Abstract
Isobutanol is a flammable compound that can be used as a biofuel due to its high energy density and suitable physical and chemical properties. In this study, we examined the capacity of engineered strains of Synechocystis PCC 6803 containing the α-ketoisovalerate decarboxylase from Lactococcus lactis and different heterologous and endogenous alcohol dehydrogenases (ADH) for isobutanol production. A strain expressing an introduced kivd without any additional copy of ADH produced 3 mg L−1 OD750−1 isobutanol in 6 days. After the cultures were supplemented with external addition of isobutyraldehyde, the substrate for ADH, 60.8 mg L−1 isobutanol was produced after 24 h when OD750 was 0.8. The in vivo activities of four different ADHs, two heterologous and two putative endogenous in Synechocystis, were examined and the Synechocystis endogenous ADH encoded by slr1192 showed the highest efficiency for isobutanol production. Furthermore, the strain overexpressing the isobutanol pathway on a self-replicating vector with the strong Ptrc promoter showed significantly higher gene expression and isobutanol production compared to the corresponding strains expressing the same operon introduced on the genome. Hence, this study demonstrates that Synechocystis endogenous AHDs have a high capacity for isobutanol production, and identifies kivd encoded α-ketoisovalerate decarboxylase as one of the likely bottlenecks for further isobutanol production.
Keywords: Cyanobacteria, Alcohol dehydrogenase, α-ketoisovalerate decarboxylase, Synechocystis PCC 6803, Isobutanol production
Highlights
-
•
Expression of kivd is the only requirement for isobutanol synthesis in Synechocystis.
-
•
ADH encoded by slr1192 is important for isobutanol production in Synechocystis.
-
•
Kivd is identified as one of the likely bottleneck for isobutanol production.
1. Introduction
CO2 emissions from the use of fossil fuels has increased significantly since 1900 and has caused an urgent demand for renewable energy alternatives (Hewitson et al., 2014). During the last decades, a lot of research has been focused on identifying environmentally sustainable methods to produce renewable biofuels for replacing traditional fossil fuels (Machado and Atsumi, 2012, Miao et al., 2017). Isobutanol is a strong candidate to be used as an alternative biofuel due to its high energy content (98% of energy content in gasoline) and lower vapor pressure compared to ethanol (Sheehan, 2009). Moreover, isobutanol has lower O2 content and lower water solubility than ethanol, which means more isobutanol can be blended into gasoline while still maintaining a low O2 content in the final product. Hence, isobutanol is an efficient and safe fuel to be used in internal combustion engines (Sheehan, 2009).
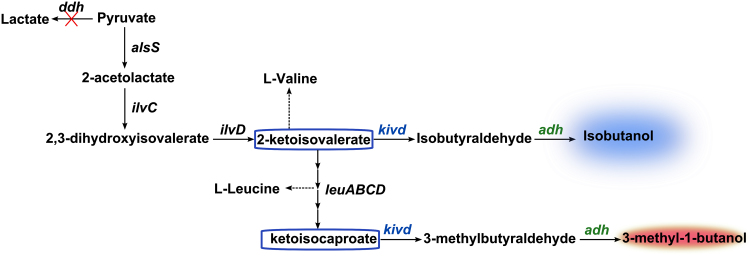
The most commonly used pathway for isobutanol biosynthesis is the 2-keto acid pathway, which shares precursor (α-ketoisovalerate) with l-valine biosynthesis pathway (Fig. 1). α-ketoisovalerate decarboxylase (Kivd) from Lactococcus lactis (L. lactis), which decarboxylates the α-keto acids to aldehydes, is an important enzyme in the 2-keto acid pathway and has been used in Escherichia coli (E. coli) (Atsumi et al., 2008; Trinh et al., 2011; Atsumi et al., 2009a; Smith and Liao, 2011; Desai et al., 2015; Liu et al., 2016), Saccharomyces cerevisiae (S. cerevisiae) (Brat and Boles, 2013; Chen et al., 2011; Kondo et al., 2012; Yuan et al., 2017), Corynebacterium glutamicum (C. glutamicum) (Smith et al., 2010, Blombach et al., 2011) and cyanobacteria for isobutyraldehyde and isobutanol production (Atsumi et al., 2009, Varman et al., 2013, Li et al., 2014) (Table 1). This enzyme was characterized as a homo-tetramer, its optimal activity was observed at 45 °C (pH 6.5) and it has a high specific activity for α-ketoisovalerate (Plaza et al., 2004). Moreover, Kivd has been engineered to utilize larger substrate in order to produce longer chain alcohols (Zhang et al., 2008, Marcheschi et al., 2012).
Fig. 1.
Overview of the isobutanol and 3-methyl-1-butanol pathways examined in this study. kivd uses α-ketoisovalerate, an important metabolite for l-Valine and l-Leucine synthesis in Synechocystis, to produce isobutanol and uses 2-ketoisocaproate to produce 3-methyl-1-butanol. kivd: gene encodes α-ketoisovalerate decarboxylase; adh: gene encodes alcohol dehydrogenase.
Table 1.
Cyanobacterial strains engineered for isobutanol production.
| Organism | Overexpressed genes | Initial OD750 | Condition | Titer | Time (days) | Reference |
|---|---|---|---|---|---|---|
| Synechococus7942 | PLlacO1:alsS-ilvC-ilvD | Nearly 1 | 50 mM NaHCO3, 55 mol photons m−2 s−1 | 450 mg L−1 | 8 | Atsumi et al. (2009b) |
| Ptrc:kivd-yqhD | ||||||
| SynechocystisPCC 6803 | Ptac:kivd-yqhD | Mid-log phase | Autotrophic (50 mM NaHCO3), 50mol photons m−2 s−1, within situtrap | 240 mg L−1 | 21 | Varman et al. (2013) |
| Mixtrophic (50 mM NaHCO3, 0.5% glucose), 50mol photons m−2 s−1, within situtrap | 298 mg L−1 | 21 | ||||
| Synechococus7942 | ΔglgC | 0.4–0.6 | 50 mM NaHCO3, 150 mol photons m−2 s−1 | 550 mg L−1 | 8 | Li et al. (2014) |
| PLlacO1:alsS-ilvC-ilvD | ||||||
| Ptrc:kivd-yqhD |
Among the different isobutanol production platforms, cyanobacteria have gained great attention due to their photosynthetic metabolism that can utilize CO2 in the atmosphere (Quintana et al., 2011). Moreover, cyanobacteria have low nutrient requirements, high tolerance to diverse environments, and uncomplicated genetic engineering capacities (Nozzi et al., 2013). The first isobutanol producing cyanobacterial strain was generated in Synechococcus elongatus PCC 7942 by overexpressing the acetolactate synthase (AlsS) from Bacillus subtilis (B. subtilis), acetohydroxy acid isomeroreductase (IlvC) and dihydroxy-acid dehydratase (IlvD) from E. coli, Kivd from L. lactis, and three different ADHs from S. cerevisiae, E. coli and L. lactis, respectively (Atsumi et al., 2009b). The engineered strain containing the E. coli ADH encoded by yqhD showed the highest isobutanol production. In the same study, increased isobutyraldehyde/isobutanol production and in vitro enzyme activities were observed after overexpressing ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). In another study from Li et al., glgC encoding glucose-1-phosphate adenylyltransferase was knocked out from the isobutanol producing strain, and the resulting ΔglgC strain exhibited 2.5 times higher isobutanol production than the control strain expressing the same isobutanol synthesis enzymes but with an intact glgC (Li et al., 2014). Furthermore, Varman et al. introduced the two key genes of the 2-keto acid pathway, kivd (from L. lactis) and adhA (from L. lactis), into Synechocystis to demonstrate the isobutanol production capacity of this strain (Varman et al., 2013). The different carbon partitioning in this engineered strain was also examined using 13C-labeled glucose and significant reduced glucose utilization was observed in the isobutanol producing strain compared to that in wild type cells.
