate in complete media that has been previously described and attributed to the preferential use of amino acids imparted by Crc (La Rosa et al., 2016, Moreno et al., 2009a). The resulting KT2440-CJ102-dervied strain in which crc had been deleted was designated KT2440-CJ238 (P. putida KT2440 ΔcatRBC::Ptac:catA ΔpcaHG::Ptac:aroY Δcrc).
2.2. Culture growth
To identify targets of Crc regulation by MS-based proteomics, cultures were grown shaking at 225 rpm, 30 °C in LB (Lennox) medium overnight, centrifuged, and resuspended in M9 minimal medium and these cells were used to inoculate 125 mL baffled flasks containing 25 mL M9 minimal media (pH 7.2) consisting of 6.78g/L Na2HPO4, 3g/L KH2PO4, 0.5g/L NaCl, 1g/L NH4Cl, 2 mM MgSO4, 100 μM CaCl2, and 18 μM FeSO4 supplemented with 20 mM p-coumaric acid (Sigma-Aldrich) or trans-ferulic acid (Sigma-Aldrich) and 10 mM glucose (Sigma-Aldrich) or 30 mM sodium acetate (Sigma-Aldrich) to an optical density at 600 nm (OD600) of 0.1, in triplicate. These cultures were incubated shaking at 225 rpm, 30 °C until reaching an OD600 of 0.6. The cultures were then centrifuged, the supernatants were aspirated, and the cells pellets were washed once in 1 mL sterile 1X M9 salts (6.78g/L Na2HPO4, 3g/L KH2PO4, 0.5g/L NaCl, 1g/L NH4Cl) and frozen at −80 °C until being analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) for protein identification.
Shake flask experiments to evaluate metabolism of p-coumarate and ferulate were also performed in triplicate as above, but rather than being harvested at OD600 0.6, the cultures were incubated further and sampled periodically to evaluate pH, growth by measurement of the OD600, and metabolite concentrations using high performance liquid chromatography (HPLC). Cultures grown on glucose were fed an additional 10 mM glucose every 12 h following sampling and the pH was adjusted back to 7.0 by the addition of NaOH when the pH was ≤ 6.0. Cultures grown on sodium acetate were fed an additional 30 mM sodium acetate after sampling at 12 h, at which point the pH was 7.3–7.7. After sampling every 12 h thereafter, the pH was found to have risen to 7.9–8.6 with the consumption of the acid, so 30 mM acetic acid was added to feed the cultures and to return the pH to 6.6–7.0.
2.3. Protein extraction and digestion
Cell pellets collected for proteomics by LC-MS/MS were suspended in lysis buffer (4% sodium deoxycholate (SDC) and 10 mM dithiothreitol (DTT) in 100 mM of NH4HCO3) and Halt protease inhibitor cocktail (Thermo Fischer Scientific) and then boiled for 5 min, sonically disrupted (30% amplitude, 10 s pulse with 10 s rest, 2 min total pulse time) and boiled for an additional 5 min. Crude protein extract was pre-cleared via centrifugation, and quantified by BCA assay (Pierce Biotechnology). Three milligrams of crude protein extract were placed on top of a 10 kDa cutoff spin column filter (Vivaspin 2, GE Health) and cysteines were blocked with iodoacetamide (30 mM) to prevent reformation of disulfide bonds. Samples were then centrifuged for 15 min at 4500g to remove small molecules. The protein extract was washed once with 100 mM NH4HCO3 and then suspended in 500 µL of 100 mM NH4HCO3 to adjust the final volume to 2% SDC. Proteins were digested via two aliquots of sequencing-grade trypsin (Promega, 1:50 [w:w]) at two different sample dilutions, 2% SDC (overnight) and subsequent 1% SDC (3 h). Following digestion, peptides were filtered through the 10 kDa cutoff spin column. The collected peptide mixture was adjusted to 0.5% formic acid to precipitate SDC. Hydrated ethyl acetate was added to each sample at a 1:1 [v:v] ratio three times to effectively remove SDC. Samples were then placed in a SpeedVac Concentrator (Thermo Fischer Scientific) to remove ethyl acetate and further concentrate the sample. The peptide-enriched flow through was quantified by BCA assay, desalted on RP-C18 stage tips (Pierce Biotechnology) and then stored at −80 °C.
