ABSTRACT
rRNAs are extensively modified during their transcription and subsequent maturation in the nucleolus, nucleus and cytoplasm. RNA modifications, which are installed either by snoRNA-guided or by stand-alone enzymes, generally stabilize the structure of the ribosome. However, they also cluster at functionally important sites of the ribosome, such as the peptidyltransferase center and the decoding site, where they facilitate efficient and accurate protein synthesis. The recent identification of sites of substoichiometric 2′-O-methylation and pseudouridylation has overturned the notion that all rRNA modifications are constitutively present on ribosomes, highlighting nucleotide modifications as an important source of ribosomal heterogeneity. While the mechanisms regulating partial modification and the functions of specialized ribosomes are largely unknown, changes in the rRNA modification pattern have been observed in response to environmental changes, during development, and in disease. This suggests that rRNA modifications may contribute to the translational control of gene expression.
Keywords: RNA modification, ribosome, snoRNA, RNA methyltransferase, methylation, ribosomopathy, acetylation, pseudouridylation, translation
Abbreviations
- A-site
aminoacylated tRNA site
- ac4C
N4-acetylcytidine
- CMC
N-cyclohexyl-N'-(2-morpholinoethyl)carbodiimidemetho-p-toluenesulfonate
- DMS
dimethyl sulfate
- E-site
exit site
- LSU
large ribosomal subunit
- m26A
N6,N6-dimethyladenosine
- m1A
N1-methyladenosine
- m1acp3ψ
N1-methyl-N3-(3-amino-3-carboxypropyl) pseudouridine
- m5C
N5-methylcytosine
- m7G
N7-methylguanosine
- m3U
N3-methyluridine
- mRNA
messenger RNA
- P-site
Peptidyl tRNA site
- PTC
peptidyltransferase center
- RNAi
RNA interference
- RNP
ribonucleoprotein complex
- rRNA
ribosomal RNA
- SAM
S-adenosylmethionine
- snoRNA
small nucleolar RNA
- snoRNP
small nucleolar RNP
- snRNA
small nuclear RNA
- SSU
small ribosomal subunit
- tRNA
transfer RNA
- Ψ
pseudouridine
- X-DC
X-linked Dyskeratosis congenita
Introduction
Nucleotide modifications are found in all 3 kingdoms of life and so far more than 140 different types of modification have been identified in cellular RNA.1-3 Such modifications expand the chemical and topological properties of the 4 standard nucleosides, thereby influencing RNA structure and regulating the biological functions of the RNAs that carry them. Recent transcriptome-wide mapping approaches have shown that RNA modifications occur on all major classes of RNA, including messenger RNAs (mRNAs).4 After transfer RNAs (tRNAs), which contain up to 17% modified nucleotides,5 ribosomal RNAs (rRNAs) are the most highly modified class of RNAs with approximately 2% of rRNA nucleotides being modified.
Ribosomes are required for the translation of all cellular proteins and in eukaryotes, cytoplasmic ribosomes are composed of 4 rRNAs and approximately 80 ribosomal proteins arranged into a small subunit (SSU) and a large subunit (LSU).6,7 During translation, the SSU mediates decoding by monitoring codon-anticodon base-pairing between the mRNA and tRNAs, while the LSU, which harbours the catalytic peptidyltransferase center, is responsible for synthesis of the nascent polypeptide chain. Ribosome production is initiated in the nucleolus by the RNA polymerase I-mediated transcription of a single rRNA precursor that contains the sequences of 3 of the 4 rRNAs (18S, 5.8S and 25S (yeast)/28S (human) rRNAs).8,9 Assembly of the ribosomal proteins to form key structural features of the mature ribosomal subunits occurs concomitantly with maturation and folding of the pre-rRNA (for a review see ref. 10). After early pre-rRNA cleavage events, the precursor (pre-)SSU and pre-LSU complexes follow separate biogenesis pathways in the nucleolus and nucleus before nuclear export and final maturation steps in the cytoplasm.9,11–13 In yeast and human cells, more than 200 trans-acting cofactors are required to orchestrate this complex and hierarchical process.9,14 These include numerous enzymatic proteins and ribonucleoprotein complexes (RNPs) that are responsible for the introduction of rRNA modifications.15,16
Despite the relatively high abundance of modified nucleotides in rRNA and the diversity of the RNA modifications observed in nature, only a limited set of different chemical modifications is found in rRNA. While the proteins that modify bacterial rRNA and many non-rRNA species in eukaryotes act mostly as stand-alone modification enzymes, some eukaryotic rRNA-modifying enzymes use RNA-guided mechanisms for recognition of their target nucleotides. In general, the rRNA modifications, which are introduced at different stages during ribosome biogenesis, serve to stabilize the secondary and tertiary structure of the rRNA scaffold,17 thereby ensuring the efficiency and accuracy of translation. It has recently emerged, however, that rRNA modifications can also represent an important source of ribosome heterogeneity that may alter ribosome function in response to environmental and developmental cues and in disease.
Modifications guided by small nucleolar RNAs
In eukaryotes, the most abundant rRNA modifications are 2′-O-methylation of the ribose, which can occur on any nucleotide, and the isomerisation of uridine to pseudouridine (Ψ). To date, 55 2′-O-methylation sites and 45 Ψ sites have been identified in the rRNAs of the yeast Saccharomyces cerevisiae and approximately 100 of each modification are reported in human rRNAs.18-21 Together these modifications outnumber base modifications almost 10-fold, which is in stark contrast to the distribution of modifications in Escherichia coli, where twice as many base methylations as ribose methylations and pseudouridylations are present. The extent of 2′-O-methylation and pseudouridylation in yeast rRNA was first estimated almost 30 years ago by 2D thin layer chromatography.22-24 Later, the use of primer extension-based assays, either under limiting dNTP conditions for the detection of 2′-O-methylation25 or after derivatization using CMC in the case of pseudouridylation,26 provided evidence for 99 specific sites of modification (Table 1).27,28 More recently, sequencing-based profiling approaches that monitor reverse transcriptase drop-off rates (Ψ) or exploit the resistance of 2′-O-methylated RNA to alkaline lysis, mass spectrometry, and nuclease protections assays combined with reversed phase high-performance liquid chromatography (RP-HPLC) have confirmed the presence of these ribose methylations and pseudouridines and have clarified the presence of a 2′-O-methylation at position G562 of the 18S rRNA (note, the numbering of residues in yeast rRNAs in this review is based on the sequences annotated in the Saccharomyces Genome Database and may therefore differ from numbers given in some of the original literature cited).18,21,29-31 Such systematic mapping methods have also been applied to human rRNA, generating an updated inventory of 2′-O-methylation sites; no evidence was found for the previously reported 18S-Gm1536 and 18S-Um1602, but modifications at 28S-A1868 and 28S-G3771 were newly identified.32 Similarly, a recent quantitative mass spectrometry-based approach has led to the identification of an additional pseudouridylation site at position U2347 of the yeast 25S rRNA (Table 1).21
Table 1.
Inventory of rRNA modifications in yeast. The rRNA, position (Posn.) and type (Mod.) of the modification and the enzyme/snoRNP(s) that introduce it are given, with information on whether partial modification was observed (✓; <85%)21 or not (x), and whether modification occurs early/chromatin-associated (E),18 in late nucleolar/nuclear (LN) particles or in the cytoplasm (C). A lack of information on the timing is indicated by “–.” Enzymes/snoRNPs targeting the same position sequentially are separated by “,” and alternative enzymes/snoRNPs that can install a single modification at a given site are separated by “/”. The information presented in this table is compiled from various references cited in the text.
| rRNA | Posn. | Mod. | Enzyme/snoRNP | Partial | Timing | rRNA | Posn. | Mod. | Enzyme/snoRNP | Partial | Timing |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 18S | 28 | Am | snR74 | x | E | 18S | 1415 | Ψ | snR83 | ✓ | — |
| 18S | 100 | Am | snR51 | ✓ | LN | 18S | 1428 | Gm | snR56 | x | E |
| 18S | 106 | Ψ | snR44 | x | — | 18S | 1572 | Gm | snR57 | x | E |
| 18S | 120 | Ψ | snR49 | x | — | 18S | 1575 | m7G | Bud23 | x | — |
| 18S | 211 | Ψ | snR49 | ✓ | — | 18S | 1639 | Cm | snR70 | ✓ | E |
| 18S | 302 | Ψ | snR49 | x | — | 18S | 1773 | ac4C | Kre33 | x | — |
| 18S | 414 | Cm | U14 | x | E | 18S | 1781 | m26A | Dim1 | x | C |
| 18S | 420 | Am | snR52 | x | E | 18S | 1782 | m26A | Dim1 | x | C |
| 18S | 436 | Am | snR87 | ✓ | E | 5.8S | 73 | Ψ | snR43 | ✓ | — |
| 18S | 466 | Ψ | snR189 | ✓ | — | 25S | 645 | m1A | Rrp8 (Bmt1) | x | — |
| 18S | 541 | Am | snR41 | x | E | 25S | 649 | Am | U18 | x | E |
| 18S | 562 | Gm | ? | ✓ | E | 25S | 650 | Cm | U18 | x | E |
| 18S | 578 | Um | snR77 | x | E | 25S | 663 | Cm | snR58 | ✓ | E |
| 18S | 619 | Am | snR47 | x | E | 25S | 776 | Ψ | snR80 | x | — |
| 18S | 632 | Ψ | snR161 | ✓ | — | 25S | 805 | Gm | snR39b | x | E |
| 18S | 759 | Ψ | snR80 | x | — | 25S | 807 | Am | snR39/snR59 | x | E |
| 18S | 766 | Ψ | snR161 | x | — | 25S | 817 | Am | snR60 | x | LN |
| 18S | 796 | Am | snR53 | x | E | 25S | 867 | Gm | snR50 | ✓ | LN |
| 18S | 974 | Am | snR54 | x | E | 25S | 876 | Am | snR72 | ✓ | LN |
| 18S | 999 | Ψ | snR31 | ✓ | — | 25S | 898 | Um | snR40 | x | LN |
| 18S | 1007 | Cm | snR79 | x | E | 25S | 908 | Gm | snR60 | x | E |
| 18S | 1126 | Gm | snR41 | x | E | 25S | 956 | m3U | Bmt5 | x | — |
| 18S | 1181 | Ψ | snR85 | x | — | 25S | 960 | Ψ | snR8 | x | — |
| 18S | 1187 | Ψ | snR36 | x | — | 25S | 966 | Ψ | snR43 | x | — |
| 18S | 1191 | m1acp3Ψ | snR35,Emg1,Tsr3 | x | E,LN,C | 25S | 986 | Ψ | snR8 | x | — |