In the present study, we expressed the α-ketoisovalerate decarboxylase encoded by kivd from L. lactis and four different ADHs either on a self-replicating vector or on Synechocystis chromosome. Gene expression and isobutanol production were compared among the resulting strains. We also demonstrated that when cultures were supplemented with isobutyraldehyde, the empty vector control strain was able to produce a relatively high level of isobutanol, indicating that Synechocystis’ endogenous ADHs are able to effectively utilize isobutyraldehyde as substrate to produce isobutanol.
2. Methods
2.1. Strains used in cloning, transformation and conjugation
For cloning and conjugation, E. coli strain DH5 and DH5 Z1 (Invitrogen) were used. The cells were grown at 37 °C in LB medium (agar or liquid) supplemented with 50 g ml−1 kanamycin (Sigma-Aldrich).
The glucose-tolerant Synechocystis PCC 6803 strain was used for this study. Cells were grown under 50 mol photons m−2 s−1 at 30 °C in BG 11 medium (Rippka et al., 1979).
2.2. Plasmid construction for gene expression
pEEK2 and pDDH were used in this study as shuttle vectors (Supplement Fig. 1).
pDDH is an integrative vector based on the recently reported pEERM series of vectors (Englund et al., 2015). The homologous recombination regions in pDDH are the 1000 bp upstream sequence and the 1000 bp downstream sequence of the slr1556 locus. The gene slr1556 encodes the D-lactate dehydrogenase, which catalyzes formation of lactate from pyruvate in Synechocystis. This could be a competing pathway for isobutanol synthesis, so we decided to delete this pathway for an improved flux of pyruvate towards isobutanol synthesis.
pEEK2 is a broad host range self-replicating vector based on the previously reported vector pPMQAK1 (Huang et al., 2010). pEEK2 was designed and constructed to utilize a bicistronic design (BCD) to avoid influence of genetic context on gene expression, and thus to make the heterologous overexpression more predictable (Mutalik et al., 2013). pEEK2 also provides a simple cloning process by only requiring a single ligation step to create a fully equipped expression plasmid. The expression in pEEK2 is driven by the strong constitutive promoter Ptrccore and the translation is initiated from the strongest BCD system from a previous report (Mutalik et al., 2013). pEEK2 has been made available through AddGene (AddGene number: 83492).
All the heterogenous genes were codon optimized and synthesized by GenScript. All the endogenous genes were amplified using specific primers from wild-type Synechocystis PCC 6803 genome using Phusion Polymerase (Thermo Fisher Scientific). Xba , BamHI, Bgl , Spe and Pst were the restriction cloning sites used to construct all the plasmids in this study. The control strains used in this study carry the corresponding empty vector.
2.3. Transformation and conjugation of Synechocystis
Synechocystis wild type cells were transformed with pDDH-based constructs by incubating 200 l concentrated (OD750 = 2.5) mid-log phase (OD750 = 0.8) wild-type cells with 2 g appropriate plasmid DNA in liquid BG11 medium. After growing under 100 mol photons m−2 s−1 at 30 °C for 4 h, the cells were spread on filters on BG11 agar plates for another 24 h incubation
For conjugation, E. coli cargo cells and E. coli HB101 helper cells with the plasmid pRL443-AmpR were grown overnight at 37 °C. The overnight cultures were centrifuged at 3000 rpm for 5 min and resuspended in fresh liquid LB medium without antibiotics. A mixture of cargo cells (1 ml), helper cells (1 ml) and wild-type Synechocystis PCC 6803 cells (200 l) was incubated under 100 mol photons m−2 s−1 at 30 °C for 1.5 h. The mixture was then spread on a filter on a BG11 agar plate for another 48 h incubation.
For colony selection and maintenance, the filters were changed onto new BG11 agar plates with 50 g ml−1 kanamycin. Colonies were analyzed by PCR using gene specific primers and DreamTaq DNA polymerase (Thermo Fisher Scientific). The correct homologous recombinants were inoculated into fresh liquid BG11 medium with 50 g ml−1 kanamycin and propagated until fully segregated. The segregation was examined by PCR using gene specific primers.
2.4. Evaporation experiment
132.5 mg L−1 of isobutanol standard solution was prepared in BG11 medium and incubated in six cotton-cap E-flasks, and six plug-sealed tissue culture flasks. The flasks were shaken at 120 rpm, under 50 mol photons m−2 s−1 at 30 °C. The isobutanol concentration was determined at days 0, 3, 4, 6 and 9.
2.5. Different cultivation conditions
Seed cultures were grown under 50 mol photons m−2 s−1 at 30 °C in BG11 with addition of 50 g ml−1 kanamycin in 100 ml Erlenmeyer flasks (VWR) until OD750 reached 2. Each seed culture was then used to inoculate 25 ml experimental cultures to OD750 = 0.1 in BioLite 25 cm2 plug-sealed tissue culture flasks (Thermo Fisher Scientific). The medium was BG11 with addition of 50 g ml−1 kanamycin and 50 mM NaHCO3 (Sigma-Aldrich). The addition of 50 mM NaHCO3 supplied abundant carbon source to compete with the exceeded oxygen level in the flask in order to maintain photosynthesis. The flasks were shaken horizontally at 120 rpm, under 50 mol photons m−2 s−1 at 30 °C.
For batch cultures, 2 ml culture was sampled from each flask every day for measurements while 2 ml fresh BG11 medium with addition of 50 g ml−1 kanamycin and 500 mM NaHCO3 was added back. The measurements continued for 7 days.
For the replenished cultures, every 4 days cells were centrifuged at 3000 rpm at room temperature and then resuspended in 25 ml fresh BG11 with addition of 50 g ml−1 kanamycin and 50 mM NaHCO3. The measurements continued for 14 days.
For the in vivo comparison of the different ADHs, 10 l isobutyraldehyde was added into each 25 ml 1-day old culture and also to 25 ml pure media as control. All the flasks were shaken at 120 rpm, under 50 mol photons m−2 s−1 at 30 °C for 24 h. Then isobutanol was extracted and quantified.
