ABSTRACT
A group of homologous nucleic acid modification enzymes called Dnmt2, Trdmt1, Pmt1, DnmA, and Ehmet in different model organisms catalyze the transfer of a methyl group from the cofactor S-adenosyl-methionine (SAM) to the carbon-5 of cytosine residues. Originally considered as DNA MTases, these enzymes were shown to be tRNA methyltransferases about a decade ago. Between the presumed involvement in DNA modification-related epigenetics, and the recent foray into the RNA modification field, significant progress has characterized Dnmt2-related research. Here, we review this progress in its diverse facets including molecular evolution, structural biology, biochemistry, chemical biology, cell biology and epigenetics.
KEYWORDS: DNA methylation, epigenetics, methylcytidine, RNA methylation, tRNA
Introduction
Dnmt2 orthologs were originally considered to be DNA methylating enzymes, based on their clear sequence homology with other enzymes from a family designated with the abbreviation “Dnmt” for DNA methyltransferase. These enzymes are known for catalyzing cytidine methylation at the carbon-5 position resulting in 5-methylcytosine, typically abbreviated as 5mC in the DNA modification field. However, robust DNA methyltransferase activity, that was comparable to other Dnmt family members, could not be observed using Dnmt2 preparations. An extensive search for target DNA sequences within different genomes led to a stunning discovery, when the Bestor group showed that Dnmt2 enzymes are actually tRNA transferases1 catalyzing the methylation of position 38 in tRNAAspGUC(1) to yield 5-methylcytosine, which is abbreviated m5C in the RNA modification field. From this discovery, many new questions arose. Is tRNA methylation the only activity, or could Dnmt2 enzymes be multifunctional and promiscuous in their substrates? Could there be other RNA targets, with or without characteristics known from tRNAs? Are these enzymes constitutively active, or do they change substrate specificity under certain conditions and upon interaction with partners? How are substrates recognized? What are the biological consequences of Dnmt2-mediated RNA methylation on protein biosynthesis and other cellular functions?
Meanwhile, various new findings and developments have excited the tRNA field, many of them relating to RNA modifications, which placed the unusual activity of Dnmt2 enzymes toward tRNA substrates well in the center of attention. For one, the view of tRNA modification has changed from that of a constitutive, and essentially static, posttranscriptional feature to include a notion of dynamic regulation of the modification status of RNAs.2 The extent of tRNA modification, and tRNA itself, were shown to respond to extrinsic stimuli such as stress, suggesting participation in the regulation of molecular responses to such insults.3 Furthermore, tRNA fragments (tRFs), long thought to be artifacts from RNA preparation procedures, were revealed to be generated by specific enzymes and to have biological impact.4-6
In the light of these issues, yet another set of questions applied to Dnmt2-related enzymes. To what stimuli do their activities respond? Do they regulate a cellular response, and if yes, how? How does tRNA methylation mediate such a response? Are tRNA substrates subject to RNA cleavage, and does tRNA methylation affect this process? What are the downstream effects of tRFs?
Over the last decade, the authors of this review article have cooperated as a group on the topic of Dnmt2 enzyme research using various organisms and experimental systems. Accordingly, we here review the field with special emphasis on the developments that have occurred since the seminal paper by Goll et al. in 2006,1 laying out the current state of knowledge with respect to the above-cited questions. Most of the recent progress on understanding Dnmt2 function was made in mammals (human and mouse), Drosophila melanogaster Dictyostelium discoideum, Entamoeba histolytica, and Schizosaccharomyces pombe. The latter 4 are also called “Dnmt2-only” organisms, as they do not encode any other enzymes of the Dnmt family. Further interesting insights into Dnmt2 function were obtained using Geobacter sulfurreducens, one of the very few bacteria containing a Dnmt2 homolog.