| 18S | 1269 | Um | snR55 | x | E | 25S | 990 | Ψ | snR49 | x | — |
| 18S | 1271 | Gm | snR40 | x | E | 25S | 1004 | Ψ | snR5 | ✓ | — |
| 18S | 1280 | ac4C | Kre33 | ✓ | — | 25S | 1042 | Ψ | snR33 | x | — |
| 18S | 1290 | Ψ | snR83 | x | — | 25S | 1052 | Ψ | snR81 | x | — |
| 25S | 1056 | Ψ | snR44 | x | — | 25S | 2351 | Ψ | snR82 | x | — |
| 25S | 1110 | Ψ | snR82 | ✓ | — | 25S | 2416 | Ψ | snR11 | x | — |
| 25S | 1124 | Ψ | snR5 | x | — | 25S | 2417 | Um | snR66 | x | E |
| 25S | 1133 | Am | snR61 | x | E | 25S | 2421 | Um | snR78 | x | LN |
| 25S | 1437 | Cm | U24 | x | E | 25S | 2619 | Gm | snR67 | x | E |
| 25S | 1449 | Am | U24 | x | E | 25S | 2634 | m3U | Bmt5 | x | — |
| 25S | 1450 | Gm | U24 | x | E | 25S | 2640 | Am | snR68 | x | LN |
| 25S | 1888 | Um | snR62 | x | — | 25S | 2724 | Um | snR67 | x | E |
| 25S | 2129 | Ψ | snR3 | x | — | 25S | 2729 | Um | snR51 | ✓ | E |
| 25S | 2133 | Ψ | snR3 | x | — | 25S | 2735 | Ψ | snR189 | x | — |
| 25S | 2142 | m1A | Bmt2 | x | — | 25S | 2791 | Gm | snR48 | x | E |
| 25S | 2191 | Ψ |
snR32 | x | — | 25S | 2793 | Gm | snR48 | x | E |
| 25S | 2197 | Cm | snR76 | x | E | 25S | 2815 | Gm | snR38 | x | E |
| 25S | 2220 | Am | snR47 | x | E | 25S | 2826 | Ψ | snR34 | x | — |
| 25S | 2256 | Am | snR63 | x | LN | 25S | 2843 | m3U | Bmt6 | x | — |
| 25S | 2258 | Ψ | snR191 | x | — | 25S | 2865 | Ψ | snR46 | x | — |
| 25S | 2260 | Ψ | snR191 | x | — | 25S | 2870 | m5C | Nop2 (Bmt4) | x | — |
| 25S | 2264 | Ψ | snR3 | x | — | 25S | 2880 | Ψ | snR34 | x | — |
| 25S | 2266 | Ψ | snR84 | x | — | 25S | 2921 | Um | snR52/Sbp1 | x | E |
| 25S | 2278 | m5C | Rcm1 (Bmt3) | x | — | 25S | 2922 | Gm | Sbp1 | x | LN |
| 25S | 2280 | Am | snR13 | x | E | 25S | 2923 | Ψ | snR10 | x | — |
| 25S | 2281 | Am | snR13 | x | E | 25S | 2944 | Ψ | snR37 | ✓ | — |
| 25S | 2288 | Gm | snR75 | x | E | 25S | 2946 | Am | snR71 | x | E |
| 25S | 2314 | Ψ | snR86 | x | — | 25S | 2948 | Cm | snR69 | x | E |
| 25S | 2337 | Cm | snR64 | x | — | 25S | 2959 | Cm | snR73 | x | E |
| 25S | 2340 | Ψ | snR9 | x | — | 25S | 2975 | Ψ | snR42 | x | — |
| 25S | 2347 | Um/Ψ/Ψm | snR65,snR9 | x | — | 5S | 50 | Ψ | Pus7 | x | — |
| 25S | 2349 | Ψ | snR82 | x | — |
These 2′-O-methylations and pseudouridines are mostly introduced by 2 classes of small nucleolar (sno)RNPs, termed box C/D and box H/ACA snoRNPs respectively.16,33–35 The rare exceptions in yeast are a pseudouridine (Ψ50) in the 5S rRNA, which is introduced by the pseudouridine synthetase Pus7 that specifically modifies targets with the RSUNΨAR consensus motif (where R is a purine, S is guanine or cytosine and N represents any nucleotide), and the stand-alone 2′-O-methyltransferase Sbp1 that modifies G2922 of the 25S rRNA.36,37 Eukaryotic snoRNPs, which are structurally and functionally conserved from archaeal sRNPs, are ribonucleoprotein complexes containing a snoRNA, which base-pairs with the pre-rRNA and directs the catalytic protein component of the snoRNP to modify a specific target residue. H/ACA box snoRNAs, so called due to the presence of a conserved H box (5′-ANANNA-3′) and an ACA sequence, form a double hairpin structure. Base-pairing with the pre-rRNA takes place in the “pseudouridylation pocket” within a hairpin, leaving the target uridine non-base-paired and hence accessible for isomerisation by the pseudouridine synthetase Cbf5 (dyskerin; Fig. 1A).38 The other protein components of H/ACA box snoRNPs, Nop10, Nhp2 and Gar1 largely perform structural functions, stabilizing the tertiary fold of the RNA and ensuring correct positioning of the target nucleotide in the Cbf5 active site.
Figure 1.
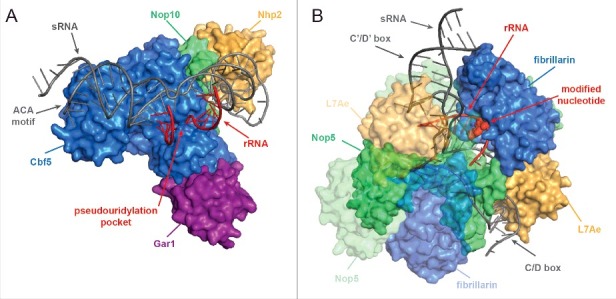
Structures of box C/D and box H/ACA sRNP complexes. (A) Structural model of an H/ACA box sRNP from Pyrococcus furiosus (PDB 3HAY)154 A surface view of the protein components is shown and sRNA and rRNA are indicated in dark gray and red, respectively. (B) Structural model of a Sulfolobus solfataricus box C/D sRNP (PDB 3PLA)155 is shown as in A. The modified nucleotide is shown in surface view. Note, that archaeal L7Ae is an ortholog of eukaryotic Snu13 and in eukaryotes, Nop56 and Nop58 are orthologous to archaeal Nop5.
Box C/D snoRNAs form a single hairpin structure through the base-pairing of conserved sequence motifs (C box, 5′- RUGAUGA-3′ and D box, 5′-CUGA-3′) at the 5′ and 3′ ends of the transcript respectively, as well as through the interaction of internal degenerate C′ and D′ boxes. Base-pairing of the pre-rRNA with the snoRNA adjacent to the D/D′ box is facilitated by the protein components of the snoRNP, Nop56, Nop58 and Snu13 (15.5K in humans). This positions the catalytic site of the methyltransferase Nop1 (fibrillarin)39 to modify its target, 5 nucleotides upstream of the D/D′ box (Fig. 1B). Box C/D snoRNAs form extensive interactions with the pre-rRNA, with guide sequences ranging from 10 to 21 nucleotides in length, and in several cases additional regions of snoRNA-rRNA complementarity close to the methylated residue have been shown to enhance methylation.40 Interestingly, recent structural analysis of archaeal sRNPs indicate that only 10 basepaired nucleotides can be accommodated during catalysis, leading to the suggestion that the additional basepairing interactions facilitate accurate and efficient snoRNP recruitment and that the subsequent unwinding, which is necessary for modification, extends the residence time of the snoRNA on the pre-rRNA to prevent premature or mis-folding of the target site.41
Although the majority of box C/D and H/ACA snoRNAs guide modification of only a single rRNA nucleotide, this is not always the case and several snoRNAs have been shown to direct modification of multiple positions using distinct mechanisms. Due to the presence of both D and D′ motifs, box C/D snoRNAs have the potential to base-pair with and modify 2 alternative sites in the pre-rRNA.34,42 Some such snoRNAs introduce 2 modifications into the same rRNA (e.g. snR60 for 25S-Am817 and 25S Gm908) while others act on both the small and the large ribosomal subunits (e.g., snR52 for 18S-Am420 and 25S-Um2921). Interestingly, snR60 guides 2 ribose methylations that are on opposite sides of a hairpin, suggesting that in addition to its catalytic function, the snoRNP may expedite formation of this rRNA secondary structure through its base-pairing interactions.43 Indeed, several other snoRNAs, including U14 and snR10, not only function in rRNA modification but also have well characterized roles in establishing long-range interactions within pre-ribosomal complexes.44-46 Notably, in several cases the sites modified by the same snoRNA are adjacent or separated by only a single nucleotide (snR13 for 25S-Am2280/1, snR48 for 25S-Gm2791/3 and U24 for 25S-Am1449/Gm1450). In these cases, an alternative mechanism involving the use of different D/D′ boxes within the same snoRNA is employed. It has been proposed that formation of a bulge within the snoRNA-rRNA duplex may lead to “sliding” of a nucleotide adjacent to the canonical fifth nucleotide into the active site of Nop1.34,42 More recent data indicates that instead, the presence of non-canonical C′/D′ boxes in snR13, snR48 and U18 allows the formation of alternative snoRNPs that differ subtly in the arrangement of their protein components and can therefore catalyze additional modifications that do not conform to the classical “5-base-pair rule.”47 The short, discontinuous base-pairing interactions formed between H/ACA box snoRNAs and their pre-rRNA targets allows an even greater degree of target flexibility and the snR3 and snR49 snoRNPs have been linked to 3 and 4 pseudouridylation events respectively.48 In addition to snoRNPs targeting multiple sites in pre-rRNAs, there are also examples of more than one snoRNP targeting the same nucleotide. This can result in redundancy between snoRNPs of the same class in mediating 2′-O-methylation of a particular site (e.g., snR39/snR59; 25S-Am807), but it has also recently been reported that the box H/ACA snoRNA snR9 and the box C/D snoRNA snR65 both target 25S-U2347 leading to the formation of Ψ, Um or Ψm at this site.21 Furthermore, 25S-U2921 can be methylated by both the snR52 snoRNP and the stand-alone methyltransferase Spb1, which additionally guides modification of the adjacent nucleotide (Gm2922), where its activity is exclusively required.
Computational predictions, often supported by experimental evidence, have enabled the vast majority of snoRNAs to be assigned to one or more modification target(s) in rRNA or small nuclear (sn)RNA (Table 1).42,49-52 In addition to the U3 and snR30 snoRNAs that play well established roles in regulating rRNA folding rather than guiding modifications, there are 3 more yeast snoRNAs, snR4, snR45 and snR190, for which no cognate rRNA modifications have been identified.34,53 In contrast, human cells contain a significant number of “orphan” snoRNAs, i.e. snoRNAs for which modification targets have not been identified (76; representing 17% of known human snoRNAs).54 It was recently suggested that some of these snoRNAs might guide modifications already attributed to other snoRNPs, implying that functional redundancy may be more prevalent than previously thought.54 The presence of orphan snoRNAs is, however, also likely to reflect the involvement of human snoRNAs in non-ribosome-associated functions, such as the regulation of alternative pre-mRNA splicing, maintaining chromatin structure, and microRNA-dependent gene silencing.55-57 Recently, some snoRNAs and snoRNA-derived fragments have also been suggested to directly regulate steady-state mRNA levels.58 Intriguingly, some of the snoRNAs implicated in such functions (e.g., SNORD27, SNORA64, SNORA75 and SNORA44) are also known to guide rRNA modifications, raising the possibility of crosstalk or co-regulation of rRNA modification with these cellular processes.
