ABSTRACT
RNA modifications are an emerging layer of posttranscriptional gene regulation in eukaryotes. N6-methyladenosine (m6A) is among the most abundant modifications in mRNAs (mRNAs) that was shown to influence many physiological processes from yeast to mammals. Like DNA methylation, m6A in mRNA is dynamically regulated. A conserved methyltransferase complex catalyzes the deposition of the methyl group on adenosine, which can be removed by specific classes of demethylases. Furthermore, YTH-domain containing proteins can recognize this modification to mediate m6A-dependent activities. Here we review the functions and mechanisms of the main m6A players with a particular focus on Drosophila melanogaster.
KEYWORDS: Drosophila, m6A, neurogenesis, RNA modifications, Sex lethal, splicing
Introduction
Epigenetic modifications regulate gene expression in response to changes in environmental cues. While the effect of DNA and chromatin modifications has been well studied, the role of RNA modifications on gene expression during organismal development and human disease is only starting to be unveiled. More than 100 modifications have so far been discovered; most of them were found on highly abundant RNAs such as transfer and rRNAs.1,2 One of the most prevalent modifications on mRNA is N6-methyladenosine (m6A). Two independent groups developed a few years ago a method called MeRIP-seq (methylated RNA immunoprecipitation sequencing) enabling for the first time a global mapping of m6A to the transcriptome.3,4 These studies revealed the presence of m6A in more than 10,000 transcripts of mRNAs and long noncoding RNAs with enrichments around stop codons, in the 3′ untranslated regions (UTRs) and within long internal exons as well as in alternatively spliced ones.3,4 A consensus sequence for m6A was derived from these and subsequent genome-wide studies, which consists of RRACH where R represents purine and H a non-guanine base.3-5
m6A formation is catalyzed by a methyltransferase complex, composed of METTL3, METTL14 and WTAP6-10 (Fig. 1). METTL3 catalyzes S-adenosyl methionine (SAM)-mediated transfer of methyl group to N6-position of adenosine base, while METTL14 is thought to be catalytically inactive with a structural role for facilitating METTL3 activity.11-13 WTAP, instead, appears essential to stabilize the interaction between the two METTL proteins.7 The three components of the core complex localize in nuclear speckles and recognize the previously derived consensus sequence RRACH.6,14 However, since only a subset of sites is methylated in vivo, additional subunits of the complex, such as KIAA1429 and RBM15, have been proposed to function in guiding the methylation to targeted sites.15
Figure 1.
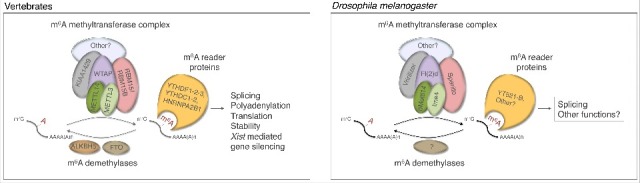
m6A mRNA pathway in vertebrates and Drosophila melanogaster. The m6A methyltransferase complex is composed of five factors. In Drosophila, the methyltransferase complex controls neural development and sex determination via its nuclear reader YT521-B. The precise functions or roles of Virilizer and its vertebrate homolog KIAA1429 remain to be identified. No demethlyase has been found so far in Drosophila.
The m6A modification was shown to modulate several physiological processes by regulating many aspects of mRNA processing, including splicing, mRNA decay and translation (for a recent review see16,17). Most of these functions are mediated by members of the YTH-domain family of proteins, which specifically recognize modified adenosines and serve as m6A readers.3,18,19 Vertebrates have five and Arabidopsis have thirteen members of YTH domain proteins, while only two members of the YTH domain family proteins exist in Drosophila; the nuclear YT521-B and the yet uncharacterized cytoplasmic protein CG6422.20-22 Both Drosophila proteins share high homology to their human counterparts in the YTH region, but low degree of similarities are observed outside the YTH domain.
m6A is dynamically regulated in mammals as it can be reverted by oxidative demethylation via the activity of two demethylases, FTO and ALKBH5. Their differential specificity toward m6A may rely on distinct temporal and spatial expression23,24 as well as on the sequence context surrounding the methylated adenosine.25 Accordingly, FTO has recently been shown to act primarily on N6-methylated adenosine that is introduced at the first position after the cap.26
Our recent work uncovers the in vivo roles of m6A in Drosophila melanogaster and identifies several major regulators. Some of these regulators bear distinct functions, suggesting that they also have m6A-independent roles. Here we summarize our current knowledge about the m6A players in Drosophila, and compare their functions with homologs in other species.