Part 1: Phylogeny, structure, and biochemistry of Dnmt2
Evolution of Dnmt2 enzymes
The group of Dnmt2 orthologs presents an unusual evolutionary history. Initially, Dnmt2 enzymes were assigned as putative DNA methyltransferase due to their sequence similarity to other mammalian and bacterial 5mC DNA methyltransferases.7,8 The Dnmt2 enzymes are highly conserved and are present in almost all eukaryotic organisms9,10 and a handful of bacterial species, which most likely acquired them via horizontal gene transfer from a eukaryote.11
Despite the high sequence and structure similarity of Dnmt2 enzymes and canonical DNA methyltransferases,12 early biochemical analyses identified at best only very weak, residual DNA methylation activity on DNA in vitro and often no activity was reported.13 This lack of robust DNA methylation activity spurred new searches for potential other substrates, which led to the identification of tRNAAspGUC C38 as a target of Dnmt2 in Arabidopsis thaliana, mouse and Drosophila melanogaster.1 It was suggested by the Bestor group that other eukaryotic DNA methyltransferases (Dnmt1 and Dnmt3) may have evolved from a Dnmt2-like RNA methyltransferase ancestor that changed its target specificity from RNA to DNA. To investigate this possibility, a phylogenetic analysis of the evolutionary relationship between the Dnmt2 family, and other known prokaryotic and eukaryotic DNA and RNA C5-pyrimidine MTases was performed.9
The phylogenetic analysis indicated that Dnmt2 is separated from Dnmt3a, Dnmt1 and Masc1 clades by numerous prokaryotic DNA methyltransferase enzymes, suggesting that these families of enzymes separately evolved from prokaryotic DNA methyltransferase ancestors. Moreover, in a clustering analysis, the RNA m5C methyltransferases formed a well-defined sequence cluster clearly separated from all the other DNA 5mC methyltransferases, indicating that neither Dnmt2 nor 5mC DNA methyltransferases are closely related to other RNA methyltransferases (Fig. 1).
Figure 1.
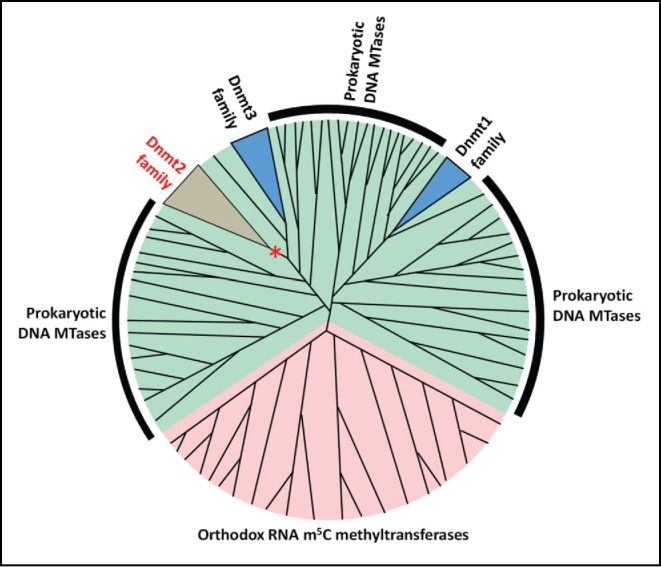
Consensus model of the phylogeny of DNA 5mC / RNA m5C methyltransferases. Dnmt1 and Dnmt2/Dnmt3 enzymes have independent origin in the prokaryotic DNA methyltransferases. The putative precursor of the Dnmt2 family changed its substrate preference from DNA to RNA (indicated with *). Green shading denotes DNA methyltransferases, pink denotes RNA methyltransferases.
Importantly, the RNA and DNA m5C/5mC methyltransferases differ in their catalytic mechanism and the residues that are employed for the catalysis. As detailed below, Dnmt2 enzymes use catalytic motifs that are characteristic for a DNA methyltransferases mechanism,14 despite the fact that they methylate RNA molecules. Taken together, the biochemical data, as well as phylogenetic and clustering analyses suggest that the ancestor of Dnmt2 was a DNA methyltransferase, which changed its substrate preference from DNA to RNA. There is no phylogenetic evidence that Dnmt2 was the precursor of the eukaryotic Dnmt1 family and the preferred hypothesis is that Dnmt2, Dnmt1 and Dnmt3 families had an independent origin in the prokaryotic DNA methyltransferases sequence space.