Base modifications introduced by stand-alone enzymes
Besides the myriad of 2′-O-methylations and pseudouridylations, eukaryotic ribosomes contain 8 different types of base modifications, which are introduced at sites distributed between the small and large ribosomal subunits (Table 1; for a review see ref. 15). In yeast, the small subunit contains an N7-methylguanosine (m7G1575) residue, 2 highly conserved N6-dimethyladenosine residues (A1781 and A1782), 2 acetylated cytosine residues (ac4C1280 and ac4C1773) and a complex N1-methyl-N3-aminocarboxypropylpseudouridine (m1acp3ψ) hypermodification at position 1191 of the 18S rRNA. Modifications are less diverse on the large subunit, which carries only 2 N1-methyladenosine (m1A645 and m1A2142), 2 N3-methyluridine (m3U2634 and m3U2843) and 2 C5-methylcytosine (m5C2278 and m5C2870) residues. While all the modifications in the 18S rRNA and both the m5C modifications in the LSU appear to be conserved in humans, only one m1A at position A1322 (equivalent to A645 in yeast) is present and a single m3U modification at position U4500 of the 28S rRNA has also been detected.20,59 It is suggested, however, that in humans both the 18S and 28S rRNAs carry additional modifications, which are not found in yeast.20
The enzymes responsible for introducing the base methylations have been identified in yeast and humans (Table 1; Fig. 2A): Bud23/WBSCR22 (m7G1575/m7G1639),60,61 Dim1/DIMT1L (A1781/A1850 and A1782/A1851),62,63 Rrp8/Bmt1/NML (m1A645/m1A1309),64,65 Bmt2 (m1A2142),66 Rcm1/Bmt3/NSUN5/WBSCR20 (m5C2278/m5C3761),67-69 Nop2/Bmt4/NSUN1/NOL1 (m5C2870/m5C4413/4),69,70 Bmt5 (m3U2634)71 and Bmt6 (m3U2843).71 These RNA methyltransferases belong to the Rossman-like fold family and use S-adenosylmethionine (SAM) as a methyl group donor. In contrast, the multi-step modification of U1191 is initiated by the H/ACA box snoRNP snR35, which catalyzes a pseudouridylation that is an essential pre-requisite for N1-methylation of this residue by the SPOUT class RNA methyltransferase Emg1/Nep1 (Fig. 2B).72-76 Recently, it has been discovered that the final step in this modification pathway, the addition of an aminocarboxypropyl group to the N3 position, is mediated by Tsr3 (Fig. 2B).77 Analysis of the crystal structure of the archaeal homolog of Tsr3 has indicated that, similar to Emg1, this enzyme has a SPOUT fold and utilises SAM, but that the distinctive mode of SAM binding ensures that the acp moiety rather than a methyl group is transferred to the m1ψ1191 substrate by Tsr3.77 Furthermore, the classical Gcn5-related acetyltransferase Kre33/Rra1 (NAT10 in humans) has recently been identified as the acetyltransferase responsible for acetylation of the cytosines 1280 and 1773 in helices 34 and 45 of the 18S rRNA.78,79 Unlike the other base-modifying enzymes, which appear to function exclusively in ribosome biogenesis, Kre33/Rra1/NAT10 also acetylates position 12 of tRNASer and tRNALeu, although it requires a specific cofactor, Tan1/THUMPD1 for this function.79
Figure 2.
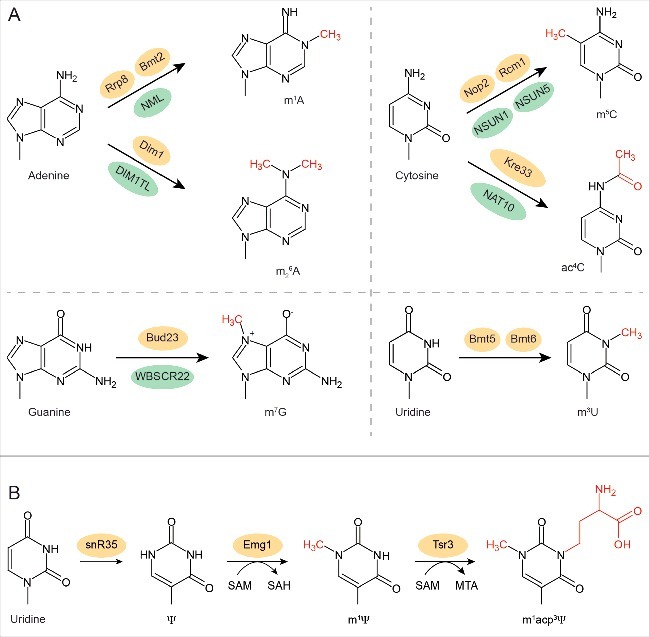
Base modifications in rRNA and the enzymes that install them. (A) Chemical structures of the 4 nucleotides and the modifications that are added in yeast rRNAs. The additional chemical groups are marked in red and the enzymes that introduce them in yeast are indicated above the arrow (yellow) and in humans below the arrow (green). (B) Three-step modification pathway for U1191 of the 18S rRNA in yeast.
Some of the enzymes required for rRNA base modifications in yeast are conserved in higher eukaryotes, while others appear to lack (Bmt2, Bmt5 and Bmt6) human counterparts.80 Interestingly, the enzymes for which human orthologues are known are essential in yeast, while those that do not have human counterparts are dispensable. This could suggest that the modifications catalyzed by the non-conserved enzymes play less important roles in ribosome function. Several of the enzymes (e.g., Dim1/DIM1TL, Emg1/EMG1, Bud23/WBSCR22) that are conserved in evolution appear, however, to have additional functions in ribosomal subunit assembly as complementation assays using catalytically defective mutants have demonstrated that the presence of these proteins in pre-ribosomal complexes, rather than their catalytic activity, is required for pre-rRNA processing.60-63,72,74 Nevertheless, this is not always the case as significant defects in pre-60S biogenesis and pre-rRNA processing were observed in yeast strains expressing a catalytically impaired mutant of Nop2.69 In both yeast and humans, Bud23/WBSCR22 is required for pre-rRNA processing steps that are necessary for nuclear export of pre-SSU complexes,60,61 while in yeast, Emg1 has been suggested to be required for the recruitment of the ribosomal protein eS19 (Rps19) into pre-SSU complexes.81 The finding that in addition to catalyzing modifications, several rRNA methyltransferases are involved in other aspects of ribosomal subunit assembly raises the possibility that the incorporation of the base modifications is coordinated with other steps in the assembly pathway, such as structural remodelling and transport of pre-ribosomal complexes.
Functions of rRNA modifications
Strikingly, neither the snoRNA-guided modifications nor the base modifications introduced by stand-alone enzymes are evenly distributed over the ribosome, but instead they cluster in functionally important regions including the decoding and tRNA binding sites (the A-, P- and E-sites), the peptidyltransferase center and the intersubunit interface (Fig. 3).6,82 This distinctive spatial distribution, which has been conserved throughout evolution,17,19 suggests that rRNA modifications play important roles in regulating ribosome function.82
Figure 3.
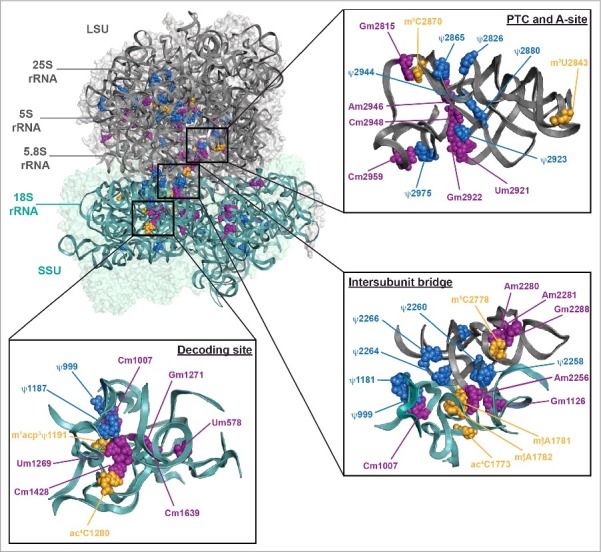
Distribution of rRNA modifications on the yeast ribosome. The S. cerevisiae 80S ribosome (PBD 4V88)6 is shown – 40S in teal and 60S in gray. The positions of 2′-O-methylations (purple), pseudouridines (blue) and base modification installed by stand-alone enzymes (orange) are indicated. Three functionally important regions of the ribosome, the peptidyltransferase center (PTC), the decoding site and the intersubunit bridge eB14, are also shown in a magnified view.
Although early studies using catalytically inactive Nop1 and Cbf5 mutants demonstrated the global importance of 2′-O-methylation and pseudouridylation for cell growth and ribosome production,39,83 loss of only a few individual modifications was found to significantly impact cell viability or ribosome function. A screen for phenotypes in cells depleted of each box C/D snoRNA individually revealed only subtle defects,84 while the presence of the H/ACA box snoRNA snR191 that guides the modification of 2 highly conserved pseudouridylations (Ψ2258 and Ψ2260) in the intersubunit bridge was found to confer a slight growth advantage compare with a deletion strain in a competition assay.85 In contrast, deletion of snR35, which initiates the hypermodification of 18S-U1191, has been shown to impair SSU biogenesis significantly.73 Similarly, expression of a catalytically inactive version of Spb1 strongly affects cell growth and ribosome structure, which is likely to reflect the importance of the Gm2922 modification, but may also be due to the redundant function of Sbp1 in formation of Um2921 as the growth defects observed in strains expressing catalytically inactive Sbp1 have been reported to be exacerbated by deletion of snR52.86
An elegant series of studies conducted by the Fournier lab and others, involving deletions of clusters of modifications in specific functional regions of the ribosome, demonstrated that the modifications instead act in a cumulative manner as significant phenotypes were often only observed upon loss of multiple modifications.45,73,86-88 Many of the strains in which multiple snoRNAs were deleted showed more severe growth defects and were more sensitivity to ribosome-targeting antibiotics than strains lacking individual snoRNAs. Furthermore, analysis of the global translation rate in these strains by monitoring 35S-labeled amino acid incorporation revealed that 2′-O-methylations and ψ modifications at the P-site and in the vicinity of A-site play an important role in maintaining efficient translation.73 Similarly, removal of 3 or 4 modifications in helix (H)69 (a part of the intersubunit bridge B2a), or 6 ψs in the PTC caused a significant decrease in the translation rate.45,88 The importance of these clusters of modifications in translation fidelity has also been investigated and lack of subsets of modifications in the decoding center was shown to impair stop-codon termination and affect reading frame maintenance.87 Base methylations have also been implicated in translation as defects in protein synthesis were observed in cells expressing a catalytically defective form of Dim1,89 the loss of Rcm1 was shown to reduce translation fidelity68 and Emg1 mutants display increased sensitivity to ribosome-targeting antibiotics that influence translation.90,91
Excitingly, in addition to maintaining the accuracy and efficiency of global protein synthesis, the functions of rRNA modifications in translation may also extend to include roles in regulating the translation of particular subsets of mRNAs. For example, alterations in rRNA pseudouridylation have been reported to affect translation initiation on specific mRNAs containing internal ribosome entry sites (IRES) by altering the affinity of the ribosome for these mRNA structures.92-94 Similarly, loss of m5C2278 modification by Rcm1 has been shown to modulate the translation of a subset of mRNAs involved in the oxidative stress response by promoting their recruitment to polysomes.68 The implications of these findings and their relevance in disease are just beginning to emerge (see below).