2.6. RNA isolation and semi-quantitative reverse transcript PCR (RT-PCR)
Total RNA was isolated from cultures (OD750 = 0.5) using RTI Reagent (Sigma-Aldrich) according to the manufacturer's instructions. Samples were digested by DNaseI (Thermo Fisher Scientific) to remove DNA and the purity of RNA was checked by PCR using RNA samples as templates and DreamTaq DNA Polymerase (Thermo Fisher Scientific). RNA concentration was measured using Nanodrop™ 2000 spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from 100 g RNA using the qScript cDNA synthesis kit (Quantabio). In the 22 cycles RT-PCR, 0.5 l of cDNA and gene specific primers were used, 16 s RNA was the control.
2.7. Crude protein extraction and SDS-PAGE/Western-immunoblot
Proteins were extracted from day 5 cultures, 1 ml of culture was harvested by centrifugation at 5000 rpm, 4 °C, for 10 min. The pellet was washed in 2 ml PBS and collected again by centrifugation (5000 rpm, 4 °C, for 10 min). Then, 200 µl PBS was used to resuspend the pellet and this mixture was frozen in − 80 °C for 10 min followed by heating at 37 °C for 10 min. After this, 2 l of 100 Protease Arrest (GBioscience) and 0.2 ml acid-washed 425–600 m diameter glass beads (Sigma-Aldrich) were mixed with the cells. Cells were disrupted using the Precellys-24 Beadbeater (Bertin Instruments), program 3 30 s. Centrifugation was performed twice at 5000 rpm, 4 °C, 15 min each, to get a clean supernatant containing soluble proteins. Protein concentration was determined by the DC protein assay (Bio-Rad).
4 µg soluble proteins in each well were separated by SDS-PAGE, using Mini-PROTEAN TGX ™ gels (Bio-Rad), and transferred to PVDF membrane (Bio-Rad). Strep-tags were detected by Anti-Strep-tag (abcam) using standard techniques. The quantification of expression level was done using Quantity One Software (Bio-Rad).
2.8. Optical density measurement and isobutanol extraction
From each flask (25 ml), 2.5 ml culture was sampled every 24 h for both OD750 measurement and isobutanol extraction. Absorbance at 750 nm was measured for 200 l culture in 96-well plates using a micro-plate reader (HIDEX, Plate Chameleon). The remaining 2.3 ml culture was centrifuged at 5000 rpm, for 10 min. Then, 1305 l of supernatant was transferred into a 15 ml screw cap tube, mixed with 45 l 3000 mg L−1 1-pentanol internal standard and 450 l dichloromethane (DCM). The mixture was shaken on Multi-Tube Vortexer VX-2500 (VWR) at maxium speed for 5 min and then centrifuged at 5000 rpm, 4 °C, for 10 min. DCM phase (bottom) was transferred into 1.5 ml clear glass gas chromatography (GC) vials (VWR).
2.9. Isobutanol quantification
The extracted samples were analyzed on a PerkinElmer GC 580 system equipped with a flame ionization detector and an Elite-WAX Polyethylene Glycol Series Capillary column, 30 m 0.25 mm 0.25 m (PerkinElmer). Nitrogen was the carrier gas, with 10 ml/min flow rate. The temperatures of injector and detector were 220 °C and 240 °C, respectively. The initial oven temperature was 50 and then raised to 100 °C with a rate of 10 °C min−1 followed by a rise to 180 with a rate of 20 °C min−1. The GC results were analyzed using TotalChrom Navigator version 6.3.2. The retention time was determined using 500 mg L−1 isobutanol, 3M1B, and 1-pentanol standards (VWR). A standard curve was made by measuring extractants from BG11 medium with different amount of isobutanol/3M1B (2.5 mg L−1, 5 mg L−1, 10 mg L−1, 25 mg L−1, 50 mg L−1) and 100 mg L−1 of the internal standard 1-pentanol. The amount of isobutanol in the sample was calculated based on the ratio of its signal peak area and that of the internal standard.
3. Results
3.1. Isobutanol evaporation examination
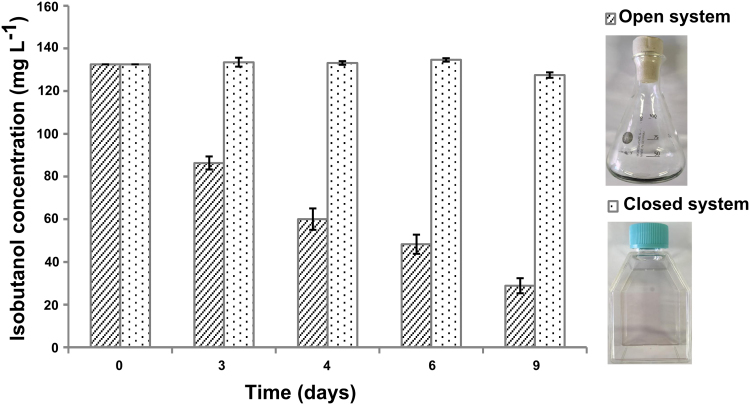
During cultivation, there is a significant risk of losing isobutanol due to evaporation through the air exchange in cotton capped E-flasks. Therefore, we firstly determined the amount of evaporation of isobutanol in both plug-sealed tissue culture flasks (closed system) and cotton capped E-flasks (open system). Isobutanol evaporation from the tissue culture flasks could barely be detected (Fig. 2) while around 64% of the isobutanol evaporated from the cotton cap E-flasks in 6 days, and only around 22% remained at day 9. Based on these observations, we grew all the engineered strains in plug-sealed tissue culture flasks.
Fig. 2.
Comparison of isobutanol evaporation in open and closed systems. Open system: cotton cap E-flasks. Closed systems: plug-sealed tissue culture flasks. The flasks were incubated for 9 days and isobutanol concentration was measured in days 0, 3, 4, 6 and 9. Result represents the mean of six technical replicates, error bars represent standard deviation.