2.4. LC–MS/MS analysis
All samples were analyzed on a Q Exactive Plus mass spectrometer (Thermo Fischer Scientific) coupled with a Proxeon EASY-nLC 1200 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated on a 75 µm inner diameter microcapillary column packed with 35 cm of Kinetex C18 resin (1.7 µm, 100 Å, Phenomenex). For each sample, a 2 μg aliquot was loaded in buffer A (0.1% formic acid, 2% acetonitrile) and eluted with a linear 60 min gradient of 2–20% of buffer B (0.1% formic acid, 80% acetonitrile), followed by an increase to 40% buffer B within 10 min and afterwards a 10 min wash at 98% buffer A. The flow rate was kept at 250 nL/min. MS data were acquired with the Thermo Xcalibur software version 4.27.19, a topN method where N could be up to 15. Target values for the full scan MS spectra were 1 × 106 charges in the 300–1500 m/z range with a maximum injection time of 25 ms. Transient times corresponding to a resolution of 70,000 at m/z 200 were chosen. A 1.6 m/z isolation window and fragmentation of precursor ions was performed by higher-energy C-trap dissociation (HCD) with a normalized collision energy of 30 eV. MS/MS scans were performed at a resolution of 17,500 at m/z 200 with an ion target value of 1 × 106 and a maximum injection time of 50 ms. Dynamic exclusion was set to 45 s to avoid repeated sequencing of peptides.
2.5. Peptide identification
MS raw data files were searched against the P. putida Uniprot FASTA database to which common contaminate proteins had been added. A decoy database, consisting of the reversed sequences of the target database, was appended in order to discern the false-discovery rate (FDR) at the spectral level. For standard database searching, the peptide fragmentation spectra (MS/MS) were searched with MyriMatch algorithm v2.2 (Tabb et al., 2007). MyriMatch was configured to derive fully-tryptic peptides with the following parameters: max 4 missed cleavages, max peptide length 75, minimum peptide length of 5 amino acids, maximum peptide mass of 10,000 Da, maximum number of charge states of 4, a precursor mass tolerance of 5 ppm (ppm), a fragment mass tolerance of 10 ppm, a static modification on cysteines (iodoacetamide; +57.0214 Da), and dynamic modifications on methionine (oxidation; 15.9949).
2.6. Protein inference and relative quantitation
Resulting peptide spectrum matches were imported, filtered and organized into protein identifications using IDPicker v.3.0 (Ma et al., 2009). Across the entire experimental dataset, proteins were required to have at least 2 distinct peptide sequences and 9 minimum spectra per protein. These filters resulted in a final peptide-level FDR < 1%.
For label-free quantification, MS1-level precursor intensities were derived from IDPicker using IDPQuantify (Chen et al., 2013). Extracted ion chromatograms were identified using the following parameters: 30 s lower and upper retention time tolerance and 10 ppm lower and upper chromatogram tolerance. Protein intensity-based values, which were calculated by summing together quantified peptides, were converted to standardized abundance values by dividing each protein abundance value by the summed sample intensity. Protein abundance values were then multiplied by a value (i.e., 108) for ease of data interpretation. The quantitative values were then log2-transformed for the purposes of the tests for differential abundance. Missing values were imputed by replacing null values with a constant value of 0.1. Using the freely available software Perseus (http://www.perseus-framework.org) (Tyanova et al., 2016), we performed pairwise t-test comparisons to identify proteins having differential abundances between conditions.
2.7. High performance liquid chromatography
HPLC analysis of samples collected during shake flask experiments to evaluate metabolism of p-coumarate and ferulate was performed using an Agilent 1100 series system equipped with a fast acid column and a diode array detector and refractive index detector as previously described (Johnson et al., 2016).