The m6A methyltransferase complex
Ime4
The corresponding homolog of METTL3 (or MTA in plants) in Drosophila is Ime4 (Inducer of meiosis 4 in yeast) (Fig. 2), which was the first subunit of the methyltransferase complex shown to bind S-Adenosyl methionine (SAM) and to transfer the methyl group from the SAM donor to N6-position of adenosine.27 A phylogenetic analysis of METTL3 revealed that many species ranging from yeast to human contain a conserved protein,28 which plays essential roles in development, but apparently is absent in fission yeast S. pombe and C. elegans.20 Sporulation is affected in budding yeast S. cerevisae that lack Ime429 and abnormal growth and seed development is observed in A. thaliana upon loss of MTA30,31 (Table 1). Knock out of METTL3 prevents naïve embryonic stem cells to differentiate and leads to early embryonic lethality in mice.32 In zebrafish, METTL3 is enriched in brain regions and its depletion leads to increased apoptosis in embryos.8 In contrast, Drosophila Ime4 knock out flies survive to adulthood, but are flightless.21,22 In addition, they display severe locomotion defects in orientation, walking speed and activity due to impaired neuronal functions. Also, defects in oogenesis due to altered Notch signaling have been reported.28 Intriguingly, female Ime4 mutant flies show altered splicing of Sxl, the master regulator of sex determination and dosage compensation in flies. In the soma, Sxl determines female physiognomy through regulation of alternative splicing of transformer (tra) and prevents dosage compensation in females by inhibition of male-specific lethal 2 (msl-2).33 In addition, Sxl is also required to initiate germ cell differentiation, but this pathway is independent of the sex determination pathway mediated by Tra. Consistently, genetic interaction between Ime4 and Sxl mutants show increased female lethality during development due to compromised dosage compensation and sexual transformations in the absence of msl-2. Furthermore, female germ cell differentiation defects were observed revealing a fine-tuning function of the m6A modification in the sex determination pathway. Initial links to the sex determination pathway have further been indicated by the interaction of Arabidopsis MTA with Fip37, the homolog of Fl(2)d in flies, previously identified to play a role in Drosophila sex determination30 (see below).
Figure 2.
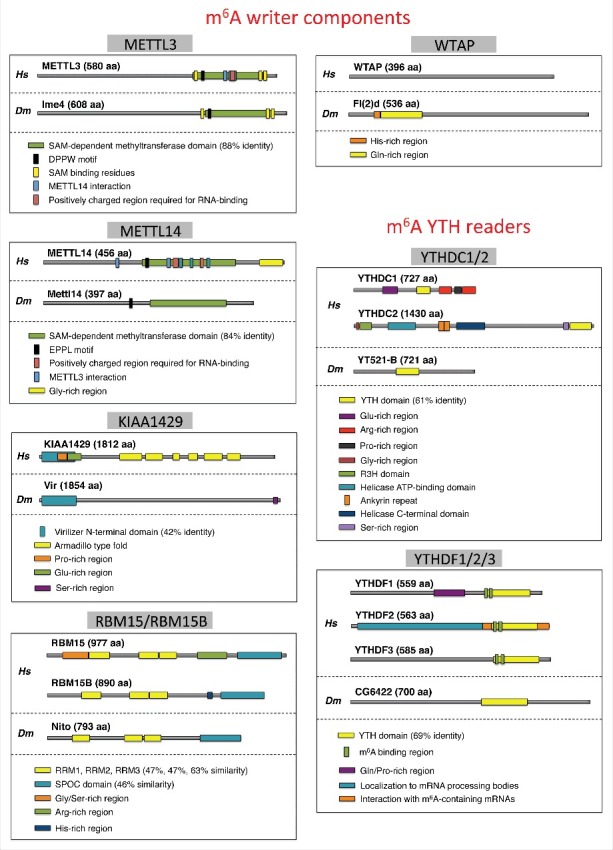
Domain-structure comparison of m6A writers and readers between Drosophila and Human. Comparison of the different proteins was based on uniprot (www.uniprot.org). Homology between similar domains was analyzed via protein BLAST from NCBI. Individual domains of Spenito were compared with RBM15; YT521-B with YTHDC1 and CG6422 with YTHDF2 (Hs - homo sapiens, Dm - Drosophila melanogaster).
Table 1.
Components of the m6A methyltransferase complex and their biologic roles.
| METTL3 | ||
|---|---|---|
| Human | METTL3 | •METTL3 KD leads to circadian clock period elongation.81 |
| •METTL3 promotes translation independently of its catalytic activity.34 | ||
| •METTL3 KD prevents differentiation of hESC.74 | ||
| Mouse | Mettl3 | •Mettl3 KO in naïve mESC leads to hyper naïve ground state, while in primed mESC boost cell differentiation.32 |
| •Mettl3 KO leads to embryonic lethality.32 | ||
| Zebrafish | METTL3 | •Morpholino depletion leads to developmental defects during embryogenesis.8 |
| A. thaliana | MTA | •MTA disruption results in embryonic lethality.30 |
| •MTA reduction leads to various developmental and organ definition defects.31 | ||
| S. cerevisiae | Ime4 | •Ime4 is required for sporulation and meiosis.64,82 |
| D. melanogaster | Ime4 | •Ime4 inactivation leads to defects during oogenesis.83 |
| •Ime4 KO affects fly locomotion due to impaired neuronal function.21,22 | ||
| •Ime4 regulates splicing of Sxl and fine tunes sex determination.21,22 | ||
| METTL14 | ||
| Human | METTL14 | •Structural component of the methyltransferase complex.7,8,11-13 |
| S. cerevisiae | Kar4 | •Transcriptional activator required for karyogamy.35,84 |
| D. melanogaster | Mettl14 | •Mettl14 KO affects fly locomotion due to impaired neuronal functions.22 |
| •Mettl14 regulates splicing of Sxl and fine tunes sex determination.21, 22 | ||
| WTAP | ||
| Human | WTAP | •Structural component of the mehyltransferase complex required for METTL3-METTL14 stabilization.7,8 |