The basis of how rRNA modifications affect ribosome function lies in their capacity to expand the topological potential of specific nucleotides. It is generally thought that the chemical modifications present in rRNA serve to stabilize its secondary and tertiary structures.82,95 In line with this, 2′-O-methylation, which is highly abundant in rRNA, stabilises helices by increasing base-stacking. Similarly, pseudouridylation, which occurs at numerous positions in rRNAs, confers greater hydrogen bonding potential than uridine and also enhances the rigidity of the sugar-phosphate backbone. The base modifications found in rRNA exert similar stabilizing effects on RNA structure but also have distinctive properties. For example, N7-methylation of guanine generates a quaternary nitrogen that increases the positive charge of the nucleotide thereby promoting ionic interactions between RNAs and proteins, methylation of uridine at the N3 position promotes hairpin formation and the introduction of m5C increases the stability of base-pairing with guanine.96-98
The effect of clusters of modifications on local rRNA structure within the ribosome has been monitored directly by chemical probing techniques. In dimethyl sulfate (DMS) structure probing experiments, lack of a methyl group on U2264 was found to affect the accessibility of A2252 with which U2264 normally base-pairs in H69 of the intersubunit bridge B2a.88 Importantly, as snoRNAs can also contribute to rRNA folding, this study also included snoRNA mutants that are able to base-pair with the rRNA sequences but which do not guide methylation, enabling the direct influence of the modifications themselves on the RNA structure to be ascertained. Similarly, loss of 6 Ψs in the PTC or the deletion of both snR52 and Spb1 alters the protection pattern observed, indicating structural changes in the corresponding regions of the PTC.45,86 Structural changes observed in ribosomes lacking both the m5C2278 base modification and the ribose methylation at G2288 have been shown to cause decreased ribosome stability and loss of ribosomal proteins from the LSU.67 This link between rRNA modification and ribosome integrity is consistent with an earlier report that loss of a cluster of modifications in H69 decreases rRNA levels as a result of increased RNA turnover.88
In addition to their functions in modulating local rRNA structure, rRNA modifications are proposed to participate in communication between distant regions of the ribosome. On one hand, this is likely to result from alterations in the dynamics of rRNA folding and ribosome assembly, as loss of an individual snoRNA can affect the extent of modification not only at its target site but also at other positions on the ribosome.18 On the other hand, long-range effects have also been suggested to arise from interactions between rRNA modifications and ribosomal proteins.15 For example, the m5C2278 modification in the LSU and the ac4C1773 and A1781- A1782 modifications in the SSU are directly contacted by the ribosomal protein eL41 (Rpl41), which forms a bridge between the LSU to the decoding center in the SSU and conducts structural information between the subunits.6 Interestingly, this ribosomal protein also acts as a pivot for 40S subunit rotation during translation, suggesting a mechanism by which monitoring for the presence of rRNA modifications is coupled with translation efficiency via ribosomal protein interactions.15,99
In summary, the minimal set of chemical modifications found in rRNA and the specific sites of rRNA modification appear to have been carefully selected during evolution to allow formation of a stable ribosome structure. A core set of modified sites, conserved from prokaryotes, likely render the ribosome capable of efficient and accurate translation, while the additional modifications found in higher organisms might not only reflect the increased size and complexity of the eukaryotic ribosome structure, but might also enable fine-tuning of the eukaryotic translation cycle to modulate gene expression.
Timing and regulation of rRNA modifications
rRNA modifications are introduced at different stages during the maturation of the ribosomal subunits. Kinetic labeling in yeast has revealed that the vast majority of 2′-O-methylations in the 18S rRNA are introduced co-transcriptionally, while such methylations are introduced both co- and post-transcriptionally into the 25S rRNA.100 A recent study comparing the 2′-O-methylation status of chromatin-associated RNA with that of total RNA supported this conclusion and also provided site-specific information about the timing of individual modifications.18 In the 18S rRNA, only one modification, Am100 occurs after release of the nascent transcript from the rDNA, while the extent of 2′-O-methylation of A817, G867, A876, A2256, U2421 and A2640 was significantly higher in the mature 25S rRNA than in the chromatin-associated RNA. Another notable exception is the 2′-O-methylation of G2922 by the stand-alone methyltransferase Sbp1, which has been reported to occur during maturation of the 27S precursor rRNA.37 Consistent with this, the recent structure of a 90S pre-ribosomal complex from Chaetomium thermophilum did not contain any snoRNPs (except the U3 snoRNP, which does not mediate rRNA modification), implying that most snoRNPs have already dissociated from the pre-rRNA transcript when it is released from the rDNA.101 The finding that snoRNP-mediated modifications are largely introduced during the early stages of ribosome biogenesis, when the pre-ribosomal complexes are thought to have a more open structure, is fitting with the snoRNA-rRNA base-pairing mechanism by which Nop1 finds its target sites and may also reflect the secondary function of snoRNAs in assisting rRNA folding. By analogy, it is assumed that H/ACA box snoRNP-mediated pseudouridylations also occur at an early stage of ribosome biogenesis but this has not been experimentally demonstrated so far. It is also not known if pseudouridylation of the 5S rRNA by Pus7 takes place before or after integration of the 5S RNP (consisting of the 5S rRNA and the ribosomal proteins uL5 (Rpl11) and uL18 (Rpl5)) into the pre-LSU.
In human cells, the majority of snoRNA-guided modifications are also likely to occur on early pre-ribosomal complexes, however, some snoRNAs have been shown to associate with later pre-SSU particles.102 Remarkably, the modifications guided by these snoRNAs lie within the binding site of the RNA helicase DDX21.102 This suggests a model in which the activity of the helicase is required for structural rearrangement of pre-ribosomal particles to enable these late-acting snoRNAs to access their pre-rRNA target sites. In yeast, the RNA helicase Prp43 has also been implicated in enabling snoRNA recruitment, as the levels of snR64 and snR67 on pre-ribosomes are reduced when Prp43 is depleted.103 Furthermore, decreased 2′-O-methylation of C2377, the target nucleotide of snR64, is observed when a catalytically inactive version of Prp43 is expressed.104
Taken together, the findings that most 2′-O-methylations and pseudouridylations occur during the short timeframe of pre-rRNA transcription, that these modifications are densely clustered, and that snoRNAs form extensive and often overlapping base-pairing interactions with the rRNA sequences, imply that in many cases individual modifications must be introduced in a stepwise manner. However, whether the association of particular snoRNAs with their pre-rRNA base-pairing sites occurs stochastically or if there is a defined hierarchy for snoRNA recruitment to pre-ribosomal complexes currently remains unclear. It has been shown, however, that knock-in of an artificial snoRNA that guides a 2′-O-methylation at the P-site and which has overlapping base-pairing sites with natural snoRNAs that guide 3 neighboring methylations reduces the adjacent modifications.18 This suggests that in some cases, snoRNAs may compete for their pre-rRNA basepairing sites and that the presence of one snoRNA can impede the modification of a nearby site guided by another snoRNA. Similarly, snR9 and snR65 have been suggested to compete for access to U2347, leading to the formation of Um, Ψ and Ψm at this site.21
In contrast to the 2′-O-methylations and pseudouridylations that are mostly introduced during the early stages of ribosomal subunit maturation, base modifications are generally thought to occur later, but the precise timing of most of them has not been determined so far. In the case of the N3-acp modification of 18S-m1Ψ1191, however, the exclusively cytoplasmic localization of the Tsr3 enzyme that installs this modification clearly identifies this as a late event in yeast.77 Furthermore, early cytoplasmic pre-40S complexes (isolated via the shuttling biogenesis factors Enp1 and Ltv1) show low levels of N3-acp modification of 18S-m1Ψ1191, while in later particles (isolated via Nob1 and Rio1), this position is acp-modified to a similar extent as the mature 18S rRNA, indicating that the final step in the hypermodification pathway of U1191 occurs just before cleavage of the 3′ end of the 18S rRNA by Nob1.105,106 Similarly, the A1781 A1782 dimethylation close to the 3′ end of the 18S rRNA is introduced by Dim1 after the export of pre-SSU complexes to the cytoplasm, but in contrast to Tsr3, Dim1 is already bound to early pre-ribosomal complexes in the nucleolus.62 It is suggested that the association of Dim1 with nucleolar pre-ribosomes acts as a surveillance mechanism in which pre-ribosomes that cannot be dimethylated, and will therefore be impaired in translation, are targeted for degradation.89 Furthermore, structural analysis of the pre-ribosomal binding site of Dim1 demonstrated that the presence of the RNA methyltransferase on the SSU inhibits its association with the LSU.107,108 This suggests that the presence of Dim1 in late pre-ribosomal complexes may serve a second quality control function by preventing premature subunit joining and immature ribosomal complexes engaging in translation. Interestingly, modification of the corresponding positions in human cells by DIMTL1 occurs in the nucleus, although the relevance of this difference in timing of the modification is currently unknown.63 While the 18S-m7G1575 modification is installed by Bud23 in the nucleus, it was recently demonstrated that this modification occurs on the 20S pre-rRNA during the later stages of pre-SSU biogenesis.109 So far, Bud23 is the only rRNA methyltransferase that has been shown to require a cofactor (Trm112) to achieve its modification. In the Bud23-Trm112 complex, rather than stimulating the catalytic activity of Bud23, the cofactor is required for stability of the methyltransferase.109,110 Furthermore, Bud23-Trm112 binds directly to the RNA helicase Dhr1111 and, similar to the proposed role of RNA helicases in enabling some snoRNAs to access their basepairing sites, Dhr1 is required for recruitment of Bud23-Trm112 to pre-ribosomal complexes.109 Efficient acetylation of the 18S rRNA by Kre33 has also been shown to involve helicase action, but in this case the activity is derived from a RecA domain within the acetyltransferase itself.79
The alternative mechanisms (snoRNA-guided and protein-only) by which rRNA modifying enzymes recognize their targets are broadly employed to introduce modifications at different stages during ribosome maturation. Although little is known about how stand-alone enzymes identify their sites of action, it is possible that sequence specificity, RNA secondary structures or multiple contacts in the broader context of the pre-ribosome are required for target identification and/or catalysis. Furthermore, an emerging principle that applies to both snoRNA-guided modifications and those installed by stand-alone enzymes is that late-acting enzymes or snoRNPs often require the coordinated action of an RNA helicase to enable target site access. The highly structured nature of pre-ribosomal complexes, compare with other RNAs/RNPs that are subjected to modification, may make this close co-ordination between modifying enzymes and helicases uniquely important for rRNA modifications.