3. Results
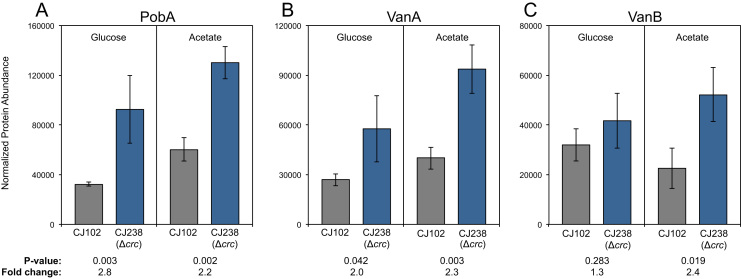
Crc regulates catabolic pathways using a multi-tier strategy targeting mRNAs that encoding transporters, transcriptional regulators, and enzymes (Hernández-Arranz et al., 2013). With respect to metabolism of 4-HBA by P. putida KT2440, transporters PcaK and PcaP as well as the hydroxylase that converts 4-HBA to protocatechuate, PobA, have been predicted to be direct targets of Crc based on the close proximity of presumptive Crc binding sites to the translation initiation sites of the mRNAs that encode these proteins. (Browne et al., 2010, Hernández-Arranz et al., 2013). PobA has also been shown to be transcriptionally regulated by Crc (Morales et al., 2004) though the nature of this regulation is unknown. In the case of vanillate, the transporter, VanP, transcriptional regulator, VanR, and one subunit of the vanillate-O-demethylase, VanA, have similarly been proposed to be direct targets of Crc (Browne et al., 2010, Hernández-Arranz et al., 2013). To determine if these proteins or other proteins in the p-coumarate and ferulate catabolic pathways are subject to Crc regulation in the presence of more preferred substrates glucose and acetate in P. putida KT2440-CJ102, which has been engineered to produce muconate (Vardon et al., 2016), we deleted the crc gene from this strain, generating KT2440-CJ238. These strains were then grown in minimal media containing either glucose or acetate and p-coumarate or ferulate and subjected to MS-based proteomics. Most of the transporters and transcriptional regulators were not identified in these experiments (Table S1, Fig. S1), likely due to low expression or membrane localization that make them less suited for evaluation using standard proteomic methods, or because the genes encoding them had been deleted in this strain to enable muconate production (CatB, CatC, PcaHG). Similarly, PobA, VanA, and VanB were either not identified or detected at very low levels in conditions in which they would not be expected to be expressed—in the presence of ferulate for PobA and in the presence of p-coumarate for VanA and VanB. PobA, however, was clearly expressed at higher levels in the strain lacking Crc, KT2440-CJ239, relative to the parent strain, KT2440-CJ102, when grown in media containing glucose or acetate and p-coumarate (Fig. 2A). Similarly, VanA was more abundant in the Δcrc strain when grown in the presence of glucose or acetate and ferulate (Fig. 2B). Expression of VanB appeared to be regulated similarly to VanA, but the difference was not statistically significant in the glucose condition (Fig. 2C). Together, these results clearly demonstrate that expression of PobA and VanAB is subject to regulation by Crc. As such, we hypothesized that deletion of Crc might enhance the conversion of both p-coumarate and ferulate to muconate in the presence of glucose or acetate.
Fig. 2.
Normalized protein abundances of PobA, VanA, and VanB as measured by MS-based proteomics. Cultures were grown in M9 minimal medium containing p-coumarate (for PobA) or ferulate (for VanA and VanB) and either glucose or acetate. Cell pellets were lysed and the proteins were digested with trypsin and subjected to analysis by LC-MS/MS. Each value represents the average of three biological replicates. The error bars represent standard deviation of the measurements. (A) Normalized abundances of PobA, (B) Normalized abundances of VanA, (B) Normalized abundances of VanB.