Structures of Dnmt2 enzymes
To date, crystal structures of 3 Dnmt2 enzymes have been deposited in the protein database (PDB), namely from human (entry 1G55),12 Spodoptera frugiperda15 (4H0N) and from Entamoeba histolytica (3QV2).16 The latter structure is shown in Fig. 2, next to a structure of tRNAAspGUC (1VTQ). All enzyme structures were obtained with SAH (S-adenosyl-homocysteine), the product resulting from the canonical cofactor SAM (S-adenosyl-methionine) after methyl transfer. However, no Dnmt2 structure includes any nucleic acid substrate. The first structure from the Cheng lab has arguably been the most influential, since it was published more than a decade earlier than the others, and concerns the human enzyme. However, in addition to lacking a defined nucleic acid binding site, a loop from catalytic motif IV (vide infra) and other parts of the catalytic center were disordered. In the unrelated DNA methyltransferase M. HhaI, the corresponding loop has been shown to interact with DNA.17 These missing features are more clearly discernible in the S. frugiperda structure, but this enzyme contains a significantly shorter loop. The active site loop (residues 80–100) is better defined in the third structure of Ehmeth, the Dnmt2 homolog of E. histolytica,16 which was solved by the Ficner and Ankri groups. Specifically, the crystal structure of Ehmeth revealed that this loop adopts an α-helical conformation. However, the conformation of the active site loop in Ehmeth is stabilized by crystal packing contacts to neighboring molecules. Therefore, it is conceivable that it might adopt a different conformation in a complex with a nucleic acid substrate. Since the sequences of the active site loops are almost identical in Dnmt2 enzymes, it is tempting to speculate that the active site loop of human Dnmt2 and Ehmeth are both flexible to accommodate either DNA or RNA substrate binding. In summary, while an understanding of the interplay of the catalytic motifs is rather advanced, the currently most burning question as to how the enzymes bind a nucleic acid substrate, still awaits clarification from structural biology approaches.
Figure 2.
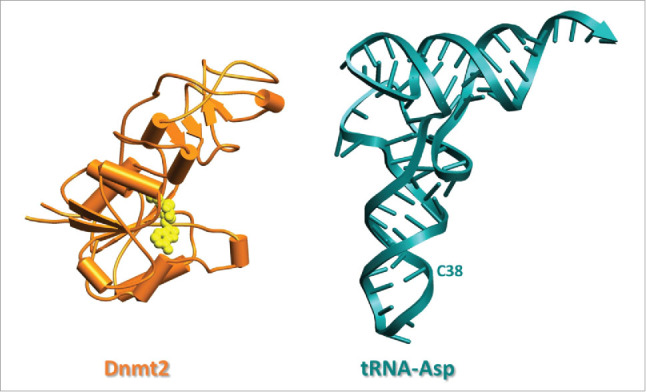
Structure of Entamoeba histolytica methyltransferase EhMeth (3QV2) and structure of tRNAAsp from S. cerevisiae (1VTQ) drawn in same scale. Dnmt2 is shown in schematic drawing in orange with SAM in yellow.
RNA substrates of Dnmt2 enzymes
With the detection of tRNAAspGUC methylation at C38 by Dnmt2 homologues,1 the search for Dnmt2 substrates (Fig. 3) took an entirely new turn. Several organisms were scrutinized for new RNA substrates, both tRNA and others. In addition to tRNAAspGUC, several other tRNA substrates were identified in flies, mice, and amoeba. An overview is given in Table 1. In addition to tRNAAspGUC, specific isoacceptors for tRNAGly, and tRNAVal are also targets for the mouse and Drosophila enzymes.5 The evolutionary trend is quite surprising: in D. melanogaster, tRNAAspGUC, tRNAGlyGCC and tRNAValAAC are methylated in vivo,5 but in S. pombe tRNAGluUUC is also methylated when the enzyme is artificially overexpressed.18 In D. discoideum, tRNAGluUUC/CUC substrates are only modified in vitro but not in vivo.19 A particular substrate choice was described for the prokaryote Geobacter sulfurreducens (vide infra).
Figure 3.
Substrate spectrum of Dnmt2. Dnmt2 has been shown to methylate tRNA-Asp and depending on the species tRNAGly, tRNAVal, and tRNAGlu. Whether other tRNAs, RNAs or DNA are methylated by this enzyme is an open question.
Table 1.