Partially modified sites in rRNA
An exciting development in the field of RNA modification arose recently with the detection of sites of partial (also termed fractional or substoichiometric) modification in rRNA, challenging the long-held belief that rRNA modifications are constitutively present.18,21,112 While under normal growth conditions all the positions carrying base modifications (except Ψ) are almost fully modified, sites of partial 2′-O-methylation and pseudouridylation have been identified.18,21,113 More specifically, it was recently reported that of the 112 known sites of modification in yeast, 18 are modified on less than 85% of ribosomes.21 Similarly, in human cell lines, approximately one-third of 2′-O-methylations appear to be substoichiometric.32 These observations highlight rRNA modification as a major source of ribosome heterogeneity and contribute to the mounting evidence for specialized ribosomes. Diversity among ribosomes on the rRNA level has long been known, because alternatively processed forms of the 5.8S rRNA are present in polysome-associated large ribosomal subunits, however, it is suggested that this is a constitutive difference rather than a dynamic one.114 Differences in ribosome composition also arise through the incorporation of alternative ribosomal protein isoforms (which exist due to ribosomal protein gene duplication) and ribosomal proteins that have undergone differential post-translational modifications such as phosphorylation, ubiquitination and acetylation (for a review see refs. 115–118).
The major question of what determines the extent of modification of these sites still remains open and it is likely that several alternative mechanisms exist. It is known that snoRNA levels can vary in cells, so one possibility is that limiting amounts of certain snoRNAs can determine the extent of 2′-O-methylation or pseudouridiylation at specific positions. Consistent with this model, the substoichiometric 2′-O-methylation of the 18S rRNA at position A100 has been shown to correlate with the low cellular levels of snR51 that guides this modification as overexpression of the snoRNA leads to complete methylation of 18S-A100.113 Another possibility is that the assembly of specific snoRNAs into functional snoRNPs is restricted as “activation steps,” including the removal of a placeholder assembly factor, have been identified in the assembly pathways of both box C/D and box H/ACA snoRNPs.114,119,120 Also, structural elements within the snoRNAs guiding these modifications may share a common feature that limits the extent of modification. For box C/D snoRNAs in HeLa cells, this possibility was examined in detail considering both the conservation of the D/D′ motifs as well as the “strength” of the guiding sequences and no clear correlations could be detected.32 As snoRNP-mediated substoichoimetric modifications mostly occur co-transcriptionally, partial modifications might arise due to changes in the accessibility of target or base-pairing sites during the highly dynamic process of ribosome maturation. Alternative rRNA folding pathways might result in modifications not taking place if particular regions of the ribosome have already acquired stable structures or ribosomal proteins have been assembled. This notion is supported by the observation that several partially modified sites are in close proximity (e.g., 18S-Am438 (73%) and 18S-Ψ468 (60%); 25S-Gm867 (78%) and 25S-Am876 (75%))21 but this finding is also in line with a model where competition between snoRNAs for base-pairing sites might limit the extent of modification in densely modified regions of the rRNA. Comparison to the recently described dynamic and reversible modification of other RNA species including mRNAs by demethylases such as FTO/ALKBH5121 also raises the possibility that some partial rRNA modifications might be the result of selective removal. To date, however, enzymes capable of “erasing” Ψ or Nm have not been identified and it is very likely that the complex tertiary structure and protein-rich surface of the ribosome would impede the access of such erasers to modified residues that are mostly buried in the functional core of the ribosome.
It is not yet clear how different rRNA modification patterns affect ribosome function. It has been proposed that the constitutively modified positions, which are generally the most conserved, serve to stabilize the core of the ribosome, while the positions that are modified in some species but not others contribute to fine-tuning of ribosome function for optimal translation accuracy.32 It will also be interesting to see if the subpopulations of differently modified ribosomes perform specific functions in the cell. For example, localized translation at the endoplasmic reticulum might be driven by a subset of ribosomes with a particular rRNA modification pattern.
Intriguingly, as well as variations in the extent of modification of specific sites in rRNAs under normal growth conditions, the degree of modification at some positions has been found to change in response to environmental changes. For example, after diauxic shift a 2-fold change in the extent of 25S-Ψ2314 was detected122 and heat-shock was observed to alter the fraction of 5S-Ψ50 that is modified by Pus7.123 The latter observation might, however, reflect a change in the subcellular localization of Pus7 or the distribution of this multifunctional enzyme between its rRNA and mRNA targets, rather than a direct role for 5S-Ψ50 in optimising ribosome function under these conditions. Interestingly, sites of inducible pseudouridylation have been identified in snRNAs upon nutrient stress,124,125 raising the possibility that additional modifications may likewise be incorporated into some rRNAs under certain physiological conditions. Together, these findings suggest that partial modification of rRNA nucleotides may not only optimise the function of different populations of ribosomes but that these modifications can also be regulated, which could allow adaption of translation and the cellular proteome in response to specific intracellular or environmental cues.
rRNA modifications in development and disease
An increasing body of evidence links defects in components of the rRNA modification machinery and changes in the extent of rRNA modifications to development, genetic diseases and cancer. For example, loss-of-function of any of 3 individual snoRNAs has been found to cause profound developmental defects in zebrafish.126 Examples of disease-associated abnormalities that affect stand-alone rRNA-modifying enzymes include a point mutation in the EMG1 gene that causes Bowen-Conradi syndrome and deletion of a chromosomal region encompassing the WBSCR22 and WBSCR20/NSUN5 genes in Williams-Beuren syndrome.127-129 In addition, NML is associated with high-fat diet-induced obesity and NSUN5 is reported to increase lifespan and stress resistance in some organisms.68,130 It is, however, unclear whether the lack of the modifications mediated by these enzymes is the cause of pathogenicity or whether these diseases could also be attributed to a general decrease in ribosome levels or deletion of other essential genes in the vicinity. Nevertheless, it is also possible that loss of the modifications introduced by these enzymes affects ribosome function, leading to changes in translation and the cellular proteome, thereby indirectly causing the disease phenotype.
Furthermore, changes in the expression levels and activity of core components of both box H/ACA and box C/D snoRNPs in disease have been reported. For example, the box C/D snoRNP component NOP56 is overexpressed in Burkitt's lymphoma-associated c-Myc mutants131 and NOP58 mRNA levels are elevated in metastatic melanoma lesions,132 making these proteins good markers of cancer progression.133,134 It is possible, however, that these changes in protein expression level might primarily reflect an upregulation of ribosome synthesis during cell proliferation. Similarly, many snoRNAs are dysregulated in cancers (for a review see refs. 135,136), but whether this actively promotes tumorigenesis remains unknown, and if so, it is unclear whether this is due to a change in rRNA modification.
Remarkably, mutations in genes encoding the catalytic snoRNP proteins and changes in their expression level have been linked to widespread changes in the rRNA modification pattern that are reported to influence ribosome function and thereby cause tumorigenesis. For example, mutations in the box H/ACA pseudouridine synthetase dyskerin are found in X-linked Dyskeratosis congenita (X-DC), a severe disorder that is characterized by bone marrow failure, lung fibrosis and increased susceptibility to cancer.137 These mutations reduce rRNA pseudouridylation138,139 and although this does not change the overall rate of protein synthesis, it is suggested to selectively influence the translation of a specific subset of mRNAs that contain IRES elements. Interestingly, in X-DC, the translation of mRNAs coding for the tumor suppressor proteins p53 and p27 as well as the anti-apoptotic factors Bcl-xL and XIAP is impaired,94,140,141 while IRES-dependent translation of the growth factor VEGF mRNA is promoted.142 Together, these finding suggest that the altered rRNA pseudouridylation profile caused by the dyskerin mutations may re-program the ribosome to promote tumorigenesis, thereby explaining the increased cancer susceptibility of X-DC patients.
Changes in the rRNA 2′-O-methylation pattern have also been linked to cancer and differences in the extent of rRNA modification at several sites have been observed in different cancer cell lines.32 Mutations in TCOF1/treacle that are found in Treacher Collins syndrome, a ribosomopathy that predisposes patients to cancer, have been reported to impair 2′-O-methylation of rRNA.143 Furthermore, in mammary epithelial cells, expression of the box C/D methyltransferase fibrillarin was suggested to be regulated by the tumor suppressor p53 and fibrillarin-dependent defects in translational fidelity were described in cells lacking p53.144 Reciprocally, depletion of fibrillarin has been proposed to cause an increase in p53 expression levels145 and surprisingly, this response was suggested to not only be mediated by the nucleolar stress response pathway involving binding of the 5S RNP to HDM2 when ribosome assembly is perturbed,146,147 but also by an increase in the cap-independent translation of the p53 mRNA.145 Also, an RNAi-based synthetic interaction screen identified the gene encoding the box C/D snoRNP assembly chaperone NOLC1/NOPP140 as a p53 target gene,148 indicating that regulation of snoRNP biogenesis might be an alternative mechanism by which p53 could regulate rRNA 2′-O-methylation and cellular proliferation.
Many studies have highlighted the central role of ribosome synthesis and function in disease.149 Ribosome assembly is regulated by key proto-oncogenes such as p14ARF and c-MYC150 and defects in ribosome assembly, including those found in ribosomopathies, regulate the tumor suppressor p53.151 Several recent reports indicate, however, that during development and disease, both global and site-specific alterations in rRNA modification may be induced by key cell signalling pathways and also that changes in the rRNA modification pattern might be important regulators of cellular transformation.
Concluding remarks
Since the first mapping of pseudouridines in eukaryotic rRNA more than 20 years ago, much has been revealed about the types of modifications present on the rRNAs, their number and positions, the enzymes that install them and their functions. Interestingly, despite the presence of modified rRNA nucleotides in all 3 phylogenetic kingdoms, the relative amounts of different classes of modification vary considerably throughout evolution. In bacteria, the majority of rRNA modifications are base methylations and a greater diversity of modifications are present in bacterial ribosomes than in ribosomes of any other taxa.152 In contrast, in thermophillic archaea (such as Sulfolobus solfataricus) and in eukarya, 2′-O-methylations exceed base methylations by almost 10-fold.21,153 This difference is likely to reflect the key role of 2′-O-methylation in stabilization of RNAs, which is important for the structural fidelity of archaeal ribosomes at higher temperatures and the larger eukaryotic ribosomes. Furthermore, the mechanisms by which 2′-O-methylations and pseudouridylations are introduced have diverged during evolution; in bacterial ribosomes they are introduced by stand-alone enzymes, whereas these modifications are mediated by sRNPs and snoRNPs in archaea and eukarya respectively. It is possible that this difference may arise due to the increase in the number of such modifications in these species and therefore the need for a strategy by which a single modifying enzyme can be directed to modify multiple specific sites within the rRNAs. To date, no rRNA modifications have been detected in the spacer fragments that separate the mature rRNA sequences in the pre-rRNA transcripts, but instead, in all species, rRNA modifications are found in the mature rRNAs at positions that form functionally relevant sites in the ribosome. Indeed, the structures of prokaryotic and eukaryotic ribosomes have enabled vizualization of the clustering of modifications in functionally important regions of the ribosome, such as the peptidyltransferase center and decoding site, paving the way for the identification of the collective and conserved role of these modifications in enabling efficient and accurate translation.