3.2. Enhancement of isobutanol production via different synthetic biology approaches
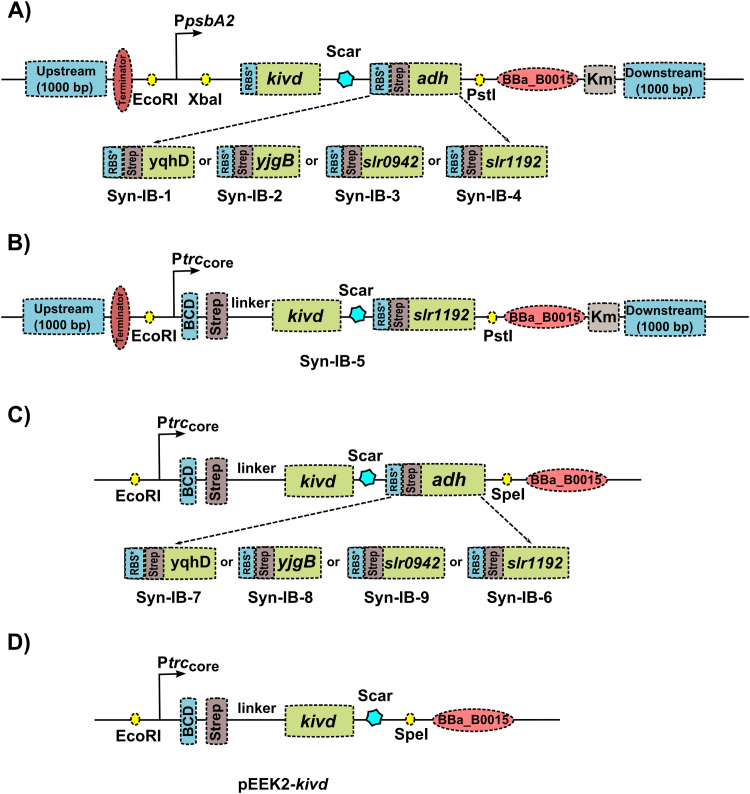
To get heterologous isobutanol production in Synechocystis, we selected the α-ketoisovalerate decarboxylase from L. lactis (Plaza et al., 2004, Atsumi et al., 2009) and four different ADHs, yqhD and yjgB from E. coli, and slr0942 and slr1192 from Synechocystis (Atsumi et al., 2009, Hewitson et al., 2014, Vidal, 2017). We initially used the pDDH vector for overexpressing the isobutanol pathway on the chromosome with simultaneous deletion of the competing lactate synthesis pathway (Fig. 1). An empty vector control strain Syn-pDDH and four isobutanol producing strains each containing kivd and a different ADH were made; Syn-IB-1 (yqhD), Syn-IB-2 (yjgB), Syn-IB-3 (slr0942) and Syn-IB-4 (slr1192) (Fig. 3A). PpsbA2 (Lindberg et al., 2010) promoter and RBS* (Heidorn et al., 2011) were used to drive the transcription and translation of the two genes, respectively. RBS* is complementary to Synechocystis’ anti-SD (Shine-Dalgarno) sequence and it has a 9 bp optimal spacing between the core SD sequence and the start codon. Thus, in Synechocystis, it shows 5 times higher strength compared to the BioBrick RBS BBa_B0034 (Heidorn et al., 2011).
Fig. 3.
Schematic presentation of all the engineered Synechocystis strains. adh: alcohol dehydrogenases. kivd: encodes α-ketoisovalerate decarboxylase (L. lactis); yqhD: encodes alcohol dehydrogenase (E. coli); yjgB: encodes alcohol dehydrogenase (E. coli); slr0942: encodes alcohol dehydrogenase (Synechocystis); slr1192: encodes alcohol dehydrogenase (Synechocystis). A) Constructs used to generate strains where the isobutanol pathway is expressed from the PpsbA2 promoter in the slr1556 locus on Synechocystis chromosome. B) Constructs used to generate strain Syn-IB-5 which has the kivd and slr1192 operon driven by Ptrccore-BCD in the slr1556 locus on Synechocystis chromosome. C) Constructs for the isobutanol pathway driven by Ptrccore-BCD on a self-replicating vector. D) Construct with only kivd overexpressed on a self-replicating vector. All alcohol dehydrogenases were Strep-tagged at the N-terminal and in B), C) and D), kivd was Strep-tagged at the N-terminal. RBS* (Heidorn et al., 2011) was added in front of each gene to allow initiation of translation.
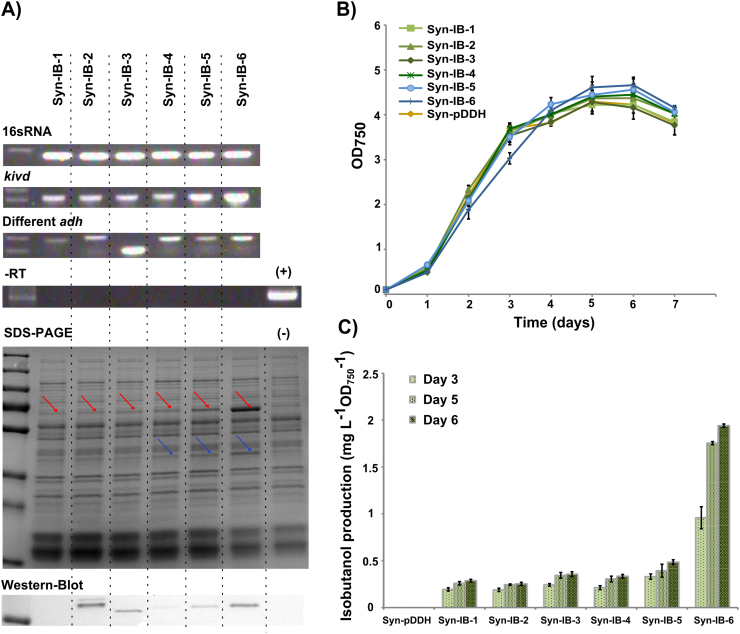
The transcription of all the genes was confirmed using RT-PCR (Fig. 4A upper) and the translation of kivd could be detected on a Coomassie stained SDS-PAGE (Fig. 4A lower). The Strep-tag Western-immunoblot showed expression of yjgB, slr0942 and slr1192, whereas the expression level of yqhD was too low to be detected. Similar growth was observed from all the strains (Fig. 4B), and isobutanol production was measured at day 3, 5 and 6. All strains produced only small amounts of isobutanol, below 0.5 mg L−1 OD−1, and no significant differences in production among the strains were detected (Fig. 4C).
Fig. 4.
Comparison of isobutanol production in engineered Synechocystis PCC 6803 strains Syn-IB-1, -2, -3, -4, -5, and -6. A) RT-PCR, SDS-PAGE and Strep-tag Western-immunoblot (top to bottom). Every dashed line separated each lane represents the results from a single engineered strain. –RT is a negative RT-PCR control using RNA as template without addition of RT enzyme to control for possible DNA contamination. The positive control is an RT-PCR carried out using the corresponding plasmid as template. Red arrows in the SDS-PAGE indicate the location of kivd, blue arrows indicate the expected location for slr1192. The Strep-tag Western-immunoblot examines the expression of all the ADHs. The negative control for SDS-PAGE and Western-immunoblot is an extract from strain Syn-pDDH. B) Growth curve during 7 days of cultivation. C) Isobutanol production at day 3, day 5 and day 6 from the different engineered strains. Results are the mean of 4 biological replicates and 3 technical replicates, error bars represent standard deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A possible explanation for the low production of isobutanol could be that the expression of kivd and ADHs was not high enough. To attempt to increase expression, we changed the PpsbA2 promoter region in Syn-IB-4 (containing kivd and slr1192) to the strong Ptrccore promoter with a BCD construct (Mutalik et al., 2013), thereby generating the strain Syn-IB-5 (Fig. 3B). Strain Syn-IB-6 (Fig. 3C) was also made, where kivd and slr1192 were placed in the self-replicative vector pEEK2 instead of in the genome, which has been reported to increase the level of expression (Ng et al., 2015). Interestingly, the transcription and translation levels of kivd were found to be step-wise increased comparing the strains Syn-IB-4, Syn-IB-5 and Syn-IB-6 (Fig. 4A) (Table 2). This pattern was also shown on the Strep-tag Western-immunoblot detecting slr1192 (Fig. 4A) (Table 2). At day 6, the isobutanol production observed in strain Syn-IB-5 was 0.48 mg L−1 OD−1, only slightly increased compared to that from Syn-IB-4. In contrast, a significant increase was shown in strain Syn-IB-6. At day 3, a production of 0.96 mg L−1 OD−1 was detected, and it reached 1.94 mg L−1 OD−1 at day 6, which was more than 4 times as much as that in Syn-IB-5 (Fig. 4C).