Confirmed tRNA substrates of Dnmt2 enzymes.
| M. musculus | D. melanogaster | D. discoideum | S. pombe | G. sufurreducens | |
|---|---|---|---|---|---|
| tRNAAspGUC | yes | yes | yes | yes, depends on Q or Pmt1 overexpression | no |
| tRNAGluUUC | no | no | in vitro only | weak, only with Pmt1 overepression | Yes |
| tRNAGlyGCC | yes | yes | in vitro only | no | no |
| tRNAValAAC | yes | yes | no | no | no |
During the search for additional Dnmt2 substrates, a variety of conventional methods was employed, and existing methods were modernized. Furthermore, entirely new approaches emerged that revolutionized not only the search for, and mapping of, m5C in RNA, but contributed to the increased interest in the RNA modification field that we see today. Among the more traditional methods, tritium incorporation assays that trace the transfer of 3H-labeled methyl groups from the SAM cofactor to the nucleic acid substrate have remained indispensable. Detection relies on scintillation-augmented detection of radioactivity of precipitated samples or in gels.19 Depending on the purity of protein and nucleic acid preparations, these assays are potentially the most sensitive, but also those most prone to erroneous interpretation, since the formation of 5mC (in DNA) or m5C (in RNA), respectively, is not confirmed by chemical or biophysical means, leaving room e.g. for inadvertent measurement of contaminating methyltransferase activities. Methods that do confirm the identity of a given modified nucleoside by biochemical or biophysical means have shifted from the use of thin-layer chromatography (TLC,20) to high performance liquid chromatography coupled to mass spectrometry (LC-MS,21). Moreover, powerful methods have emerged that use the specific chemical properties of m5C to detect its occurrence in a transcriptome-wide sequence context. Foremost among these is the adaptation of bisulfite sequencing from DNA to RNA by Schäfer and Lyko,22 an accomplishment that has opened up the field to several such methods targeted to the high throughput detection of other RNA modifications, including e.g., m1A23-26 and pseudouridine.27 With RNA bisulfite sequencing meanwhile having been applied to numerous RNA preparations (e.g.28-30), it appears that consistent methylation signals are typically obtained from abundant RNAs such as tRNA and rRNA, while signals in reads from low abundance RNAs tend to be close to noise levels (deamination artifacts). Although m5C candidate sites outside tRNA and rRNA were reported in transcriptome-wide studies,31,30 including also alleged Dnmt2 dependent m5C sites28,30,32 validation by independent methods or by other groups using the same approach has yet remained elusive. Another transcriptome-wide approach for the mapping of m5C sites utilizes azacytidine, a potent inhibitor of m5C:methyltransferases, which forms complexes between a potential methylation target cytidine and the cognate methyltransferase, allowing an enrichment of target sequences by immunoprecipitation and subsequent identification of turned-over residues by misincorporation in the cDNA sequences.32 As expected, the known tRNA substrates of Dnmt2 were detected by this method. Interestingly, while 2 potential non-tRNA, Dnmt2-dependent sites were reported, these do not overlap with any of the previously reported ones.28
Experiments using cross-linking immunoprecipitation (CLIP) of the Dnmt2 homolog in D. discoideum, DnmA, yielded sequence fragments mapping to tRNAAspGUC as well as to tRNAGluCUC/UUC and tRNAGlyGCC. This prompted the suggestion that DnmA may have additional, non-enzymatic functions which are exerted by binding and not necessarily by target methylation.19 Further sequences included snoRNAs, some intergenic non-coding RNAs and the spliceosomal RNAs U1, U2 and U4. U2 is known to contain 2 tRNA like stem-loop structures which contain a C38 equivalent in a sequence context similar to tRNAAspGUC. Out of several candidates tested, U2 was the only one which could be weakly methylated by DnmA and human Dnmt2 in vitro. In vivo methylation by bisulfite sequencing was not detected. Like for tRNAGluUUC and tRNAGlyGCC, it may be that the protein binds to U2 in vivo but has a different function than methylation of the substrate.33 Hence, currently validated in vivo Dnmt2 substrates remain restricted to tRNAs.