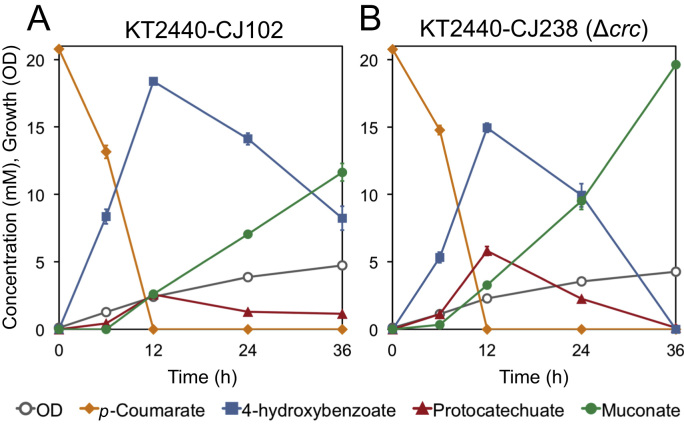
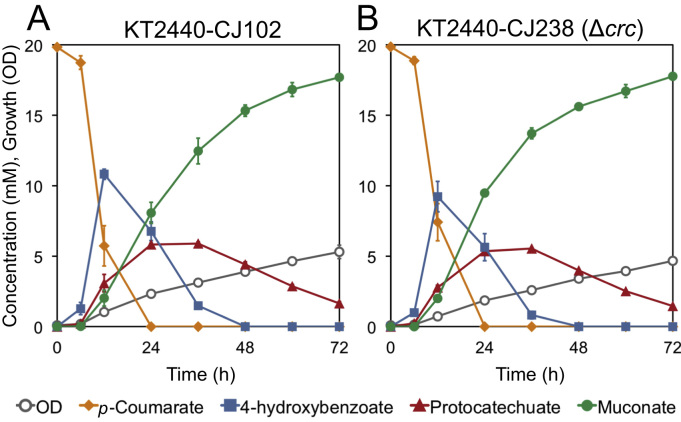
To test this hypothesis, we compared the performance of the Δcrc to its parent in shake flask experiments. We first examined the effect of crc deletion on conversion of 20 mM p-coumarate to muconate in the presence of glucose fed periodically to provide energy and carbon for growth. The deletion of crc in KT2440-CJ238 reduced the accumulation of the intermediate 4-HBA and subsequently increased muconate production, which reached a yield of 94.6 ± 0.6% (mol/mol) after 36 h, at which point the parent strain, KT2440-CJ102 yielded only 56.0 ± 3.0% (mol/mol) (Fig. 3). More rapid metabolism of 4-HBA also resulted in greater accumulation of PCA, a bottleneck Sonoki et al. (2014) described a means of overcoming previously, which we have demonstrated to be quite effective in P. putida KT2440-derived strains engineered to produce muconate (Johnson et al., 2016). The metabolic engineering strategies to overcome the PCA bottleneck were not incorporated in the strains examined here; the combination of these strains along with additional metabolic improvements will be reported in a forthcoming study reporting titer, rate, and yield optimization of muconate production in bioreactors.
Fig. 3.
Shake-flask evaluation of the effect of crc deletion on muconate production from p-coumarate by engineered P. putida KT2440 strains in the presence of glucose. Cultures were grown in M9 minimal medium containing p-coumarate and fed glucose periodically as a source of carbon and energy for growth and sampled to evaluate culture growth by OD600 and the concentration of metabolites in the medium by HPLC. Each value represents the average of three biological replicates. The error bars represent standard deviation of the measurements. (A) KT2440-CJ102, (B) KT2440-CJ238, the Δcrc derivative of KT2440-CJ102.
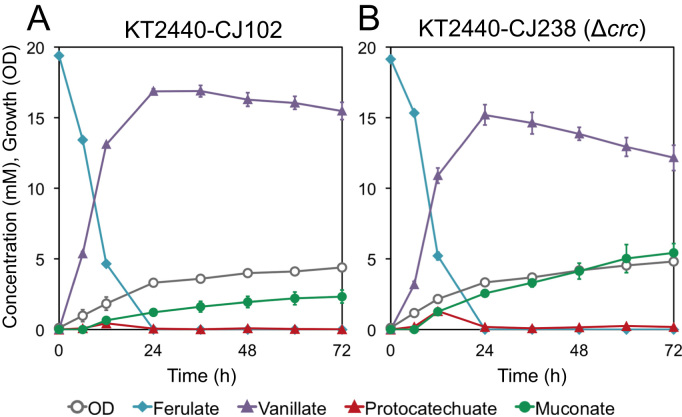
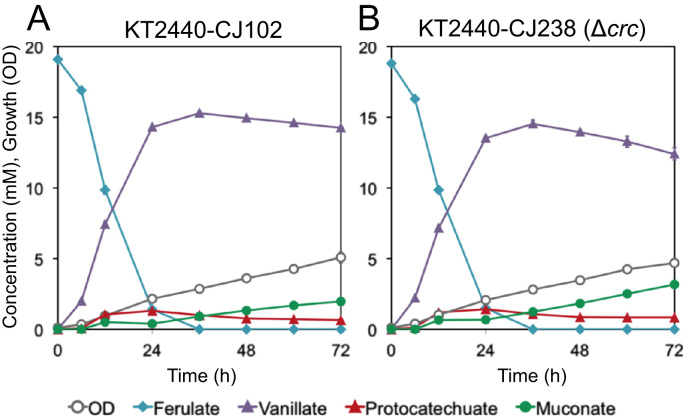
We then evaluated this strain in a similar shake flask experiment, feeding ferulate as a substrate for conversion to muconate in the presence of glucose. As with p-coumarate, we found that the conversion of ferulate to muconate was enhanced with deletion of crc as a result of more rapid metabolism of an intermediate, in this case vanillate, resulting in a 28.3 ± 3.3% (mol/mol) yield of muconate by KT2440-CJ238 after 72 h, compared to just 12.0 ± 2.3% (mol/mol) in the case of KT2440-CJ102 (Fig. 4).