Table 2.
Expression quantification for Kivd and ADHs. The expression level of each protein is presented by the corresponding band intensity per mm2 on SDS-PAGE.
| Stain | Kivd expression (intensity mm2−1) | ADH (slr1192) expression ( intensity mm2−1) |
|---|---|---|
| Syn-IB-4 | 1768.58 | 1792.85 |
| Syn-IB-5 | 4758.77 | 3541.15 |
| Syn-IB-6 | 11,767.73 | 5102.74 |
| Syn-IB-7 | 11,621.05 | 14,390.42 |
| Syn-IB-8 | 12,196.91 | 56,637.99 |
| Syn-IB-9 | 11,572.82 | 23,948.11 |
| pEEK2-kivd | 16,405.52 | – |
3.3. Comparison of different ADH in vivo
Based on the improvement on expression and isobutanol production when using a self replicative vector with a stronger promoter, we decided to express Kivd and remaining ADHs from the pEEK2 vector, resulting in the strains Syn-IB-7 (yqhD), Syn-IB-8 (yjgB) and Syn-IB-9 (slr0942) (Fig. 3C). At the same time, we constructed the strain pEEK2-kivd (Fig. 3D), which only expressed Kivd without any additional copy of ADH.
All these engineered strains showed stronger transcription and translation of kivd and ADHs compared to strains Syn-IB-1, -2, -3, -4 where the cassettes were placed on the chromosome (Fig. 5A). The expression level of the four different ADHs varied significantly and yjgB was the highest expressing one (Fig. 5A lower). The initial growth from all the strains was similar, but the empty vector control strain Syn-pEEK2 and strain Syn-IB-8 reached a higher final OD750 (Fig. 5B). Isobutanol production from Syn-IB-6, 7, -8, and -9 were similar to each other, reaching around 2 mg L−1 OD750−1 after six days of cultivation (Fig. 5C). Surprisingly, strain pEEK2-kivd gave slightly higher isobutanol production than the other strains, indicating that endogenous Synechocystis ADHs are enough to fully catalyze the formation of isobutanol from isobutyraldehyde, and that additional ADH expression in strains Syn-IB-6, -7, -8, -9 does not contribute to increased levels of isobutanol production.
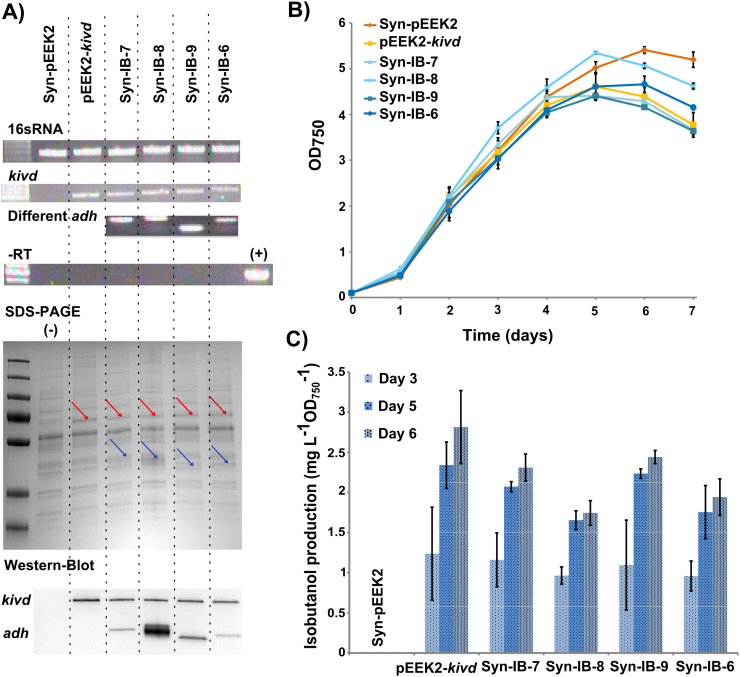
Fig. 5.
Comparison of growth, gene transcription, translation and isobutanol production in engineered strains Syn-pEEK2, pEEK2-kivd, Syn-IB-7, -8, -9 and -6. A) RT-PCR, SDS-PAGE and Strep-tag Western-immunoblot (top to bottom). Each lane represents the results from a single engineered strain. –RT is a negative RT-PCR control using RNA as template without addition of RT enzyme to control for possible DNA contamination. The positive control is an RT-PCR carried out using the corresponding plasmid as template. Red arrows indicate the location of kivd, blue arrows indicate the expected location for different AHDs. The first row of the Strep-tag Western-immunoblot shows the expression of kivd and the second row shows the expression of all the ADHs. The negative control for protein gel and Western-immunoblot is the empty vector strain Syn-pEEK2. B) Growth curve during 7 days of cultivation. C) Isobutanol production at day 3, day 5 and day 6 from all the engineered strains. Results are the mean of 4 biological replicates and 3 technical replicates, error bars represent standard deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
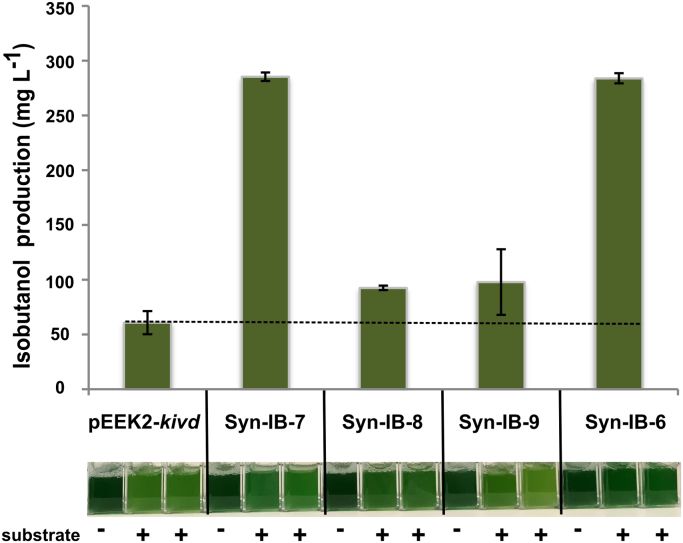
In order to be able to examine in vivo activity of all the ADHs, we supplied 316 mg L−1 external isobutyraldehyde, the substrate of ADH for isobutanol synthesis, to cultures on day 1 and measured isobutanol production after 24 h. By comparing a culture grown with isobutyraldehyde to a control sample without any cells, we could clearly show that the cells were able to uptake the external isobutyraldehyde, and isobutanol was produced (Supplement Fig. 2). Production from strain pEEK2-kivd, 60.8 mg L−1 in 24 h at OD750 around 0.8, represents the capacity of Synechocystis’ endogenous ADH/ADHs to utilize isobutyraldehyde for isobutanol production in this condition, and it was considered as the background value for the comparison among the other 4 strains (Fig. 6). Furthermore, we observed significant differences in the isobutanol production among the strains where different ADHs were overexpressed in the cells. Syn-IB-7 and Syn-IB-6 produced 285 mg L−1 and 283 mg L−1 isobutanol, respectively. Meanwhile, Syn-IB-8 and Syn-IB-9 produced approximately 95 mg L−1 isobutanol, which is one third of the production from Syn-IB-6 and Syn-IB-7. However, strain Syn-IB-6, expressing extra copies of the endogenous slr1192, showed less bleaching than the other strains (Fig. 6).