The development of sensitive, unbiased, systematic methods for the identification of rRNA modifications has yielded updated inventories of the sites of rRNA modification in eukaryotes. These methods, combined with the advanced computational predictions of snoRNA base-pairing sites and yeast genetics, have provided a comprehensive picture of the stand-alone enzymes and snoRNP complexes responsible for installing these modifications in yeast. We have also gained substantial insights into the diverse timing of rRNA modifications and an understanding of how some modifying enzymes are regulated. Notably, enzymes that install modifications during the later stages of ribosome assembly often require the coordinated action of an RNA helicase, presumably to facilitate access to their target sites on a compact pre-ribosome in which many complex rRNA folds present in the mature ribosome have already been established.
Most recently, the identification of sites of partial modification has overturned the view that all rRNA modifications are constitutive features and highlighted them as an important source of ribosome diversity, alongside differential protein composition and alternative pre-rRNA processing pathways. The ribosome heterogeneity arising from partial modifications, together with the emerging concept that alterations in the rRNA modification profile may selectively adapt the ribosome to translate specific subsets of mRNAs, could suggest the existence of a new layer of translational control of gene expression. This additional level of regulation of gene expression may be particularly relevant in higher eukaryotes as during evolution there has been relative increase in the number of pseudouridylations and 2′-O-methylations, which can be substoichiometric, compare with base modifications that, at least under optimal growth conditions, appear to be constitutive. In the future, it will be very interesting to see whether the ribosome sub-populations characterized by particular rRNA modification patterns perform specific functions within the cell, for example in localized translation or in promoting/repressing translation of particular subsets of mRNAs. It will also be important to explore potential changes in the rRNA modification pattern in different cell types (in particular in primary cells and tissues), in response to environmental and developmental cues, and in disease, as this will provide a basis for understanding the role of rRNA modifications in regulating translation in order to adapt the cellular proteome.
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1784: BO3442/2-1 to M.T.B., EN13/12-1 and EN134/13-1 to K.D.E.; and SFB860 to M.T.B.), the Alexander von Humboldt Foundation (post-doctoral fellowship to K.E.S. and M.T.B), the European Molecular Biology Organisation (post-doctoral fellowship ALTF644-2014 to S.S.), the Université Libre de Bruxelles (ULB), the Fonds National de la Recherche (F.R.S/FNRS), the Walloon Region (DGO6), the Fédération Wallonie-Bruxelles, and the European Research Development Fund (ERDF) (to D.L.J.L.), the State of Hesse and Goethe University Frankfurt (to K.-D.E.) and the Faculty of Medicine, Georg-August-University Göttingen (M.T.B).
References
- 1.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res 2011; 39:D195-201; PMID:21071406; https://doi.org/23118484 10.1093/nar/gkq1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al.. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; https://doi.org/ 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011; 2:611-31; PMID:21823225; https://doi.org/ 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- 4.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: Form, distribution, and function. Science 2016; 352:1408-12; PMID:27313037; https://doi.org/ 10.1126/science.aad8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev RNA 2013; 4:35-48; PMID:23139145; https://doi.org/ 10.1002/wrna.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 2011; 334:1524-9; PMID:22096102; https://doi.org/ 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 7.Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature 2015; 520:640-5; PMID:25901680; https://doi.org/ 10.1038/nature14427 [DOI] [PubMed] [Google Scholar]
- 8.Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 2015; 6:225-42; PMID:25346433; https://doi.org/ 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woolford JL Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013; 195:643-81; PMID:24190922; https://doi.org/ 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Cruz J, Karbstein K, Woolford JL Jr. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annual Rev Biochem 2015; 84:93-129; PMID:25706898; https://doi.org/24817020 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerhardy S, Menet AM, Pena C, Petkowski JJ, Panse VG. Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma 2014; 123:327-44; PMID:24817020; https://doi.org/ 10.1007/s00412-014-0463-z [DOI] [PubMed] [Google Scholar]
- 12.Sloan KE, Gleizes PE, Bohnsack MT. Nucleocytoplasmic Transport of RNAs and RNA-Protein Complexes. J Mol Biol 2016; 428:2040-59; PMID:26434509; https://doi.org/ 10.1016/j.jmb.2015.09.023 [DOI] [PubMed] [Google Scholar]
- 13.Thomson E, Ferreira-Cerca S, Hurt E. Eukaryotic ribosome biogenesis at a glance. J Cell Sci 2013; 126:4815-21; PMID:24172536; https://doi.org/ 10.1242/jcs.111948 [DOI] [PubMed] [Google Scholar]
- 14.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 2013; 51:539-51; PMID:23973377; https://doi.org/ 10.1016/j.molcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 15.Sharma S, Lafontaine DL. ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem Sci 2015; 40:560-75; PMID:26410597; https://doi.org/ 10.1016/j.tibs.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 16.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012; 3:397-414; PMID:22065625; https://doi.org/ 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 17.Polikanov YS, Melnikov SV, Soll D, Steitz TA. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 2015; 22:342-4; PMID:25775268; https://doi.org/ 10.1038/nsmb.2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem 2015; 54:451-5; PMID:20174677; https://doi.org/16381836 10.1002/anie.201408362 [DOI] [PubMed] [Google Scholar]
- 19.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 2006; 34:D158-62; PMID:16381836; https://doi.org/ 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res 2008; 36:D178-83; PMID:17947322; https://doi.org/ 10.1093/nar/gkm855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taoka M, Nobe Y, Yamaki Y, Yamauchi Y, Ishikawa H, Takahashi N, Nakayama H, Isobe T. The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res 2016; pii:gkw564; PMID:27325748; https://doi.org/3783688 10.1093/nar/gkw564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maden BE. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J Mol Biol 1986; 189:681-99; PMID:3783688; https://doi.org/ 10.1016/0022-2836(86)90498-5 [DOI] [PubMed] [Google Scholar]
- 23.Maden BE. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J Mol Biol 1988; 201:289-314; PMID:3418702; https://doi.org/7599273 10.1016/0022-2836(88)90139-8 [DOI] [PubMed] [Google Scholar]
- 24.Maden EH, Wakeman JA. Pseudouridine distribution in mammalian 18 S ribosomal RNA. A major cluster in the central region of the molecule. Biochem J 1988; 249:459-64; PMID:3342024; https://doi.org/7599273 10.1042/bj2490459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 1995; 77:22-9; PMID:7599273; https://doi.org/ 10.1016/0300-9084(96)88100-4 [DOI] [PubMed] [Google Scholar]
- 26.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 1993; 32:9754-62; PMID:8373778; https://doi.org/ 10.1021/bi00088a030 [DOI] [PubMed] [Google Scholar]
- 27.Bakin A, Lane BG, Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry 1994; 33:13475-83; PMID:7947756; https://doi.org/ 10.1021/bi00249a036 [DOI] [PubMed] [Google Scholar]
- 28.Bakin A, Ofengand J. Mapping of the 13 pseudouridine residues in Saccharomyces cerevisiae small subunit ribosomal RNA to nucleotide resolution. Nucleic Acids Res 1995; 23:3290-4; PMID:7545286; https://doi.org/ 10.1093/nar/23.16.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Incarnato D, Anselmi F, Morandi E, Neri F, Maldotti M, Rapelli S, Parlato C, Basile G, Oliviero S. High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. Nucleic Acids Res 2016; pii:gkw810; PMID:27614074; https://doi.org/27302133 10.1093/nar/gkw810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res 2016; 44:e135; PMID:27302133; https://doi.org/ 10.1093/nar/gkw547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Sharma S, Kotter P, Entian KD. Identification of a new ribose methylation in the 18S rRNA of S. cerevisiae. Nucleic Acids Res 2015; 43:2342-52; PMID:25653162; https://doi.org/9182768 10.1093/nar/gkv058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogh N, Jansson MD, Hafner SJ, Tehler D, Birkedal U, Christensen-Dalsgaard M, Lund AH, Nielsen H. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res 2016; 44(16):7884–7895; PMID:27257078; https://doi.org/9182768 10.1093/nar/gkw482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997; 89:799-809; PMID:9182768; https://doi.org/ 10.1016/S0092-8674(00)80263-9 [DOI] [PubMed] [Google Scholar]
- 34.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 1996; 85:1077-88; PMID:8674114; https://doi.org/ 10.1016/S0092-8674(00)81308-2 [DOI] [PubMed] [Google Scholar]
- 35.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997; 89:565-73; PMID:9160748; https://doi.org/ 10.1016/S0092-8674(00)80238-X [DOI] [PubMed] [Google Scholar]
- 36.Decatur WA, Schnare MN. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Mol Cell Biol 2008; 28:3089-100; PMID:18332121; https://doi.org/ 10.1128/MCB.01574-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol Cell 2004; 16:663-9; PMID:15546625; https://doi.org/ 10.1016/j.molcel.2004.10.022 [DOI] [PubMed] [Google Scholar]
- 38.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev 1998; 12:527-37; PMID:9472021; https://doi.org/ 10.1101/gad.12.4.527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 1993; 72:443-57; PMID:8431947; https://doi.org/ 10.1016/0092-8674(93)90120-F [DOI] [PubMed] [Google Scholar]
- 40.van Nues RW, Granneman S, Kudla G, Sloan KE, Chicken M, Tollervey D, Watkins NJ. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J 2011; 30:2420-30; PMID:21556049; https://doi.org/ 10.1038/emboj.2011.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z, Lin J, Ye K. Box C/D guide RNAs recognize a maximum of 10 nt of substrates. Proc Natl Acad Sci 2016; pii:201604872; PMID:27625427; https://doi.org/10024243 10.1073/pnas.1604872113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science 1999; 283:1168-71; PMID:10024243; https://doi.org/ 10.1126/science.283.5405.1168 [DOI] [PubMed] [Google Scholar]
- 43.Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, et al.. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci 2014; 111:10251-6; PMID:24982194; https://doi.org/ 10.1073/pnas.1407205111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enright CA, Maxwell ES, Eliceiri GL, Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 1996; 2:1094-9; PMID:8903340. [PMC free article] [PubMed] [Google Scholar]
- 45.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 2003; 11:425-35; PMID:12620230; https://doi.org/ 10.1016/S1097-2765(03)00040-6 [DOI] [PubMed] [Google Scholar]