| Mouse | WTAP | •WTAP KO results in early embryonic lethality.39 |
| Zebrafish | WTAP | •WTAP morpholinos display defects in head and brain development.8 |
| S. cerevisiae | Mum2 | •Mum2 is required for meiotic mRNA methylation as part of the MIS complex (Mum2, Ime4, Slz1).82 |
| D. melanogaster | Fl(2)d | •Fl(2)d is required for splicing of Sxl and its KO leads to embryonic lethality.41 |
| •Fl(2)d controls retinal development.44 | ||
| •Structural component of the methyltransferase complex, required for Ime4-Mettl14 stabilization.22 | ||
| RBM15, RBM15B | ||
| Human | RBM15,RBM15B | •RBM15 fusion with MKL1 is associated with acute megakaryoblastic leukemia.50 |
| •RBM15 and RBM15B are components of the methyltransferase complex, responsible for complex recruitment to targeted sites.15 | ||
| Mouse | RBM15, RBM15B | •Loss of RBM15 leads to embryonic lethality.47 |
| •RBM15 controls hematopoiesis, B-cell and megakaryocyte differentiation.47–49 | ||
| D. melanogaster | Nito | •Nito regulates wingless signaling and photoreceptor development.51,52 |
| •Nito is required for splicing of Sxl.53 | ||
| •Component of the methyltransferase complex.22 | ||
Ime4 is a 68 kilodalton (kDa) protein with a nuclear localization signal. Ime4 co-localizes extensively with RNA Pol II on Drosophila polytene chromosomes suggesting global co-transcriptional deposition of m6A.21 However, m6A levels in Drosophila mRNAs measured by dot blot and LC-MS/MS analyses are low, indicating that more methyl groups might initially be incorporated but are then removed by intron removal or demethylation. Alternatively, the methyltransferase complex, despite being widely present on chromosomes, might be active only under certain conditions. MeRIP-seq analysis with mRNA from Drosophila S2 embryonic cells also points toward lower m6A levels compared to vertebrates as around one thousand putative m6A sites were identified in Drosophila, while at least 10,000 sites were identified in various vertebrate cells.3,4,22 Whether m6A modification is more abundant and plays more prominent roles in Drosophila neuronal cells remains to be investigated. Nevertheless the consensus RRACH was present in most of the m6A peaks, and enrichment near start and stop codons was also observed, suggesting conserved functions and regulatory mechanisms.
Intriguingly, a novel function for mammalian METTL3, independent of its m6A catalytic activity, was recently found. A fraction of METTL3 localizes in the cytoplasm and was shown to promote translation of a subset of RNAs containing m6A peaks in 3′UTRs by interaction with the eIF3b subunit of the translation initiation complex.34 Whether Drosophila Ime4 has a similar role in the cytoplasm remains to be investigated.
Mettl14
The corresponding homolog of METTL14 in Drosophila is Mettl14 (Fig. 2). Mettl14 is an essential component of the methyltransferase complex and contains, like Ime4, a catalytic domain.7-10 However, recent structural studies revealed that steric constraints from side groups near the putative SAM binding pocket prevent METTL14 to accommodate SAM and is therefore catalytically inactive.11-13 Rather, METTL14 was shown to stabilize the interaction between the methyltransferase complex and RNA by forming a charged grove at the interface of METTL3 and METTL14 for RNA accommodation.11-13 These studies therefore indicate that METTL3 requires METTL14 for its activity. Drosophila Mettl14 shares a 62% identity with human ortholog and sequence comparison shows that side chains, which prevent accommodation of SAM in METTL14 are conserved. Both proteins lack aromatic residues that interact with acceptor adenine as well as residues that enable formation of hydrogen bonds with SAM in METTL3 protein. Loss of function of Mettl14 in Drosophila is reminiscent to the loss of function of Ime4, suggesting that they act together21,22 (Table 1). Furthermore, quantitative mass spectrometry analysis revealed an interaction at a one to one ratio and show that the stability of both proteins depends on each other.22 Interestingly, the ortholog of METTL14 in yeast, Kar4, can bind DNA and possesses transcriptional activity.35 Whether this function is related to m6A methylation and whether similar functions linked to transcription exist for METTL14 in other species is currently unknown.