Function and specificity of the Dnmt2 homolog found in Geobacter species
As described above, Dnmt2 is widely distributed in eukaryotes, but only very few Dnmt2 homologs were identified in bacteria, one of them in Geobacter species.9 An investigation of the activity of the Geobacter sulfurreducens Dnmt2 homolog (GsDnmt2), which was conducted by in vitro methylation analyses, showed that GsDnmt2 methylates tRNAAspGUC from flies and mice.11 Unexpectedly, it had only a weak activity toward the Geobacter tRNAAspGUC but instead methylated Geobacter tRNAGluUUC with good activity. In agreement with this result, RNA bisulfite methylation analysis showed that tRNAGluUUC was methylated in Geobacter cells while the methylation was absent in tRNAAspGUC. The activities of Dnmt2 enzymes from Homo sapiens, D. melanogaster, Schizosaccharomyces pombe and Dictyostelium discoideum for methylation of the Geobacter tRNAAspGUC and tRNAGluUUC were determined showing that all these enzymes preferentially methylate tRNAAspGUC. Hence, the GsDnmt2 enzyme has a uniquely swapped tRNA specificity. This finding can be interpreted in the context of the acquisition of Dnmt2 by Geobacter through horizontal gene transfer. After this event, a coevolution of Geobacter Dnmt2, tRNAAspGUC and tRNAGluUUC might have occurred, which for currently unknown reasons resulted in a situation where tRNAAspGUC is no longer methylated but tRNAGluUUC has become a very good substrate of Dnmt2 in Geobacter sulfurreducens.
By comparing substrate and non-substrate tRNAs of GsDnmt2, it became apparent that all non-substrates contain a GG dinucleotide in the variable loop as a characteristic feature. Indeed, introduction of this sequence into murine tRNAAspGUC and Geobacter tRNAGluUUC drastically reduced their methylation by GsDnmt2. This result strongly suggested that the variable loop in GsDnmt2 has a role as an important specificity determinant. Interestingly, the same loss of activity was observed with human Dnmt2, indicating that the variable loop might function as a specificity determinant for tRNA recognition by Dnmt2 enzymes. However, introduction of a favorable variable loop into murine tRNAGluUUC did not increase its methylation, indicating that Dnmt2 uses further tRNA-specific sequence determinants in the anticodon stem/loop for RNA recognition.
Dnmt2-mediated DNA methylation
Until the description of tRNA methylation by Dnmt2 enzymes, Dnmt2-dependent DNA methylation was found to be very low or absent both in vitro and in vivo.1,34,35 In D. melanogaster and D. discoideum, retrotransposons were reported to be methylated but the data are disputed.36-39 Curiously, the issue whether Dnmt2 enzymes are also DNA methyltransferases is not yet ultimately clarified, even as of today. Recent reports of Spodoptera frugiperda Dnmt2-dependent DNA methylation by Dnmt2 in vitro,15 or the trace detection of 5mC in Dnmt2-only organisms still fuel the ongoing debate.35,40,41 One possible explanation for the contradictory reports of Dnmt2-dependent DNA methylation might lie in the low expression levels of Dnmt2 enzymes in organisms lacking reproducibly detectable DNA methylation. Indeed, lab strains of D. discoideum, D. melanogaster and E. histolytica were found to lose Dnmt2 expression upon prolonged culture under laboratory conditions(19 and G.R., A.S. unpublished results). As an example, the D. discoideum wild type strain NC4 produces approximately 4-fold more DnmA mRNA compared to the NC4 derived laboratory strain AX2.19 With the exception of zebrafish, which display developmental perturbations42 upon DNMT2 depletion by an antisense technique, Dnmt2 knock-outs or gene disruptions in most organisms studied this far cause mutant phenotype which are indiscernible by superficial analysis on the macroscopic level.1,34 Only upon closer and prolonged inspection of both macroscopic features and molecular details, subtle differences were detected in Dnmt2 mutant organisms (vide infra). From these combined observations it may be concluded that Dnmt2 enzymes are not required for normal development under laboratory conditions. In contrast, the ever-changing conditions in variable environments might make the functions, including RNA and even DNA methylation, of these ancient enzymes advantageous for organismal survival, thus explaining their evolutionary conservation.