- 46.Martin R, Hackert P, Ruprecht M, Simm S, Bruning L, Mirus O, Sloan KE, Kudla G, Schleiff E, Bohnsack MT. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA 2014; 20:1173-82; PMID:24947498; https://doi.org/ 10.1261/rna.044669.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Nues RW, Watkins NJ. Unusual C′/D' motifs enable box C/D snoRNPs to modify multiple sites in the same rRNA target region. Nucleic Acids Res 2016; PMID:27651461; https://doi.org/15306656 10.1093/nar/gkw842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schattner P, Decatur WA, Davis CA, Ares M Jr., Fournier MJ, Lowe TM. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res 2004; 32:4281-96; PMID:15306656; https://doi.org/ 10.1093/nar/gkh768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kehr S, Bartschat S, Stadler PF, Tafer H. PLEXY: efficient target prediction for box C/D snoRNAs. Bioinformatics 2011; 27:279-80; PMID:21076148; https://doi.org/ 10.1093/bioinformatics/btq642 [DOI] [PubMed] [Google Scholar]
- 50.Schattner P, Barberan-Soler S, Lowe TM. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA 2006; 12:15-25; PMID:16373490; https://doi.org/ 10.1261/rna.2210406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tafer H, Kehr S, Hertel J, Hofacker IL, Stadler PF. RNAsnoop: efficient target prediction for H/ACA snoRNAs. Bioinformatics 2010; 26:610-6; PMID:20015949; https://doi.org/ 10.1093/bioinformatics/btp680 [DOI] [PubMed] [Google Scholar]
- 52.Yoshihama M, Nakao A, Kenmochi N. snOPY: a small nucleolar RNA orthological gene database. BMC Res Notes 2013; 6:426; PMID:24148649; https://doi.org/ 10.1186/1756-0500-6-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci 2011; 108:10010-5; PMID:21610164; https://doi.org/ 10.1073/pnas.1017386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, Zavolan M, Gruber AR. An updated human snoRNAome. Nucleic Acids Res 2016; 44:5068-82; PMID:27174936; https://doi.org/ 10.1093/nar/gkw386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, Nevo Y, Eyras E, Sperling R, Stamm S. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci 2016; 113:E1625-34; PMID:26957605; https://doi.org/ 10.1073/pnas.1519292113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA–novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Let 2013; 340:201-11; PMID:23376637; https://doi.org/23022379 10.1016/j.canlet.2012.11.058 [DOI] [PubMed] [Google Scholar]
- 57.Schubert T, Pusch MC, Diermeier S, Benes V, Kremmer E, Imhof A, Langst G. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol Cell 2012; 48:434-44; PMID:23022379; https://doi.org/ 10.1016/j.molcel.2012.08.021 [DOI] [PubMed] [Google Scholar]
- 58.Sharma E, Sterne-Weiler T, O'Hanlon D, Blencowe BJ. Global Mapping of Human RNA-RNA Interactions. Mol Cell 2016; 62:618-26; PMID:27184080; https://doi.org/ 10.1016/j.molcel.2016.04.030 [DOI] [PubMed] [Google Scholar]
- 59.Klagsbrun M. An evolutionary study of the methylation of transfer and ribosomal ribonucleic acid in prokaryote and eukaryote organisms. J Biol Chem 1973; 248:2612-20; PMID:4633356. [PubMed] [Google Scholar]
- 60.Haag S, Kretschmer J, Bohnsack MT. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 2015; 21:180-7; PMID:25525153; https://doi.org/18332120 10.1261/rna.047910.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol Cell Biol 2008; 28:3151-61; PMID:18332120; https://doi.org/ 10.1128/MCB.01674-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev 1995; 9:2470-81; PMID:7590228; https://doi.org/ 10.1101/gad.9.20.2470 [DOI] [PubMed] [Google Scholar]
- 63.Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgue-Hamard V, Lafontaine DL. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol Biol Cell 2015; 26:2080-95; PMID:25851604; https://doi.org/ 10.1091/mbc.E15-02-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peifer C, Sharma S, Watzinger P, Lamberth S, Kotter P, Entian KD. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res 2013; 41:1151-63; PMID:23180764; https://doi.org/ 10.1093/nar/gks1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waku T, Nakajima Y, Yokoyama W, Nomura N, Kako K, Kobayashi A, Shimizu T, Fukamizu A. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J Cell Sci 2016; 129:2382-93; PMID:27149924; https://doi.org/ 10.1242/jcs.183723 [DOI] [PubMed] [Google Scholar]
- 66.Sharma S, Watzinger P, Kotter P, Entian KD. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res 2013; 41:5428-43; PMID:23558746; https://doi.org/ 10.1093/nar/gkt195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gigova A, Duggimpudi S, Pollex T, Schaefer M, Kos M. A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA 2014; 20:1632-44; PMID:25125595; https://doi.org/ 10.1261/rna.043398.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al.. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun 2015; 6:6158; PMID:25635753; https://doi.org/ 10.1038/ncomms7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma S, Yang J, Watzinger P, Kotter P, Entian KD. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res 2013; 41:9062-76; PMID:23913415; https://doi.org/ 10.1093/nar/gkt679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bourgeois G, Ney M, Gaspar I, Aigueperse C, Schaefer M, Kellner S, Helm M, Motorin Y. Eukaryotic rRNA Modification by Yeast 5-Methylcytosine-Methyltransferases and Human Proliferation-Associated Antigen p120. PloS one 2015; 10:e0133321; PMID:26196125; https://doi.org/ 10.1371/journal.pone.0133321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma S, Yang J, Duttmann S, Watzinger P, Kotter P, Entian KD. Identification of novel methyltransferases, Bmt5 and Bmt6, responsible for the m3U methylations of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res 2014; 42:3246-60; PMID:24335083; https://doi.org/ 10.1093/nar/gkt1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leulliot N, Bohnsack MT, Graille M, Tollervey D, Van Tilbeurgh H. The yeast ribosome synthesis factor Emg1 is a novel member of the superfamily of alpha/beta knot fold methyltransferases. Nucleic Acids Res 2008; 36:629-39; PMID:18063569; https://doi.org/ 10.1093/nar/gkm1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009; 15:1716-28; PMID:19628622; https://doi.org/ 10.1261/rna.1724409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, et al.. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic Acids Res 2011; 39:1526-37; PMID:20972225; https://doi.org/ 10.1093/nar/gkq931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor AB, Meyer B, Leal BZ, Kotter P, Schirf V, Demeler B, Hart PJ, Entian KD, Wohnert J. The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site. Nucleic Acids Res 2008; 36:1542-54; PMID:18208838; https://doi.org/ 10.1093/nar/gkm1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kotter P, Engels JW, Heckel A, Karas M, Entian KD, et al.. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res 2010; 38:2387-98; PMID:20047967; https://doi.org/ 10.1093/nar/gkp1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meyer B, Wurm JP, Sharma S, Immer C, Pogoryelov D, Kotter P, Lafontaine DL, Wohnert J, Entian KD. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res 2016; 44:4304-16; PMID:27084949; https://doi.org/ 10.1093/nar/gkw244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, Suzuki T. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol Chem 2014; 289:35724-30; PMID:25411247; https://doi.org/ 10.1074/jbc.C114.602698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, Lafontaine DL. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 2015; 43:2242-58; PMID:25653167; https://doi.org/ 10.1093/nar/gkv075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ebersberger I, Simm S, Leisegang MS, Schmitzberger P, Mirus O, von Haeseler A, Bohnsack MT, Schleiff E. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res 2014; 42:1509-23; PMID:24234440; https://doi.org/ 10.1093/nar/gkt1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buchhaupt M, Meyer B, Kotter P, Entian KD. Genetic evidence for 18S rRNA binding and an Rps19p assembly function of yeast nucleolar protein Nep1p. Molecular genetics and genomics : MGG 2006; 276:273-84; PMID:16721597; https://doi.org/ 10.1007/s00438-006-0132-x [DOI] [PubMed] [Google Scholar]
- 82.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci 2002; 27:344-51; PMID:12114023; https://doi.org/ 10.1016/S0968-0004(02)02109-6 [DOI] [PubMed] [Google Scholar]
- 83.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol 1999; 19:7461-72; PMID:10523634; https://doi.org/ 10.1128/MCB.19.11.7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Esguerra J, Warringer J, Blomberg A. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA 2008; 14:649-56; PMID:18256246; https://doi.org/ 10.1261/rna.845808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Badis G, Fromont-Racine M, Jacquier A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 2003; 9:771-9; PMID:12810910; https://doi.org/ 10.1261/rna.5240503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PloS one 2007; 2:e174; PMID:17245450; https://doi.org/ 10.1371/journal.pone.0000174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res 2009; 37:7665-77; PMID:19820108; https://doi.org/ 10.1093/nar/gkp816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 2007; 28:965-77; PMID:18158895; https://doi.org/ 10.1016/j.molcel.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 89.Lafontaine DL, Preiss T, Tollervey D. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol 1998; 18:2360-70; PMID:9528805; https://doi.org/ 10.1128/MCB.18.4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eschrich D, Buchhaupt M, Kotter P, Entian KD. Nep1p (Emg1p), a novel protein conserved in eukaryotes and archaea, is involved in ribosome biogenesis. Curr Genetics 2002; 40:326-38; PMID:11935223; https://doi.org/11694595 10.1007/s00294-001-0269-4 [DOI] [PubMed] [Google Scholar]
- 91.Liu PC, Thiele DJ. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol Biol Cell 2001; 12:3644-57; PMID:11694595; https://doi.org/ 10.1091/mbc.12.11.3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Basu A, Das P, Chaudhuri S, Bevilacqua E, Andrews J, Barik S, Hatzoglou M, Komar AA, Mazumder B. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol Cell Biol 2011; 31:4482-99; PMID:21930789; https://doi.org/ 10.1128/MCB.05804-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al.. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 2011; 44:660-6; PMID:22099312; https://doi.org/ 10.1016/j.molcel.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 2006; 312:902-6; PMID:16690864; https://doi.org/ 10.1126/science.1123835 [DOI] [PubMed] [Google Scholar]
- 95.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 2006; 34:721-33; PMID:16452298; https://doi.org/ 10.1093/nar/gkj471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agris PF, Sierzputowska-Gracz H, Smith C. Transfer RNA contains sites of localized positive charge: carbon NMR studies of [13C]methyl-enriched Escherichia coli and yeast tRNAPhe. Biochem 1986; 25:5126-31; PMID:3533144; https://doi.org/11574682 10.1021/bi00366a022 [DOI] [PubMed] [Google Scholar]