Fig. 4.
Shake-flask evaluation of the effect of crc deletion on muconate production from ferulate by engineered P. putida KT2440 strains in the presence of glucose. Cultures were grown in M9 minimal medium containing p-coumarate and fed glucose periodically as a source of carbon and energy for growth and sampled to evaluate culture growth by OD600 and the concentration of metabolites in the medium by HPLC. Each value represents the average of three biological replicates. The error bars represent standard deviation of the measurements. (A) KT2440-CJ102, (B) KT2440-CJ238, the Δcrc derivative of KT2440-CJ102.
We next examined the effect of crc deletion on conversion of p-coumarate and ferulate to muconate in the presence of acetate, which represents an alternative to glucose as a cost-effective substrate that could be provided for energy and growth to P. putida strains engineered to produce muconate from aromatic molecules. We observed similar effects of crc deletion on conversion of both p-coumarate (Fig. 5) and ferulate (Fig. 6) to muconate when acetate was fed, although the effects were less pronounced than observed with glucose, perhaps due to differences in the induction of Crc by glucose and acetate and/or the difference in reducing power generated during their catabolism. KT2440-CJ238 yielded 47.7 ± 0.6% (mol/mol) from p-coumarate after 24 h compared to 40.7 ± 3.6% (mol/mol) by KT2440-CJ102 due to reduced accumulation of 4-HBA (Fig. 5), although yields were roughly equivalent later because of a buildup of PCA in both cultures. From ferulate, KT2440-CJ238 ultimately yielded 16.9 ± 1.4% (mol/mol) after 72 h compared to 10.3 ± 0.1% by KT244-CJ102 due to more rapid metabolism of the vanillate (Fig. 6).
Fig. 5.
Shake-flask evaluation of the effect of crc deletion on muconate production from p-coumarate by engineered P. putida KT2440 strains in the presence of acetate. Cultures were grown in M9 minimal medium containing p-coumarate and fed acetate periodically as a source of carbon and energy for growth and sampled to evaluate culture growth by OD600 and the concentration of metabolites in the medium by HPLC. Each value represents the average of three biological replicates. The error bars represent standard deviation of the measurements. (A) KT2440-CJ102, (B) KT2440-CJ238, the Δcrc derivative on KT2440-CJ102.
Fig. 6.
Shake-flask evaluation of the effect of crc deletion on muconate production from ferulate by engineered P. putida KT2440 strains in the presence of acetate. Cultures were grown in M9 minimal medium containing ferulate and fed acetate periodically as a source of carbon and energy for growth and sampled to evaluate culture growth by OD600 and the concentration of metabolites in the medium by HPLC. Each value represents the average of three biological replicates. The error bars represent standard deviation of the measurements. (A) KT2440-CJ102, (B) KT2440-CJ238, the Δcrc derivative on KT2440-CJ102.
4. Discussion and conclusions
The impact that microbial production processes have had on humanity is a testament to the power and diversity of microbial metabolism and physiology. While many traits that have enabled the biological success of microbes in their native environments are undoubtedly central to such processes, the elimination of superfluous cellular processes could be beneficial to modern microbial cell factories, but must be weighed carefully. To this end, one approach has been to reduce the size of the genome. The recent report of the design and synthesis of a dramatically minimized
Mycoplasma mycoides genome found that there was ultimately a tradeoff between genome size and growth rate (Hutchison et al., 2016) and similar effects have been observed upon genome reduction in E. coli (Kurokawa et al., 2016). Genome reduction may also eliminate other traits that are beneficial both in nature and in biotechnological applications. Successfully enhancing microbial cell factories by eliminating unnecessary features of a genome without introducing deleterious effects has been demonstrated, though. Most notably with regard to P. putida KT2440, it was shown that deletion of 4.3% of the genome encoding 300 genes, including those required for the flagellar machinery, reduced growth lag and sensitivity to oxidative stress while increasing growth rate, biomass yield (Martínez-García et al., 2014), and heterologous gene expression (Lieder et al., 2015).