Fl(2)d
Female-Lethal(2)D is the ortholog of mammalian WTAP (Wilms' tumor 1 associated protein), a nuclear protein that was found to interact with splicing factors and other proteins involved in RNA processing36 (Fig. 2). Its localization to nuclear speckles depends on the presence of BCLAF1 and THRAP3. WTAP was initially found in a yeast two-hybrid screen to identify interactors of Wilms' tumor 1 protein (WT1).14 WT1 encodes several protein isoforms that can either interact with DNA and act in transcription, or bind RNA and colocalize with splicing factors.37 Intriguingly, isoforms binding to RNA are required for sex determination in mice, since male mutants lacking these isoforms undergo sex reversal due to reduced levels of sex-determining region Y (SRY) protein.38 Whether m6A and WTAP are involved in this function remains to be addressed. WTAP knock out mice show embryonic lethality and defects in cell cycle progression36,39 (Table 1). Drosophila Fl(2)d also colocalizes with several splicing factors in the nucleus and regulates m6A levels.22,40 Accordingly, its expression pattern strictly correlates with the level of m6A during development, supporting the notion that, in Drosophila, m6A metabolism is primarily dependent on the presence of a functional methyltransferase complex, and less so from potential demethylases (see below). Its depletion strongly compromises the interaction between Ime4 and Mettl14.22
Fl(2)d was among the first proteins identified to be required for sex-specific alternative splicing of Sxl and tra.41,42 In contrast to Ime4 and Mettl14, fl(2)d is essential during development and analysis of sexual mosaics showed male somatic transformations in females, which is also observed in transheterozygous Ime4, sxl female mutants made viable by the lack of msl-2.17 Lethality of fl(2)d mutant females thus suggests other roles, independent of its activity within the methyltransferase complex. In line with these observations, depletion of WTAP in zebrafish also causes more severe developmental defects compared with the loss of METTL3.8 Furthermore, gel filtration experiments indicate that human WTAP co-fractions with METTL3 and METTL14 at a size of 300 kDa, but is also present at a higher molecular weight, supporting its association in distinct complexes.7
fl(2)d encodes for two isoforms generated via alternative 5′ splice site selection in the 5′UTR. A long isoform contains an N-terminal histidine and glutamine rich region, found in many transcription factors.43 This isoform interacts with Sine Oculis (So) to control retinal development.44 Interestingly, fl(2)d splicing is regulated via m6A located near the proximal splice site. Depletion of methyltransferase complex components leads to increased usage of the distal splice site and formation of the long protein isoform, but whether this impacts on m6A methylosome activity awaits further investigation.22
Virilizer
Virlizer (Vir) is the ortholog of KIAA1429, which was found in a mass spectrometry-based approach as an interacting protein of the core components of the methyltransferase complex (Fig. 2). Its depletion severely reduces m6A levels on mRNA.9,22 Vir is a large nuclear protein of 1854 amino acids, and like Fl(2)d has essential functions as null mutants are lethal. Like Fl(2)d, Vir is also required for female specific alternative splicing of Sxl45,46 (Table 1). In vir2F female mutants, ectopic expression of Sxl is sufficient to rescue female lethality. The precise role of Vir and its human homolog in the context of m6A biogenesis awaits further investigation.
Spenito
Spenito (Nito) has two orthologs in mammals: RBM15 and RBM15B (Fig. 2). Mice lacking RBM15 die at embryonic day 9.5 and display defects in heart, spleen, vasculature as well as in hematopoiesis, B-cell and megakaryocyte differentiation47-49 (Table 1). A well-characterized chromosomal aberration involving RBM15 and Megakaryoblastic leukemia 1 (MKL1) is associated with acute megakaryoblastic leukemia,50 demonstrating also the pivotal role of RBM15 in cancer. In Drosophila, Nito promotes Wingless signaling51 and its overexpression in the eye leads to defects in photoreceptor development,52 while its depletion in ovaries results in stem cell tumor appearance.53 The loss of Nito affects Sxl splicing and gives rise to male somatic transformations, which is in agreement with the role of other m6A components in sex determination.53 Furthermore, Nito interacts with subunits of the methyltransferase complex and its depletion drastically decreases m6A levels.22 Interestingly, RBM15 was also recently found to regulate m6A levels in human cells and to control X-chromosome inactivation for dosage compensation in female cells via m6A-methylation of XIST, which in turn promotes transcriptional repression of the inactive X chromosome.15 RBM15 binds near m6A sites on XIST mRNA and on other transcripts and is predicted to recruit the methyltransferase complex to its target transcripts. RBM15 interacts with RNA directly via its RRM domains and was also shown to interact with the Setd1b protein, an H3K4me3 histone methlyltransferase, via its SPOC domain.85 The importance of the individual motif for RBM15/Nito function in regards to m6A activity awaits further investigation.
Demethylases
In vertebrates, methylation of adenosine is reversible due to the activity of two demethylases, namely FTO and ALKBH5.24,54 FTO demethylates m6A through N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A) intermediates.55,56 A recent study showed that FTO preferentially acts on N6,2′-O-dimethyladenosine (m6Am) modification adjacent to mRNA cap, which in turn negatively affects mRNA stability.26 FTO loss of function leads to postnatal growth retardation, altered locomotor activity, defects of signaling in dopaminergic neurons and reduced fat mass.23,57-62 Likewise, overexpression of FTO results in obesity.63 Another m6A demethylase, ALKBH5, was later found to play a role in male fertility.24 In contrast to vertebrates, the specificity of m6A occupancy in yeast seems to be determined by the restricted expression of the methyltransferase complex.64 Similarly, alignment of FTO and ALKBH5 nucleotide sequences to the Drosophila genome failed to identify homologs in flies. FTO appearance is concomitant to the vertebrate clade, with the exception of homologs present in diverse marine eukaryotic algae.65 Despite the fact that ALKBH5 is also absent, additional members of the ALKBH family that localize into the cytoplasm are present. However, depletion of these candidates, either individually or in combination, has no consequence on the m6A/A ratio (Lence et al, unpublished), indicating that these factors are not functional in flies in regards to their ability to demethylate m6A on mRNA. Additional studies will be necessary to address whether other unknown demethylases are required to fine-tune m6A levels in this organism.