Catalytic mechanism of Dnmt2
As described above, the amino acid sequence and 3D structure of Dnmt2 closely resembles DNA (cytosine-C5) methyltransferases12,43,44 Stimulated by this, it was studied how an enzyme that looks like a DNA methyltransferase could methylate RNA.14 Mechanistically, the C5 methylation of cytosines is not a trivial reaction, because cytosine is an electron-poor heterocyclic aromatic ring and the C5 of cytosine cannot easily perform a nucleophilic attack on the methyl group of SAM without activation. Therefore, the reactions catalyzed by RNA- and DNA (cytosine-C5) methyltransferases follow a reaction pathway involving activation by a Michael addition. DNA methylation is initiated by a nucleophilic attack of a catalytic cysteine residue located in a conserved amino acid sequence motif (motif IV PCQ) on the C6 position of the target cytosine.44 The enzyme facilitates the nucleophilic attack by a transient protonation of the cytosine ring at the endocyclic nitrogen atom N3, which is stabilized by the glutamate residue from another highly conserved motif (motif VI ENV). The attack of the Cys on the cytosine C6 leads to the formation of a covalent protein-DNA complex. Thereby, the C5 position of the cytosine is activated and performs a nucleophilic attack on the SAM methyl group. The covalent protein-DNA complex is resolved by deprotonation at the C5 position, which leads to the elimination of the cysteine SH group and the reestablishment of aromaticity. Then, the methylated base together with the cofactor product, S-adenosyl-L-homocysteine, is released. In addition to the residues already mentioned, a second arginine residue in motif VIII (RXR) plays an important role in the catalytic mechanism of DNA (cytosine-C5) methyltransferases.45 Unexpectedly, Liu and Santi showed that RNA (cytosine-C5) methyltransferases do not use a cysteine from motif IV for the initial attack on the base, but instead use one located in motif VI (TCS in RNA MTases).46 Moreover, instead of using the glutamate residue located in motif VI of DNA methyltransferases (ENV), RNA methyltransferases use an aspartate residue from motif IV (DAPC) to stabilize the transition state of the reaction.47
Dnmt2 proteins contain all the conserved residues that are used for catalysis by DNA-(cytosine C5)-methyltransferases. Using a radioactive in vitro tRNA methylation assay and tRNA isolated from biological sources as well as in vitro transcribed tRNAAspGUC the mechanism of human Dnmt2 was studied.14 Using site directed mutagenesis, it was shown that Dnmt2 has a DNA methyltransferase-like mechanism, because similar residues from motifs IV, VI and VIII are involved in catalysis as identified in DNA-(cytosine C5)-methyltransferases. Exchange of C292, which is located in a CFT motif conserved among Dnmt2 proteins, reduced the catalytic activity of Dnmt2, but it showed clear residual activity, suggesting that this residue has no direct role in catalysis. Additional cysteine residues in Dnmt2 had no role in catalysis. These results showed that Dnmt2 represents the first example of an RNA methyltransferase using a DNA methyltransferase type of mechanism. It relates to the evolutionary background of Dnmt2 as outlined above.
Mapping of the tRNA binding site
Since there is no tRNA/Dnmt2 co-crystal structure available, it was attempted to map the tRNA binding site of human Dnmt2 by systematically mutating 20 surface exposed lysine and arginine residues to alanine (as shown in Fig. 4) and studying the tRNA methylation activity and binding of the corresponding variants.48 The rationale behind this approach was that nucleic acid binding pockets in proteins usually contain basic residues that are engaged in electrostatic interactions with the phosphodiester backbone. Eight basic residues were identified that caused a strong (more than 4-fold) reduction in catalytic activity but no large changes in SAM binding (R95, R84, R275, K367, R371, R289, K122, and K295, in the order of increasing residual activity). These residues cluster within and next to a cleft on the surface of Dnmt2, which is located next to the SAM binding pocket, and the catalytic residues of Dnmt2 are positioned at its walls. Manual placing of tRNA into the structure suggests that Dnmt2 mainly interacts with the anticodon stem-loop, which is in agreement with other data, for example the recognition of the queuosine base at position 34 of tRNAAspGUC by Dnmt2 proteins from some species (vide infra). Also, a role of the extra loop described above is in accordance with this model.
Figure 4.