To maximize yield and productivity, the ideal biocatalyst would rapidly and simultaneously metabolize any and all carbon sources with which it is provided. CCR is generally inconsistent with this ideal and is of special concern if the aim is to utilize complex, heterogeneous feedstocks such as those often derived from biomass. With this in mind, here we have demonstrated that reducing CCR in P. putida KT2440 is beneficial for the conversion of less preferred substrates in the presence of substrates that are more preferred. The metabolism of both p-coumarate and ferulate were enhanced, leading to a subsequent increase in muconate production in the presence of glucose and acetate, co-feeds that are necessary for growth but, based on the data presented here, clearly induce Crc-mediated regulation that is detrimental to the conversion of these lignin monomers. Based on the presence of the AANAANAA Crc-binding motif near their translation initiating ATGs, both PobA and VanA, as well as several transcriptional regulators (VanR) and transporters (VanP, PcaK, PcaP) that could affect expression and activity of PobA and VanAB, have been proposed as putative targets of Crc repression (Browne et al., 2010, Hernández-Arranz et al., 2013). pobA has also been demonstrated to be transcriptionally regulated by Crc (Morales et al., 2004). In the present study, we have confirmed the regulation of PobA and VanAB by Crc using MS-based proteomics (Fig. 2). We were unable to identify the transcriptional regulators or transporters involved in the metabolism of 4-HBA or vanillate with the exception of the transcriptional regulator VanR, the expression of which did not obviously reflect regulation by Crc (Table S1, Fig. S1). However, we believe PobA and VanAB are likely subject to multi-tier regulation by Crc as proposed by Hernández-Arranz et al. (2013), the details of which could not be elucidated in the present study based on the conditions examined and methods employed. Similarly, we are unable to confirm or refute other proposed targets of Crc in the relevant pathways (Fig. 1) based on our data (Table S1, Fig. S1). In that light, it is striking that of proteins involved in these pathways that we were able to quantify in our proteomics data (Table S1, Fig. S1), PobA, VanA, and VanB exhibit some of the clearest and most substantial regulation by Crc, which could suggest that these proteins represent the primary targets by which Crc regulates the pathways for catabolism of p-coumarate and ferulate. Interestingly, PobA and VanAB are the only enzymes in these pathways that require reducing equivalents for their activity. Because the purpose of carbon catabolite repression is to metabolize substrates judiciously based on the carbon and energy they yield, it might be beneficial to regulate these enzymes more tightly.
It should also be noted that while we believe the increase in muconate demonstrated here is attributed to de-repression of 4-HBA and vanillate metabolism, it is likely that other enzymes in these pathways may have also been de-repressed. The effect of de-repressing other enzymes, however, could be masked in the presence of larger bottlenecks such as those caused by PobA and VanAB, but may become relevant if further engineering is performed to remove these bottlenecks, an endeavor we are currently pursuing. CatB, CatC, and PcaHG have been demonstrated to be targets of Crc by RT-PCR (Morales et al., 2004), but the genes that encode these enzymes are deleted in the strains examined here to enable muconate production, so any impact of this regulation can not be evaluated using these strains. Thus, the full effect of global de-repression may be difficult to fully appreciate in any single experiment or condition, including those examined here. While the enhancement in muconate production with the deletion of crc is relatively modest in the shake flask experiments demonstrated here, such effects could be substantial in bioreactor cultivations. We believe it is worthwhile to consider whether the elimination of CCR might be beneficial in other biocatalytic systems. Indeed, many attempts to eliminate or rewire CCR in other systems have been described (Reviewed in Vinuselvi et al. (2012)). While some of these have been successful, impaired growth resulting from metabolic de-optimization and/or the crosstalk between CCR and other cellular physiology is a pervasive complication. One of the unique aspects of Crc in pseudomonads is its apparent lack of involvement in other cellular processes critical for growth; as we have observed here and others have shown previously, growth on substrates such as glucose, succinate, acetate or benzoate is only marginally affected in mutants lacking Crc (Hernández-Arranz et al., 2013, La Rosa et al., 2015, Moreno et al., 2014, Sonnleitner et al., 2012). In addition, deletion of crc was shown to double the amount of ATP and NADPH in strains grown on glucose and succinate by redirecting flux through central metabolism (La Rosa et al., 2015). These findings and those described here make the deletion of crc an attractive target for those interested in engineering pseudomonads for efficient biocatalysis.