m6A binding YTH proteins
YT521-B
YT521-B is the closest ortholog of YTHDC1 (Fig. 2). The YTH domain was initially found as an RNA binding domain recognizing the hexanucleotide GCAUAC sequence, based on in vitro SELEX experiments.18 More recently, proteins of the YTH-domain family were recognized as specific binders of m6A RNA modification.3 A number of crystal structures revealed the mechanism of this binding by the hydrophobic pocket and aromatic residues.19,66-68 A 50-fold increase in binding to methylated in comparison to non methylated residues was observed.19 YTHDC1 is localized in the nucleus and is involved in splicing regulation via m6A-in long exons. This mechanism involves the YTHDC1-mediated recruitment of the splicing regulator SRSF3 and the exclusion of SRSF10.69 YTHDC1 was also recently shown to induce X chromosome inactivation in human via binding to m6A on Xist RNA.15 The precise mechanism of YTHDC1 in this process is currently unclear. Interactome studies indicate its association with members of Polycomb group complexes, suggesting that YTHDC1, via its ability to recognize m6A, may facilitate the binding of gene-silencing proteins to Xist RNA. The sub-nuclear distribution of YTHDC1 is controlled via its association with the KH-domain containing Sam68 protein and this interaction is abolished upon YTHDC1 phosphorylation by p59fyn kinase.70,71 In Drosophila, YT521-B is enriched in the embryonic neuroectoderm and in heads of adult flies.22 It localizes to the nucleus and specifically binds m6A-modified transcripts.21,22 In particular, YT521-B assists Sxl in repressing inclusion of the male-specific alternative exon by binding to nearby intronic m6A sites.21 Consistent with the observation in mammals, Drosophila YT521-B regulates most of m6A-dependent splicing events (about 60 to 70% overlap with Ime4). Intron retention and alternative splicing in 5′UTRs are overrepresented. This regulation influences the number of upstream AUGs in 5′UTRs, suggesting that m6A-regulated alternative splicing affects translation. Furthermore, YT521-B appears to be the main mediator of m6A-dependent processes in vivo, as flies lacking a functional YT521-B resemble the phenotypes observed in mutants for methyltransferase complex components.21,22 Using SILAC-based proteomic analysis of YT521-B, several potential interactors were found, which includes many predicted mRNA binding proteins, such as the KH-domain containing Quaking related-family proteins, Hrb27C, Syp, Imp and others. The relevance of these interactions, however, awaits further validations.
CG6422
The closest vertebrate ortholog of CG6422 is YTHDF2 (Fig. 2). YTHDF2 is a cytoplasmic protein that belongs to the YTH-domain containing family of proteins. It was identified as an m6A binding protein in a pull-down experiment with a methylated probe.3 Its role in mRNA decay was the first study providing a functional mechanism of m6A modification in the mRNA life cycle.72 YTHDF2 binds m6A predominantly in the 3′UTR of mRNAs via its C-terminally located YTH domain. Its N-terminal Glutamine/Proline-rich region interacts directly with CNOT1, a component of the CCR4-NOT deadenylase complex, guiding methylated mRNAs to processing bodies.73 This function appears important in human embryonic cells to degrade mRNA encoding pluripotency factors, eventually allowing for differentiation.32,74 Likewise, in zebrafish, YTHDF2 is required for maternal mRNA clearance during the maternal to zygotic transition.75 Interestingly, YTHDF2 was shown to re-localize to the nucleus under stress conditions and to specifically bind m6A in the 5′UTRs of the newly transcribed mRNAs. This binding protects m6A sites from FTO-mediated demethylation, which in turn enhances cap-independent translation of heat shock responsive transcripts.56 Further m6A can also serve on its own to direct cap-independent translation. eIF3 was shown to directly bind m6A in the 5′UTR of mRNA upon UV-irradiation or heat-shock conditions, allowing the recruitment of the 43S pre-initiation complex independently of the m7G cap modification.76 Hence, translation of stress-induced transcripts is enabled by this mechanism when translation of other cellular transcripts is shut down. YTHDF1 is another cytoplasmic YTH-domain containing protein in vertebrates that binds m6A around stop codon and in the 3′UTR. YTHDF1 was shown to enhance protein production by direct interaction with eIF3.76 Recently, a third protein, YTHDF3, was shown to act cooperatively with YTHDF1 and YTHDF2, to promote translation and mRNA decay, respectively.77,78 In Drosophila, CG6422 localizes in the cytoplasm but does not re-localize to the nucleus upon heat shock or under UV-irradiation (Lence et al, unpublished). Consistent also with a possible role in the maternal to zygotic transition its expression is high in the first two hours of embryogenesis and decreases during development.22 Its function in the context of m6A and potential roles in translation and/or mRNA decay await further investigation.
Conclusion and future directions
With the advance of novel techniques, m6A was found on thousands of mRNA sites in several species. Several recent studies demonstrated its role in nearly all aspects of mRNA processing, via a group of YTH-domain family proteins, but also by altering the binding of some RNA interacting proteins to their recognition sites via m6A-mediated changes in RNA secondary structure or “RNA switches”.10,79 The players that catalyze, remove and recognize the modification are conserved across evolution, although with exceptions.
While the precise molecular function of the core methyltransferase complex, including METTL3, METTL14 and WTAP is almost solved; the role of the other co-factors remains less understood. KIAA1429 and its fly homolog Vir, interact with other components of the methlyltransferase complex and regulate m6A levels, but their molecular functions are currently unknown. RBM15 binds near methylated sites and its absence prevents recruitment of the methlytransferase complex to its targeted sites. While this model provides an elegant explanation on why only a subset of m6A sites is methylated, it remains to be explored whether RBM15 is sufficient to recruit the complex and to induce m6A or whether other players are also involved? Intriguingly, RBM15 was shown to interact with chromatin binding proteins via its SPOC domain and has the ability to influence histone marks. Whether RBM15 provides a link between methylated RNA and the chromatin state is therefore an attractive possibility. In fact, it is likely that m6A deposition happens co-transcriptionally, as m6A regulates splicing and several sites were found in introns.3,21 Thus, future research will reveal the dynamics of m6A deposition and its distribution in relation with transcription and chromatin features.