Fig. 6.
Isobutanol production and images showing the different cultures at the same time point from strains pEEK2-kivd, Syn-IB-6, -7, 8, and -9 after the addition of 316 mg L−1 isobutyraldehyde. The cultures were inoculated as OD750 = 0.1 and cultivated for 1 day. Then, 316 mg L−1 isobutyraldehyde was added into each flask and isobutanol production was measured after 24 h. The dashed line indicates the likely isobutanol production level from naturally existing Synechocystis endogenous ADH in each strain. The culture pictures were taken when isobutanol samples were taken from the flasks (at day 2), and a culture without the additional isobutyraldehyde was used as a control for each strain. Results are the mean of 2 biological replicates and 3 technical replicates, error bars represent standard deviation.
3.4. Replenished culture to enhance isobutanol productivity
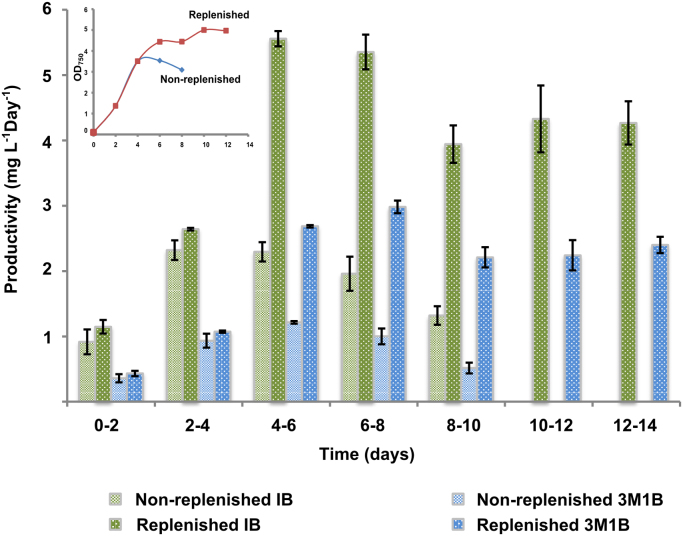
In all the previous experiments, we observed that the growth of all the strains started to decrease after 4 days when we cultivated the cells in BG11 with addition of 50 mM NaHCO3 and 2 ml of 500 mM NaHCO3 was supplied every second day. Therefore, we grew strain pEEK2-kivd and changed the media every 4 days in order to remove potential salt stress from accumulated NaHCO3 and to examine for how long the cells can keep the capacity to synthesize the products. During the 14 days of cultivation with the media refreshed every four days, both isobutanol and 3-methyl-1-butanol were measured. The latter is a by-product from isobutanol synthesis (Fig. 1). By continuously replenishing with fresh media, the cells could reach a higher cell density while still maintaining the productivity for both products (Fig. 7). To calculate the total production of isobutanol and 3-methyl-1-butanol in the replenished cultures, we summarized the amount of the two products respectively, and normalized them by cell dry weight at the last measurement day. 16.8 mg g-1 DCW isobutanol and 9.8 mg g-1 DCW 3-methyl-1-butanol were produced in the replenished cultures during 14 days and 7.1 mg g-1 DCW isobutnaol and 4.3 mg g-1 DCW 3-methyl-1-butanol were produced in the non-replenished cultures during 10 days.
Fig. 7.
Growth and isobutanol/3-methyl-1-butanol productivity from the replenished cultures and the non-replenished cultures of strain pEEK2-kivd. Not replenished cultures were monitored for 8 days while the replenished cultures were monitored for 14 days. Isobutanol and 3-methyl-1-butanol was measured every second day and the specific productivity was calculated by dividing the production (mg L−1) with time (2 days). Results are the mean of 3 biological replicates and 2 technical replicates, error bars represent standard deviation.
4. Discussion
Synechocystis has been used to produce several alcohols, e.g. ethanol (Englund et al., 2015, Hewitson et al., 2014), 1-butanol (Anfelt et al., 2015), and isobutanol (Varman et al., 2013). Extra copies of heterologous or endogenous ADH were always co-expressed with the other genes in the biosynthesis pathways. In the present study, we have examined the isobutanol producing capacity of naturally occurring Synechocystis ADHs for the first time. The comparison of isobutanol production from strain pEEK2-kivd with and without the addition of isobutyraldehyde indicates that the ADH substrate produced by overexpressed kivd in this study is not enough to saturate Synechocystis endogenous ADHs. This is the reason why we could not observe any significant difference in isobutanol production among the strains harboring different ADHs without the addition of external isobutyraldehyde to the media. Hence, when the substrate for ADHs is limited, the step-wise increased isobutanol production we observed from strains Syn-IB-4, -5, -6 was not related to the increased expression level of slr1192 but kivd (Fig. 4) (Table 2). In addition, the slightly higher isobutanol production from strain pEEK2-kivd is also positively related to the higher expression level of kivd in this strain compared to strains Syn-IB-6, -7, 8, -9 (Fig. 5) (Table 2). Thus, kivd is one of the likely bottleneck of this isobutanol pathway at these levels of production.
Moreover, compared to using a stronger promoter on the Synechocystis chromosome, overexpressing genes on a self-replicating vector gave a higher increase in the relative amount of mRNA, protein expression and isobutanol production (Fig. 4). This may be due to the different copy number of pEEK2 and Synechocystis chromosome. pEEK2 carries the RSF1010 replicon which have been shown to have a copy number of between 10 and 30 in Synechocystis (Marraccini et al., 1993, Ng et al., 2000). The copy number of Synechocystis chromosome depends on the growth condition and normally is around 12 (Labarre et al., 1989). Therefore, a possible explanation for the higher expression from a plasmid based system, is that it provides a higher gene dosage during our 6 days cultivation period.