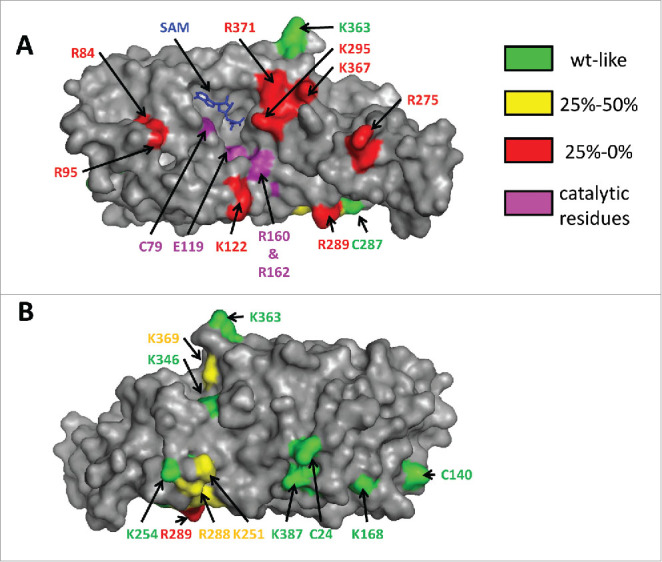
Different views of DNMT2 in surface representation. The residues subjected to mutagenesis are colored according to their residual activity. The cofactor product S-adenosyl-l-homocysteine is colored blue. Panel A shows that residues which strongly interfere with catalysis (colored red here) cluster on the “front” face of the enzyme and surround a cleft that also contains the SAM binding pocket and the catalytic residues. Panel B represents a view of the “back side” of the enzyme after a 180° rotation of the view shown in panel A about the vertical axis. Reproduced from48 with permission. Copyright (2012) American Chemical Society.
Another interesting result from this study was that many of the Dnmt2 variants with strongly reduced catalytic activity showed only a weak loss of tRNA binding or bound even better to tRNA than wild-type Dnmt2, indicating that these residues do not have a major role in ground-state tRNA binding. As the kinetic data implicate an important role of these residues in transition state binding, this result suggests that the enzyme induces conformational changes of the tRNA substrate during the catalytic cycle.
Part 2: Effects of tRNA methylation by Dnmt2 enzymes
The role of Dnmt2 in protein translation
As outlined above (Table 1), Dnmt2-dependent tRNA methylation is largely specific for the C38 residue of 3 tRNAs: tRNAAspGUC, tRNAGlyGCC and tRNAValAAC in human and Drosophila cells.5,39 As such, Dnmt2-mediated tRNA methylation is targeted to a position that is close to the wobble base,49 which is subject to various modifications that affect protein translation.50 Together, these findings raised the possibility that Dnmt2 might also be involved in the regulation of protein translation.
First insight into the function of Dnmt2 in protein translation came from the analysis of transgenic mice that lacked both Dnmt2 and NSun2, another cytosine-5 tRNA methyltransferase in mammals.51 tRNA methylation analysis demonstrated complementary target-site specificities for Dnmt2 and NSun2 and a complete loss of cytosine-5 tRNA methylation in double-knockout mice.52 While mRNA levels were not detectably changed, steady-state levels of unmethylated tRNAs were substantially reduced, resulting in significantly reduced rates of protein synthesis.52 These findings demonstrated that a complete loss of cytosine-5 tRNA methylation has dramatic consequences for the stability of certain tRNAs and established a role of tRNA methylation in protein translation.
Interestingly, two more recent studies revealed additional mechanistic details about the interplay between Dnmt2-mediated C38 methylation and protein translation. As the addition of methyl groups often has a role in modulating the specificity of protein-nucleic acid interactions, the role of C38 methylation in the charging of tRNAAspGUC was investigated in vitro.53 This approach was based on the fact that C38 has been identified as a key identity element of aspartyl-tRNA-synthetase (AspRS) in bacteria, archea and eukaryotes.54 Using recombinant mouse AspRS and synthetic tRNAAspGUC which was methylated or unmethylated, respectively, at C38 for in vitro aminoacylation assays, it was shown that AspRS had a clear preference for the C38 methylated tRNAAspGUC (Fig. 5A). In agreement with this, the charging level of tRNAAspGUC in Dnmt2 knockout murine embryonic fibroblast cells was reduced. An expression analysis with fluorescent reporter proteins fused to an N-terminal poly-Asp sequence showed that protein synthesis of poly-Asp tagged reporter proteins was markedly reduced in Dnmt2 KO cells. The same effect was observed with endogenous proteins containing poly-Asp sequences indicating that Dnmt2 mediated C38 methylation of tRNAAspGUC regulates the translation of proteins containing poly-Asp sequences.