Loss of components of the Drosophila methyltransferase leads to neuronal and sex determination defects, but how m6A precisely regulates these processes remains to be determined. It will be interesting to examine whether other m6A functions could be revealed upon stress conditions or in a sensitive context when the dosage of important players involved in specific physiological processes is reduced? Intriguingly, the loss of Ime4 and Mettl14 gives rise to milder phenotypes compared with the depletion of the other complex components such as Nito, Fl(2)d and Vir. This suggests m6A-independent functions for these subunits, or else, they may work with distinct m6A methyltransferases.
Once the RNA is methylated it can be recognized by a different set of reader proteins and/or the methylation can be removed by the activity of demethylases. Several questions on how selectivity is achieved remain open. Likely, competition exists at individual m6A sites for various proteins to bind and protect the m6A or to remove it. In addition, binding of other RNA binding proteins such as hnRNPC or ELAV/Hu family proteins might be regulated by m6A RNA switches and/or concomitant binding of YTH proteins.3,79,80 Therefore, sequence context for m6A sites is a key to which proteins will bind and which regulatory program will be initiated; as for example, distinct members of the YTHDF proteins can initiate translation or mRNA decay. Overall, the fate of each m6A-modified mRNA should take into consideration the “place and time” including the m6A position within mRNA, the sequence context, the expression levels of m6A regulators as well as the cell type and its developmental stage. Therefore, we foresee that the study of single gene reporters will bring additional mechanistic insights into these questions.
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Funding
T. Lence and J-Y Roignant are supported by the Marie Curie CIG (334288), the Deutsche Forschungsgemeinschaft (DFG) SPP1935 Grant (RO 4681/4–1) and the European COST action (CA16120). M. Soller is supported by the Biotechnology and Biological Science Research Council (BBSRC).
References
- 1.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdisciplinary Rev RNA 2011; 2:611; PMID:21823225; http://dx.doi.org/ 10.1002/wrna.79 [DOI] [PubMed] [Google Scholar]
- 2.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al.. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262; PMID:3531130; http://dx.doi.org/ 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201; PMID:22575960; http://dx.doi.org/ 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 4.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635; PMID:3383396; http://dx.doi.org/ 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 2015; 12:767;PMID:4487409; http://dx.doi.org/ 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna 1997; 3:1233; PMID:1369564 [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014; 10:93; PMID:3911877; http://dx.doi.org/ 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al.. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014; 24:177; PMID:3915904; http://dx.doi.org/ 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al.. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Reports 2014; 8:284; PMID:4142486; http://dx.doi.org/ 10.1016/j.celrep.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014; 16:191; PMID:24394384; http://dx.doi.org/ 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 2016; 5:e18434; PMID:5023411; http://dx.doi.org/ 10.7554/eLife.18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 2016; 1:16011; PMID:27572442; http://dx.doi.org/ 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al.. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016; 534:575; PMID:27281194; http://dx.doi.org/ 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- 14.Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum Mol Genetics 2000; 9:2231; PMID:24100041; http://dx.doi.org/ 10.1074/jbc.M113.500397 [DOI] [PubMed] [Google Scholar]
- 15.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016; 537:369; PMID:27602518; http://dx.doi.org/ 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zhao JC. Update: Mechanisms Underlying N6-Methyladenosine Modification of Eukaryotic mRNA. Trends Genetics: TIG 2016; 32:763; PMID:5123927; http://dx.doi.org/ 10.1016/j.tig.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2016; 1:31; PMID:27808276; http://dx.doi.org/ 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH, et al.. The YTH domain is a novel RNA binding domain. J Biol Chem 2010; 285:14701; PMID:2863249; http://dx.doi.org/ 10.1074/jbc.M110.104711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theler D, Dominguez C, Blatter M, Boudet J, Allain FH. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res 2014; 42:13911; PMID:4267619; http://dx.doi.org/ 10.1093/nar/gku1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dezi V, Ivanov C, Haussmann IU, Soller M. Nucleotide modifications in messenger RNA and their role in development and disease. Biochem Soc Transactions 2016; 44:1385; PMID:27911721; http://dx.doi.org/ 10.1042/BST20160110 [DOI] [PubMed] [Google Scholar]
- 21.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016; 540:301; PMID:27919081; http://dx.doi.org/ 10.1038/nature20577 [DOI] [PubMed] [Google Scholar]
- 22.Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, et al.. m6A modulates neuronal functions and sex determination in Drosophila. Nature 2016; 540:242; PMID:27919077; http://dx.doi.org/ 10.1038/nature20568 [DOI] [PubMed] [Google Scholar]
- 23.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC, Rüther U. Inactivation of the Fto gene protects from obesity. Nature 2009; 458:894; PMID:19234441; http://dx.doi.org/ 10.1038/nature07848 [DOI] [PubMed] [Google Scholar]
- 24.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013; 49:18; PMID:3646334; http://dx.doi.org/ 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou S, Toh JD, Wong KH, Gao YG, Hong W, Woon EC. N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Scientific Reports 2016; 6:25677; PMID:4860565; http://dx.doi.org/ 10.1038/srep25677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al.. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 2017; 541:371; PMID:28002401; http://dx.doi.org/ 10.1038/nature21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem 1994; 269:17697; PMID:8021282 [PubMed] [Google Scholar]