To compare the activity of the different ADHs in vivo, we added isobutyraldehyde to each 1-day old culture to be able to saturate Synechocystis endogenous ADHs and to show differences among the overexpressed ADHs. The addition of 316 mg L−1 isobutyraldehyde did not have any major effect on cell growth, although some retardation of growth was observed compared to the cultures without isobutyraldehyde (Fig. 6). Among the 4 different ADHs we overexpressed in this study, yqhD (E. coli) and slr1192 (Synechocystis) in Syn-IB-7 and Syn-IB-6, respectively, contributed the most towards isobutanol production within 24 h while the yjgB (E. coli) in Syn-IB-8 and slr0942 (Synechocystis) in Syn-IB-9 seemed to contribute only slightly more isobutanol than the background strain pEEK2-kivd. Furthermore, yjgB showed significantly higher expression than the other ADHs, 3.9 times higher than yqhD and 11.1 times higher than slr1192 (Table 2). This indicates that the yjgB encoded ADH probably has low catalytic efficiency in Synechocystis or this enzyme was not in active form for the most part. Nevertheless, the ADHs encoded by yqhD and slr1192 seem to have better catalytic efficiency on reducing isobutyraldehyde into isobutanol. Interestingly, after 24 h in the presence of isobutyraldehyde, strain Syn-IB-6 grew till OD 750 = 1.16, which was higher than OD750 = 0.73 from strain Syn-IB-7 (data not shown), and showed less bleaching (Fig. 6). One explanation could be that slr1192 encoded ADH has higher efficiency for aldehyde reduction than for alcohol oxidation (Vidal et al., 2009), hence, it was able to lower the concentration of isobutyraldehyde fast, resulting in better growth since isobutyraldehyde may have slightly higher toxicity than isobutanol to cyanobacteria (Atsumi et al., 2009b). Moreover, the expression of slr1192 has been shown to be enhanced when the cells are exposed to different environmental stresses, e.g. salt stress and osmotic stress (Vidal et al., 2009). slr1192 encoded ADH also has been shown to play an important role in ethanol tolerance and production in Synechocystis. Upon exposure to external ethanol in the media, the lack of slr1192 resulted in reduced survival and a more bleaching phenotype compared to wild type. Furthermore, a strain overexpressing slr1192 showed higher internal ethanol production and tolerance compared to an slr1192 deficient strain overexpressing an ADH encoded by adhB from Z. mobilis (Vidal, 2017). Similarly, in the present study, another explanation for the better growth and less bleaching phenotype could be that the addition of isobutyraldehyde caused stress responses in the cells and the balance of reduction power was disturbed. The over-expression of slr1192-encoded ADH in strain Syn-IB-6 efficiently helped maintain an adequate intracellular NAD(H):NADP(H) ratio through its activity towards both NAD and NADP, thereby enhancing the tolerance to external isobutyraldehyde and internally synthesized isobutanol.
When we cultivated the cells in BG11 with 50 mM NaHCO3 and 2 ml 500 mM NaHCO3 was supplemented to the culture every second day, the approximately 200 mM Na+ thus added after 8 days potentially resulting in a salinity stress in the culture which can lead to the death of the cells. By changing the media every 4 days, we removed the potential salt stress as well as provided enough fresh nutrients to the cells, which resulted in around 2.3 times isobutanol and 3-methyl-1-butanol production increase compared to the non-replenished cultures, and the production time was prolonged to at least 14 days.
5. Conclusion
In the present study, we found that α-ketoisovalerate decarboxylase is the only heterologous enzyme that needs to be introduced into Synechocystis to enable isobutanol production, and that this enzyme may be one of the bottleneck in the pathway. We extended our understanding and confirmed the high catalytic efficiency of Synechocystis ADH encoded by slr1192 and E. coli ADH encoded by yqhD in the reduction of isobutyraldehyde toward isobutanol. Moreover, this study indicated that using the self-replicating vector pEEK2 to overexpress genes could result in increased transcription, translation and product formation in Synechocystis.
Authors’ contributions
RM and XFL performed bioinformatics research and designed part of the constructs together. RM designed most of the experiments and performed all the experiments, analyzed the data and wrote the manuscript. EE designed and made the pEEK2 vector and revised the manuscript. PiL supervised the project and revised the manuscript. PeL is the main supervisor for this project and revised the manuscript. All the authors read and approved the final version of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by the Knut and Alice Wallenberg Foundation (project MoSE, #2011.0067), the Swedish Energy Agency (#11674-5), and the European Union Horizon 2020 Framework Programme under the Grant agreement no. 640720 (Photofuel).
Acknowledgement
The authors thank Michele Bedin from Molecular inorganic chemistry, Uppsala University, for help in the design of the isobutanol extraction method.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2017.07.003.
Contributor Information
Rui Miao, Email: rui.miao@kemi.uu.se.
Xufeng Liu, Email: xufeng.liu@kemi.uu.se.
Elias Englund, Email: Elias.Englund@Kemi.Uu.Se.
Pia Lindberg, Email: pia.lindberg@kemi.uu.se.
Peter Lindblad, Email: peter.lindblad@kemi.uu.se.