Acknowledgements
The authors wish to acknowledge Dr. Eduardo Díaz for helpful conversations regarding Crc regulation. We thank the U.S. Department of Energy Bioenergy Technologies Office for funding this work via Contract No. DE-AC36-08GO28308 with the National Renewable Energy Laboratory. CWJ, PEA, RLH, and GTB, acknowledge funding for the proteomics analyses from the BioEnergy Science Center, a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research, Genome Sciences program, in the DOE Office of Science. The U.S. Government retains and the publisher, by accepting the article for publication, acknowledges that the U.S. Government retains a nonexclusive, paid up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for U.S. Government purposes.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2017.05.002.
Appendix A. Supplementary material
Supplementary material
.
References
- Beckham G.T., Johnson C.W., Karp E.M., Salvachúa D., Vardon D.R. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016;42:40–53. doi: 10.1016/j.copbio.2016.02.030. [DOI] [PubMed] [Google Scholar]
- Blomfield I.C., Vaughn V., Rest R.F., Eisenstein B.I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- Browne P., Barret M., O'Gara F., Morrissey J.P. Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria. BMC Microbiol. 2010;10:300. doi: 10.1186/1471-2180-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-Y., Chambers M.C., Li M., Ham A.-J.L., Turner J.L., Zhang B., Tabb D.L. IDPQuantify: combining precursor intensity with spectral counts for protein and peptide quantification. J. Proteome Res. 2013;12:4111–4121. doi: 10.1021/pr400438q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-H., Kumar A., Schweizer H.P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Görke B., Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Hernández-Arranz S., Moreno R., Rojo F. The translational repressor Crc controls the Pseudomonas putida benzoate and alkane catabolic pathways using a multi-tier regulation strategy. Environ. Microbiol. 2013;15:227–241. doi: 10.1111/j.1462-2920.2012.02863.x. [DOI] [PubMed] [Google Scholar]
- Hutchison C.A., Chuang R.-Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L., Pelletier J.F., Qi Z.-Q., Richter R.A., Strychalski E.A., Sun L., Suzuki Y., Tsvetanova B., Deerinck K.S., Smith H.O., Glass J.I., Merryman C., Gibson D.G., Venter J.C. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Beckham G.T. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab. Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Salvachúa D., Khanna P., Smith H., Peterson D.J., Beckham G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016;3:111–119. doi: 10.1016/j.meteno.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp E.M., Donohoe B.S., O’Brien M.H., Ciesielski P.N., Mittal A., Biddy M.J., Beckham G.T. Alkaline pretreatment of corn stover: bench-scale fractionation and stream characterization. ACS Sustain. Chem. Eng. 2014;2:1481–1491. [Google Scholar]
- Karp E.M., Nimlos C.T., Deutch S., Salvachúa D. Quantification of acidic compounds in complex biomass-derived streams. Green Chem. 2016;18:4750–4760. [Google Scholar]
- Karp E.M., Resch M.G., Donohoe B.S., Ciesielski P.N., O’Brien M.H., Nill J.E., Mittal A., Biddy M.J., Beckham G.T. Alkaline pretreatment of switchgrass. ACS Sustain. Chem. Eng. 2015;3:1479–1491. [Google Scholar]
- Kurokawa M., Seno S., Matsuda H., Ying B.-W. Correlation between genome reduction and bacterial growth. DNA Res. 2016 doi: 10.1093/dnares/dsw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa R., Behrends V., Williams H.D., Bundy J.G., Rojo F. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ. Microbiol. 2016;18:807–818. doi: 10.1111/1462-2920.13126. [DOI] [PubMed] [Google Scholar]
- La Rosa R., Nogales J., Rojo F. The Crc/CrcZ-CrcY global regulatory system helps the integration of gluconeogenic and glycolytic metabolism in Pseudomonas putida. Environ. Microbiol. 2015;17:3362–3378. doi: 10.1111/1462-2920.12812. [DOI] [PubMed] [Google Scholar]