- 97.Micura R, Pils W, Hobartner C, Grubmayr K, Ebert MO, Jaun B. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res 2001; 29:3997-4005; PMID:11574682; https://doi.org/ 10.1093/nar/29.19.3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Behrmann E, Loerke J, Budkevich TV, Yamamoto K, Schmidt A, Penczek PA, Vos MR, Burger J, Mielke T, Scheerer P, et al.. Structural snapshots of actively translating human ribosomes. Cell 2015; 161:845-57; PMID:25957688; https://doi.org/ 10.1016/j.cell.2015.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Behrmann E, Loerke J, Budkevich TV, Yamamoto K, Schmidt A, Penczek PA, Vos MR, Burger J, Mielke T, Scheerer P, et al.. Structural snapshots of actively translating human ribosomes. Cell 2015; 161:845-57; PMID:25957688; https://doi.org/ 10.1016/j.cell.2015.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 2010; 37:809-20; PMID:20347423; https://doi.org/ 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Kos-Braun I, Kos M, Thoms M, Berninghausen O, Beckmann R, et al.. Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell 2016; 166:380-93; PMID:27419870; https://doi.org/ 10.1016/j.cell.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 102.Sloan KE, Leisegang MS, Doebele C, Ramirez AS, Simm S, Safferthal C, Kretschmer J, Schorge T, Markoutsa S, Haag S, et al.. The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res 2015; 43:553-64; PMID:25477391; https://doi.org/ 10.1093/nar/gku1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell 2009; 36:583-92; PMID:19941819; https://doi.org/ 10.1016/j.molcel.2009.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol 2006; 26:513-22; PMID:16382143; https://doi.org/ 10.1128/MCB.26.2.513-522.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fatica A, Oeffinger M, Dlakic M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol 2003; 23:1798-807; PMID:12588997; https://doi.org/ 10.1128/MCB.23.5.1798-1807.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hector RD, Burlacu E, Aitken S, Le Bihan T, Tuijtel M, Zaplatina A, Cook AG, Granneman S. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res 2014; 42:12138-54; PMID:25200078; https://doi.org/ 10.1093/nar/gku815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J 2010; 29:2026-36; PMID:20453830; https://doi.org/ 10.1038/emboj.2010.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, Karbstein K 3rd, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 2011; 333:1449-53; PMID:21835981; https://doi.org/ 10.1126/science.1208245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Letoquart J, Huvelle E, Wacheul L, Bourgeois G, Zorbas C, Graille M, Heurgue-Hamard V, Lafontaine DL. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc Natl Acad Sci 2014; 111:E5518-26; PMID:25489090; https://doi.org/ 10.1073/pnas.1413089111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DL, Heurgue-Hamard V. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol Cell Biol 2012; 32:2254-67; PMID:22493060; https://doi.org/ 10.1128/MCB.06623-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sardana R, Zhu J, Gill M, Johnson AW. Physical and functional interaction between the methyltransferase Bud23 and the essential DEAH-box RNA helicase Ecm16. Mol Cell Biol 2014; 34:2208-20; PMID:24710271; https://doi.org/ 10.1128/MCB.01656-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Andersen TE. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA 2004; 10:907-13; PMID:15146074; https://doi.org/ 10.1261/rna.5259404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Buchhaupt M, Sharma S, Kellner S, Oswald S, Paetzold M, Peifer C, Watzinger P, Schrader J, Helm M, Entian KD. Partial methylation at Am100 in 18S rRNA of baker's yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PloS one 2014; 9:e89640; PMID:24586927; https://doi.org/ 10.1371/journal.pone.0089640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 2015; 22:11-9; PMID:25565028; https://doi.org/ 10.1038/nsmb.2939 [DOI] [PubMed] [Google Scholar]
- 115.Dinman JD. Pathways to Specialized Ribosomes: The Brussels Lecture. J Mol Biol 2016; 428:2186-94; PMID:26764228; https://doi.org/ 10.1016/j.jmb.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci 2011; 36:127-32; PMID:21242088; https://doi.org/ 10.1016/j.tibs.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shi Z, Barna M. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu Rev Cell Dev Biol 2015; 31:31-54; PMID:26443190; https://doi.org/ 10.1146/annurev-cellbio-100814-125346 [DOI] [PubMed] [Google Scholar]
- 118.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 2012; 13:355-69; PMID:22617470; https://doi.org/ 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leulliot N, Godin KS, Hoareau-Aveilla C, Quevillon-Cheruel S, Varani G, Henry Y, Van Tilbeurgh H. The box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. J Mol Biol 2007; 371:1338-53; PMID:17612558; https://doi.org/ 10.1016/j.jmb.2007.06.031 [DOI] [PubMed] [Google Scholar]
- 120.Rothe B, Back R, Quinternet M, Bizarro J, Robert MC, Blaud M, Romier C, Manival X, Charpentier B, Bertrand E, et al.. Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Res 2014; 42:2015-36; PMID:24234454; https://doi.org/ 10.1093/nar/gkt1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014; 15:293-306; PMID:24662220; https://doi.org/ 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 122.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; 515:143-6; PMID:25192136; https://doi.org/ 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al.. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159:148-62; PMID:25219674; https://doi.org/ 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 2011; 30:79-89; PMID:21131909; https://doi.org/ 10.1038/emboj.2010.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu YT, Meier UT. RNA-guided isomerization of uridine to pseudouridine–pseudouridylation. RNA Biol 2014; 11:1483-94; PMID:25590339; https://doi.org/ 10.4161/15476286.2014.972855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Higa-Nakamine S, Suzuki T, Uechi T, Chakraborty A, Nakajima Y, Nakamura M, Hirano N, Suzuki T, Kenmochi N. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res 2012; 40:391-8; PMID:21908402; https://doi.org/ 10.1093/nar/gkr700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Armistead J, Khatkar S, Meyer B, Mark BL, Patel N, Coghlan G, Lamont RE, Liu S, Wiechert J, Cattini PA, et al.. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am J Hum Genet 2009; 84:728-39; PMID:19463982; https://doi.org/ 10.1016/j.ajhg.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Doll A, Grzeschik KH. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet Cell Genet 2001; 95:20-7; PMID:11978965; https://doi.org/ 10.1159/000057012 [DOI] [PubMed] [Google Scholar]
- 129.Warda AS, Freytag B, Haag S, Sloan KE, Gorlich D, Bohnsack MT. Effects of the Bowen-Conradi syndrome mutation in EMG1 on its nuclear import, stability and nucleolar recruitment. Hum Mol Genet 2016; PMID:27798105; https://doi.org/ 10.1093/hmg/ddw351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oie S, Matsuzaki K, Yokoyama W, Tokunaga S, Waku T, Han SI, Iwasaki N, Mikogai A, Yasuzawa-Tanaka K, Kishimoto H, et al.. Hepatic rRNA transcription regulates high-fat-diet-induced obesity. Cell Rep 2014; 7:807-20; PMID:24746822; https://doi.org/ 10.1016/j.celrep.2014.03.038 [DOI] [PubMed] [Google Scholar]
- 131.Cowling VH, Turner SA, Cole MD. Burkitt's lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis. Oncogene 2014; 33:3519-27; PMID:24013231; https://doi.org/ 10.1038/onc.2013.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nakamoto K, Ito A, Watabe K, Koma Y, Asada H, Yoshikawa K, Shinomura Y, Matsuzawa Y, Nojima H, Kitamura Y. Increased expression of a nucleolar Nop5/Sik family member in metastatic melanoma cells: evidence for its role in nucleolar sizing and function. Am J Path 2001; 159:1363-74; PMID:11583964; https://doi.org/ 10.1016/S0002-9440(10)62523-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, et al.. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Brit J Cancer 2011; 104:1168-77; PMID:21407217; https://doi.org/ 10.1038/sj.bjc.6606076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z, Jiang F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer 2010; 9:198; PMID:20663213; https://doi.org/ 10.1186/1476-4598-9-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta 2012; 1826:121-8; PMID:22498252; https://doi.org/ 10.1016/j.bbcan.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer 2012; 12:84-8; PMID:22257949; https://doi.org/ 10.1038/nrc3195 [DOI] [PubMed] [Google Scholar]
- 137.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 1998; 19:32-8; PMID:9590285; https://doi.org/ 10.1038/ng0598-32 [DOI] [PubMed] [Google Scholar]
- 138.Bellodi C, McMahon M, Contreras A, Juliano D, Kopmar N, Nakamura T, Maltby D, Burlingame A, Savage SA, Shimamura A, et al.. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep 2013; 3:1493-502; PMID:23707062; https://doi.org/ 10.1016/j.celrep.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 2003; 299:259-62; PMID:12522253; https://doi.org/ 10.1126/science.1079447 [DOI] [PubMed] [Google Scholar]
- 140.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J 2010; 29:1865-76; PMID:20453831; https://doi.org/ 10.1038/emboj.2010.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res 2010; 70:6026-35; PMID:20587522; https://doi.org/ 10.1158/0008-5472.CAN-09-4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rocchi L, Pacilli A, Sethi R, Penzo M, Schneider RJ, Trere D, Brigotti M, Montanaro L. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res 2013; 41:8308-18; PMID:23821664; https://doi.org/ 10.1093/nar/gkt587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gonzales B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum Mol Genet 2005; 14:2035-43; PMID:15930015; https://doi.org/ 10.1093/hmg/ddi208 [DOI] [PubMed] [Google Scholar]
- 144.Marcel V, Ghayad SE, Belin S, Therizols G, Morel AP, Solano-Gonzalez E, Vendrell JA, Hacot S, Mertani HC, Albaret MA, et al.. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 2013; 24:318-30; PMID:24029231; https://doi.org/ 10.1016/j.ccr.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang TH, Thompson I, Yuan ZM. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014; 33:1348-58; PMID:23542174; https://doi.org/ 10.1038/onc.2013.89 [DOI] [PubMed] [Google Scholar]
- 146.Bursac S, Brdovcak MC, Pfannkuchen M, Orsolic I, Golomb L, Zhu Y, Katz C, Daftuar L, Grabusic K, Vukelic I, et al.. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci 2012; 109:20467-72; PMID:23169665; https://doi.org/ 10.1073/pnas.1218535109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 2013; 5:237-47; PMID:24120868; https://doi.org/ 10.1016/j.celrep.2013.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Krastev DB, Slabicki M, Paszkowski-Rogacz M, Hubner NC, Junqueira M, Shevchenko A, Mann M, Neugebauer KM, Buchholz F. A systematic RNAi synthetic interaction screen reveals a link between p53 and snoRNP assembly. Nat Cell Biol 2011; 13:809-18; PMID:21642980; https://doi.org/ 10.1038/ncb2264 [DOI] [PubMed] [Google Scholar]
- 149.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol BioSys 2010; 6:481-93; PMID:20174677; https://doi.org/21543223 10.1039/b919670f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stumpf CR, Ruggero D. The cancerous translation apparatus. Curr Opin Genet Dev 2011; 21:474-83; PMID:21543223; https://doi.org/ 10.1016/j.gde.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bursac S, Brdovcak MC, Donati G, Volarevic S. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim Biophys Acta 2014; 1842:817-30; PMID:24514102; https://doi.org/ 10.1016/j.bbadis.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 152.Marbaniang CN, Vogel J. Emerging roles of RNA modifications in bacteria. Curr Opin Microbiol 2016; 30:50-7; PMID:26803287; https://doi.org/ 10.1016/j.mib.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 153.Noon KR, Bruenger E, McCloskey JA. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J Bact 1998; 180:2883-8; PMID:9603876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 2009; 34:427-39; PMID:19481523; https://doi.org/ 10.1016/j.molcel.2009.05.005 [DOI] [PubMed] [Google Scholar]
- 155.Lin J, Lai S, Jia R, Xu A, Zhang L, Lu J, Ye K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 2011; 469:559-63; PMID:21270896; https://doi.org/ 10.1038/nature09688 [DOI] [PubMed] [Google Scholar]