- 28.Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J Mol Evolution 2002; 55:431; PMID:12355263; http://dx.doi.org/ 10.1007/s00239-002-2339-8 [DOI] [PubMed] [Google Scholar]
- 29.Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol 1992; 12:1078; PMID:369539; http://dx.doi.org/ 10.1128/MCB.12.3.1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008; 20:1278;PMID:2438467; http://dx.doi.org/ 10.1105/tpc.108.058883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, May S, Fray RG. Adenosine Methylation in Arabidopsis mRNA is associated with the 3′ End and reduced levels cause developmental defects. Frontiers Plant Sci 2012; 3:48; PMID:22639649; http://dx.doi.org/ 10.3389/fpls.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor Y, et al.. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015; 347:1002; PMID:25569111; http://dx.doi.org/ 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- 33.Schutt C, Nothiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 2000; 127:667; PMID:10648226 [DOI] [PubMed] [Google Scholar]
- 34.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 2016; 62:335; PMID:4860043; http://dx.doi.org/ 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurihara LJ, Stewart BG, Gammie AE, Rose MD. Kar4p, a karyogamy-specific component of the yeast pheromone response pathway. Mol Cell Biol 1996; 16:3990; PMID:231395; http://dx.doi.org/ 10.1128/MCB.16.8.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 2013; 288:33292; PMID:3829175; http://dx.doi.org/ 10.1074/jbc.M113.500397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 1995; 81:391; PMID:7736591 [DOI] [PubMed] [Google Scholar]
- 38.Hammes A, Guo JK, Lutsch G, Leheste JR, Landrock D, Ziegler U, Gubler MC, Schedl A. Two splice variants of the Wilms' tumor 1 gene have distinct functions during sex determination and nephron formation. Cell 2001; 106:319; PMID:11509181 [DOI] [PubMed] [Google Scholar]
- 39.Horiuchi K, Umetani M, Minami T, Okayama H, Takada S, Yamamoto M, Aburatani H, Reid PC, Housman DE, Hamakubo T, et al.. Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci U S A 2006; 103:17278; PMID:1634838; http://dx.doi.org/ 10.1073/pnas.0608357103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penn JK, Graham P, Deshpande G, Calhoun G, Chaouki AS, Salz HK, Schedl P. Functioning of the Drosophila Wilms'-tumor-1-associated protein homolog, Fl(2)d, in Sex-lethal-dependent alternative splicing. Genetics 2008; 178:737; http://dx.doi.org/ 10.1534/genetics.107.081679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granadino B, Campuzano S, Sanchez L. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. EMBO J 1990; 9:2597; PMID:552292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Granadino B, Penalva LO, Sanchez L. The gene fl(2)d is needed for the sex-specific splicing of transformer pre-mRNA but not for double-sex pre-mRNA in Drosophila melanogaster. Mol General Genetics: MGG 1996; 253:26; PMID:9003283; http://dx.doi.org/ 10.1007/s004380050292 [DOI] [PubMed] [Google Scholar]
- 43.Penalva LO, Ruiz MF, Ortega A, Granadino B, Vicente L, Segarra C, Valcárcel J, Sánchez L. The Drosophila fl(2)d gene, required for female-specific splicing of Sxl and tra pre-mRNAs, encodes a novel nuclear protein with a HQ-rich domain. Genetics 2000; 155:129; PMID:1461084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson AM, Weasner BP, Weasner BM, Kumar JP. The Drosophila Wilms Tumor 1-Associating Protein (WTAP) homolog is required for eye development. Dev Biol 2014; 390:170; PMID:4063124; http://dx.doi.org/ 10.1016/j.ydbio.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilfiker A, Amrein H, Dubendorfer A, Schneiter R, Nothiger R. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development 1995; 121:4017; PMID:8575302 [DOI] [PubMed] [Google Scholar]
- 46.Niessen M, Schneiter R, Nothiger R. Molecular identification of virilizer, a gene required for the expression of the sex-determining gene Sex-lethal in Drosophila melanogaster. Genetics 2001; 157:679; PMID:1461513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raffel GD, Mercher T, Shigematsu H, Williams IR, Cullen DE, Akashi K, Bernard OA, Gilliland DG. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc Natl Acad Sci U S A 2007; 104:6001; PMID:1851606; http://dx.doi.org/ 10.1073/pnas.0609041104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niu C, Zhang J, Breslin P, Onciu M, Ma Z, Morris SW. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood 2009; 114:2087; PMID:2744570; http://dx.doi.org/ 10.1182/blood-2009-01-197921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raffel GD, Chu GC, Jesneck JL, Cullen DE, Bronson RT, Bernard OA, Gilliland DG. Ott1 (Rbm15) is essential for placental vascular branching morphogenesis and embryonic development of the heart and spleen. Mol Cell Biol 2009; 29:333; PMID:2612519; http://dx.doi.org/ 10.1128/MCB.00370-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Z, Morris SW, Valentine V, Li M, Herbrick JA, Cui X, Bouman D, Li Y, Mehta PK, Nizetic D, et al.. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genetics 2001; 28:220; PMID:11431691; http://dx.doi.org/ 10.1038/90054 [DOI] [PubMed] [Google Scholar]
- 51.Chang JL, Lin HV, Blauwkamp TA, Cadigan KM. Spenito and Split ends act redundantly to promote Wingless signaling. Dev Biol 2008; 314:100; PMID:18174108; http://dx.doi.org/ 10.1016/j.ydbio.2007.11.023 [DOI] [PubMed] [Google Scholar]