Appendix A. Supplementary material
Supplementary material
References
- Anfelt J., Kaczmarzyk D., Shabestary K., Renberg B., Rockberg J., Nielsen J., Uhlen M., Hudson E.P. Genetic and nutrient modulation of acetyl-CoA levels in Synechocystis for n-butanol production. Microb. Cell Factor. 2015;14:167. doi: 10.1186/s12934-015-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S., Hanai T., Liao J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Atsumi S., Wu T.-Y., Eckl E.-M., Hawkins S.D., Buelter T., Liao J.C. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 2009;85:651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S., Higashide W., Liao J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- Blombach B., Riester T., Wieschalka S., Ziert C., Youn J.-W., Wendisch V.F. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat D., Boles E. Isobutanol production from d-xylose by recombinant Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13:241–244. doi: 10.1111/1567-1364.12028. [DOI] [PubMed] [Google Scholar]
- Chen X., Nielsen K.F., Borodina I., Kielland-Brandt M.C., Karhumaa K. Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol. Biofuels. 2011;4:21. doi: 10.1186/1754-6834-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S.H., Rabinovitch-Deere C.A., Fan Z., Atsumi S. Isobutanol production from cellobionic acid in Escherichia coli. Microb. Cell Factor. 2015;14:52. doi: 10.1186/s12934-015-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter J., Fu P. Metabolic engineering of cyanobacteria for ethanol production. Energy Environ. Sci. 2009;2:857–864. [Google Scholar]
- Englund E., Andersen-Ranberg J., Miao R., Hamberger B., Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synth. Biol. 2015;4:1270–1278. doi: 10.1021/acssynbio.5b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Zhao H., Li Z., Tan X., Lu X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 2012;5:9857–9865. [Google Scholar]
- Hewitson B., Janetos A.C., Carter T.R., Giorgi F., Jones R.G., Kwon W.-T., Mearns L.O., Schipper E.L.F., van Aalst M. Regional context. In: Barros V.R., Field C.B., Dokken D.J., Mastrandrea M.D., Mach K.J., Bilir T.E., Chatterjee M., Ebi K.L., Estrada Y.O., Genova R.C., Girma B., Kissel E.S., Levy A.N., MacCracken S., Mastrandrea P.R., White L.L., editors. Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part B: Regional Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press; Cambridge, United Kingdom and New York, NY, USA: 2014. pp. 1133–1197. [Google Scholar]
- Heidorn T., Camsund D., Huang H.-H., Lindberg P., Oliveira P., Stensjö K., Lindblad P. Synthetic biology in cyanobacteria engineering and analyzing novel functions. Methods Enzymol. 2011;497:539–579. doi: 10.1016/B978-0-12-385075-1.00024-X. [DOI] [PubMed] [Google Scholar]
- Huang H.-H., Camsund D., Lindblad P., Heidorn T. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Tezuka H., Ishii J., Matsuda F., Ogino C., Kondo A. Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J. Biotechnol. 2012;159:32–37. doi: 10.1016/j.jbiotec.2012.01.022. [DOI] [PubMed] [Google Scholar]
- Labarre J., Chauvat F., Thuriaux P. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the Cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 1989;171:3449–3457. doi: 10.1128/jb.171.6.3449-3457.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Shen C.R., Liao J.C. Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942. Photosynth. Res. 2014;120:301–310. doi: 10.1007/s11120-014-9987-6. [DOI] [PubMed] [Google Scholar]
- Lindberg P., Park S., Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 2010;12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Liu Z., Liu P., Xiao D., Zhang X. Improving isobutanol production in metabolically engineered Escherichia coli by co-producing ethanol and modulation of pentose phosphate pathway. J. Ind. Microbiol. Biotechnol. 2016;43:851–860. doi: 10.1007/s10295-016-1751-9. [DOI] [PubMed] [Google Scholar]
- Machado I.M.P., Atsumi S. Cyanobacterial biofuel production. J. Biotechnol. 2012;162:50–56. doi: 10.1016/j.jbiotec.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Marcheschi R.J., Li H., Zhang K., Noey E.L., Kim S., Chaubey A., Houk K.N., Liao J.C. A synthetic recursive “+1” pathway for carbon chain elongation. ACS Chem. Biol. 2012;7:689–697. doi: 10.1021/cb200313e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraccini P., Bulteau S., Cassier-Chauvat C., Mermet-Bouvier P., Chauvat F. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 1993;23:905–909. doi: 10.1007/BF00021546. [DOI] [PubMed] [Google Scholar]
- Miao R., Wegelius A., Dural C., Liang F., Khanna N., Lindblad P. Engineering cyanobacteria for biofuel production. In: Hallenbeck P., editor. Modern Topics in the Phototrophic Prokaryotes, Environmental and Applied Aspects. Part II: Bioremediation, Secondary Metabolites and Other Applied Aspects. 1st edition. Springer International Publishing; Switzerland: 2017. pp. 351–393. [Google Scholar]
- Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.A., Tran A.B., Paull M., Keasling J.D., Arkin A.P., Endy D. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013;10(4):354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- Ng A.H., Berla B.M., Pakrasi H.B. Fine-tuning of photoautotrophic protein production by combining promoters and neutral sites in the Cyanobacterium Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2015;81:6857–6863. doi: 10.1128/AEM.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W.-O., Zentella R., Wang Y., Taylor J.-S.A., Pakrasi H.B. phrA, the major photoreactivating factor in the Cyanobacterium Synechocystis sp. strain PCC 6803 codes for a cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch. Microbiol. 2000;173:412–417. doi: 10.1007/s002030000164. [DOI] [PubMed] [Google Scholar]
- Nozzi N.E., Oliver J.W.K., Atsumi S. Cyanobacteria as a platform for biofuel production. Synth. Biol. 2013;1:7. doi: 10.3389/fbioe.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza M., de la, Palencia P.F., de, Peláez C., Requena T. Biochemical and molecular characterization of α-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol. Lett. 2004;238:367–374. doi: 10.1016/j.femsle.2004.07.057. [DOI] [PubMed] [Google Scholar]
- Quintana N., Van der Kooy, F., Van de Rhee M.D., Voshol G.P., Verpoorte R. Renewable energy from Cyanobacteria: energy production optimization by metabolic pathway engineering. Appl. Microbiol. Biotechnol. 2011;91:471–490. doi: 10.1007/s00253-011-3394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology. 1979;111:1–61. [Google Scholar]
- Sheehan J. Engineering direct conversion of CO2 to biofuel: genetically engineered cyanobacteria harvest light energy to directly produce isobuteraldehyde and isobutanol. Nat. Biotechnol. 2009;27:1128. doi: 10.1038/nbt1209-1128. [DOI] [PubMed] [Google Scholar]
- Smith K.M., Liao J.C. An evolutionary strategy for isobutanol production strain development in Escherichia coli. Metab. Eng. 2011;13:674–681. doi: 10.1016/j.ymben.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Smith K.M., Cho K.-M., Liao J.C. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 2010;87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh C.T., Li J., Blanch H.W., Clark D.S. Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl. Environ. Microbiol. 2011;77:4894–4904. doi: 10.1128/AEM.00382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varman A.M., Xiao Y., Pakrasi H.B., Tang Y.J. Metabolic engineering of Synechocystis sp. Strain PCC 6803 for isobutanol production. Appl. Environ. Microbiol. 2013;79:908–914. doi: 10.1128/AEM.02827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R. Alcohol dehydrogenase AdhA plays a role in ethanol tolerance in model Cyanobacterium Synechocystis sp. PCC 6803. Appl. Microbiol. Biotechnol. 2017;101:3473–3482. doi: 10.1007/s00253-017-8138-3. [DOI] [PubMed] [Google Scholar]
- Vidal R., López-Maury L., Guerrero M.G., Florencio F.J. Characterization of an alcohol dehydrogenase from the Cyanobacterium Synechocystis sp. Strain PCC 6803 that responds to environmental stress conditions via the Hik34-Rre1 two-component system. J. Bacteriol. 2009;191:4383–4391. doi: 10.1128/JB.00183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Chen X., Mishra P., Ching C.-B. Metabolically engineered Saccharomyces cerevisiae for enhanced isoamyl alcohol production. Appl. Microbiol. Biotechnol. 2017;101:465–474. doi: 10.1007/s00253-016-7970-1. [DOI] [PubMed] [Google Scholar]
- Zhang K., Sawaya M.R., Eisenberg D.S., Liao J.C. Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. USA. 2008;105:20653–20658. doi: 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.