- Lieder S., Nikel P.I., de Lorenzo V., Takors R. Genome reduction boosts heterologous gene expression in Pseudomonas putida. Microb. Cell Fact. 2015;14:23. doi: 10.1186/s12934-015-0207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Lu F., Chen J., Yu W., Huang Q., Zhang J., Xu J. Production of diethyl terephthalate from biomass-derived muconic acid. Angew. Chem. Int. Ed. 2015;55:249–253. doi: 10.1002/anie.201509149. [DOI] [PubMed] [Google Scholar]
- Ma Z.-Q., Dasari S., Chambers M.C., Litton M.D., Sobecki S.M., Zimmerman L.J., Halvey P.J., Schilling B., Drake P.M., Gibson B.W., Tabb D.L. IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García E., Nikel P.I., Aparicio T., de Lorenzo V. Pseudomonas 2.0: genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Fact. 2014;13:159. doi: 10.1186/s12934-014-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales G., Linares J.F., Beloso A., Albar J.P., Martínez J.L., Rojo F. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 2004;186:1337–1344. doi: 10.1128/JB.186.5.1337-1344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R., Hernández-Arranz S., La Rosa R., Yuste L., Madhushani A., Shingler V., Rojo F. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ. Microbiol. 2014;17:105–118. doi: 10.1111/1462-2920.12499. [DOI] [PubMed] [Google Scholar]
- Moreno R., Martínez-Gomariz M., Yuste L., Gil C., Rojo F. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics. 2009;9:2910–2928. doi: 10.1002/pmic.200800918. [DOI] [PubMed] [Google Scholar]
- Moreno R., Marzi S., Romby P., Rojo F. The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res. 2009;37:7678–7690. doi: 10.1093/nar/gkp825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M.S., Karp E.M., Nimlos C.T., Salit M., Beckham G.T. Gradient elution moving boundary electrophoresis enables rapid analysis of acids in complex biomass-derived streams. ACS Sustain. Chem. Eng. 2016;4:7175–7185. [Google Scholar]
- Ragauskas A.J., Beckham G.T., Biddy M.J., Chandra R., Chen F., Davis M.F., Davison B.H., Dixon R.A., Gilna P., Keller M., Langan P., Naskar A.K., Saddler J.N., Tschaplinski T.J., Tuskan G.A., Wyman C.E. Lignin valorization: improving lignin processing in the biorefinery. Science. 2014;344 doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- Rojo F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 2010:1–27. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- Rorrer N.A., Dorgan J.R., Vardon D.R., Martinez C.R., Yang Y., Beckham G.T. Renewable unsaturated polyesters from muconic acid. ACS Sustain. Chem. Eng. 2016;4:6867–6876. [Google Scholar]
- Rorrer N.A., Vardon D.R., Dorgan J.R., Gjersing E.J., Beckham G.T. Biomass-derived monomers for performance-differentiated fiber reinforced polymer composites. Green Chem. 2017;311 [Google Scholar]
- Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E., Abdou L., Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2009;106:21866–21871. doi: 10.1073/pnas.pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E., Valentini M., Wenner N., Haichar F.E.Z., Haas D., Lapouge K. Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoki T., Morooka M., Sakamoto K., Otsuka Y., Nakamura M., Jellison J., Goodell B. Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J. Biotechnol. 2014;192:71–77. doi: 10.1016/j.jbiotec.2014.10.027. [DOI] [PubMed] [Google Scholar]
- Tabb D.L., Fernando C.G., Chambers M.C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardon D.R., Franden M.A., Johnson C.W. Adipic acid production from lignin. Energy Environ. Sci. 2015;8:617–628. [Google Scholar]
- Vardon D.R., Rorrer N.A., Salvachúa D., Settle A.E., Johnson C.W., Menart M.J., Cleveland N.S., Ciesielski P.N., Steirer K.X., Dorgan J.R., Beckham G.T. cis, cis-Muconic acid: separation and catalysis to bio-adipic acid for nylon-6, 6 polymerization. Green Chem. 2016 [Google Scholar]
- Vinuselvi P., Kim M.-K., Lee S.-K., Ghim C.-M. Rewiring carbon catabolite repression for microbial cell factory. BMB Rep. 2012;45:59–70. doi: 10.5483/BMBRep.2012.45.2.59. [DOI] [PubMed] [Google Scholar]
- Zakzeski J., Bruijnincx P.C.A., Jongerius A.L., Weckhuysen B.M. The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 2010;110:3552–3599. doi: 10.1021/cr900354u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material