- 52.Jemc J, Rebay I. Characterization of the split ends-like gene spenito reveals functional antagonism between SPOC family members during Drosophila eye development. Genetics 2006; 173:279; PMID:1461450; http://dx.doi.org/ 10.1534/genetics.106.055558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan D, Perrimon N. spenito is required for sex determination in Drosophila melanogaster. Proc Natl Acad Sci U S A 2015; 112:11606; http://dx.doi.org/ 10.1073/pnas.1515891112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885; PMID:3218240; http://dx.doi.org/ 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al.. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Communications 2013; 4:1798; PMID:23653210; http://dx.doi.org/ 10.1038/ncomms2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015; 526:591; PMID:26458103; http://dx.doi.org/ 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et al.. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genetics 2007; 39:724; PMID:17496892; http://dx.doi.org/ 10.1038/ng2048 [DOI] [PubMed] [Google Scholar]
- 58.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al.. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007; 316:889; PMID:2646098; http://dx.doi.org/ 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al.. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007; 316:1341; PMID:3214617; http://dx.doi.org/ 10.1126/science.1142382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orrú M, Usala G, et al.. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genetics 2007; 3:e115; PMID:1934391; http://dx.doi.org/ 10.1371/journal.pgen.0030115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, Meyre D, Golzio C, Molinari F, Kadhom N, et al.. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genetics 2009; 85:106; PMID:2706958; http://dx.doi.org/ 10.1016/j.ajhg.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Brönneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, et al.. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci 2013; 16:1042; PMID:23817550; http://dx.doi.org/ 10.1038/nn.3449 [DOI] [PubMed] [Google Scholar]
- 63.Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Brüning JC, Nolan PM, Ashcroft FM, et al.. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genetics 2010; 42:1086; PMID:3018646; http://dx.doi.org/ 10.1038/ng.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al.. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013; 155:1409; PMID:3956118; http://dx.doi.org/ 10.1016/j.cell.2013.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robbens S, Rouzé P, Cock JM, Spring J, Worden AZ, Van de Peer Y. The FTO gene, implicated in human obesity, is found only in vertebrates and marine algae. J Mol Evolution 2008; 66:80; PMID:18058156; http://dx.doi.org/ 10.1007/s00239-007-9059-z [DOI] [PubMed] [Google Scholar]
- 66.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 2014; 10:927; PMID:25242552; http://dx.doi.org/ 10.1038/nchembio.1654 [DOI] [PubMed] [Google Scholar]
- 67.Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B homology domain family of proteins. J Biol Chem 2015; 290:24902; PMID:4598999; http://dx.doi.org/ 10.1074/jbc.M115.680389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo S, Tong L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc Natl Acad Sci U S A 2014; 111:13834; PMID:25201973; http://dx.doi.org/ 10.1073/pnas.1412742111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al.. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell 2016; 61:507; PMID:26876937; http://dx.doi.org/ 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 70.Hartmann AM, Nayler O, Schwaiger FW, Obermeier A, Stamm S. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol Biol Cell 1999; 10:3909; PMID:25688; http://dx.doi.org/ 10.1091/mbc.10.11.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rafalska I, Zhang Z, Benderska N, Wolff H, Hartmann AM, Brack-Werner R, Stamm S. The intranuclear localization and function of YT521-B is regulated by tyrosine phosphorylation. Hum Mol Genetics 2004; 13:1535; PMID:15175272; http://dx.doi.org/ 10.1093/hmg/ddh167 [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505:117; http://dx.doi.org/ 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Communications 2016; 7:12626; PMID:27558897; http://dx.doi.org/ 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al.. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014; 15:707; PMID:4278749; http://dx.doi.org/ 10.1016/j.stem.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017; 542:475; PMID:5323276; http://dx.doi.org/ 10.1038/nature21355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015; 163:999; PMID:4695625; http://dx.doi.org/ 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al.. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 2017; 27(3):444-7; PMID:28106076; http://dx.doi.org/ 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C, et al.. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 2017; 27(3):315-328; PMID:28106072; http://dx.doi.org/ 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015; 518:560; PMID:4355918; http://dx.doi.org/ 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zaharieva E, Haussmann IU, Brauer U, Soller M. Concentration and Localization of Coexpressed ELAV/Hu Proteins Control Specificity of mRNA Processing. Mol Cell Biol 2015; 35:3104; PMID:4539368; http://dx.doi.org/ 10.1128/MCB.00473-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al.. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793; PMID:24209618; http://dx.doi.org/ 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- 82.Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genetics 2012; 8:e1002732; PMID:22685417; http://dx.doi.org/ 10.1371/journal.pgen.1002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci U S A 2011; 108:14855; PMID:21873203; http://dx.doi.org/ 10.1073/pnas.1111577108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lahav R, Gammie A, Tavazoie S, Rose MD. Role of transcription factor Kar4 in regulating downstream events in the Saccharomyces cerevisiae pheromone response pathway. Mol Cell Biol 2007; 27:818; PMID:17101777; http://dx.doi.org/ 10.1128/MCB.00439-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J-H, Skalnik DG. Rbm15-Mkl1 Interacts with the Setd1b Histone H3-Lys4 Methyltransferase via a SPOC Domain That Is Required for Cytokine-Independent Proliferation. PLoS ONE. 7(8):e42965. http://dx.doi.org/ 10.1371/journal.pone.0042965 [DOI] [PMC free article] [PubMed] [Google Scholar]