Figure 5.
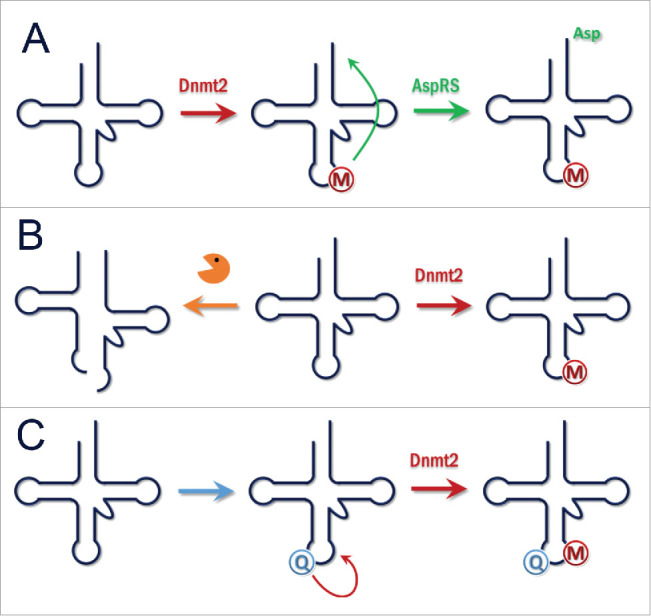
(A) Function of Dnmt2. Methylation of tRNAAspGUC enhances its charging by AspRS. This stimulates the translation of Asp-rich proteins. (B) Function of Dnmt2. Methylation of tRNAs protects them from cleavage into tRNA fragments and increases overall translation. Massive production of tRNA fragments after loss or downregulation of Dnmt2 affects Dicer dependent processing of small RNAs. C) Function of Dnmt2. Depending on nutrition or other signals, Queuosine is incorporated into tRNAAspGUC at position 34. This stimulates Dnmt2 mediated methylation and triggers downstream effects.
The selective reduction in translational efficiency of poly-Asp proteins observed in that study represented a novel mechanism of post-transcriptional regulation mediated by methylation of tRNAAspGUC. Gene ontology analyses showed that poly-Asp proteins tend to have nuclear localization and that they have roles in gene expression and transcription regulation. Based on these results, it was proposed that Dnmt2 mediated tRNAAspGUC methylation represents a novel mechanism of translational fine tuning of groups of proteins containing poly-Asp sequences. Moreover, homo-polymeric stretches of amino acids other than poly-Asp are widespread in the human proteome and could have a similar regulatory role. This translational regulation pathway could participate in the Dnmt2-mediated stress response together with other Dnmt2 mediated effects on protein biosynthesis. A similar concept emerged from an independent study which showed that systematic differences in protein expression of Dnmt2-deficient mice were due to specific codon mistranslation by tRNAs lacking Dnmt2-dependent methylation.55 These findings demonstrated a novel and highly interesting function of C38 tRNA methylation in the discrimination of near-cognate codons and in the maintenance of accurate polypeptide synthesis.55
Of note, several post-transcriptional modifications in the anticodon loop of tRNAs have been shown to be important for the fine-tuning of protein translation and for the maintenance of proteome integrity in yeast and in C. elegans.56-58 In Dnmt2-deficient mice, ribosome profiling showed a decrease of codon occupancy in the ribosome A-site for all Dnmt2 target codons.55 This was explained by a model where the absence of C38 methylation in the anticodon loop reduces the available time for the discrimination between the near-cognate tRNAAspGUC and tRNAGluUUC codons, thereby reducing translational fidelity. Dnmt2-mediated tRNA methylation could therefore play an adaptive role in codon usage, thus functioning as a modulator of protein translation. This hypothesis is further supported by recent findings that link Dnmt2-mediated C38 methylation to queosinylation, a nutrient-dependent modification of the wobble base,59 which will be discussed below in more detail.
Taken together, these findings suggest a direct mechanism for nutrient-, or stress-dependent recoding of entire genomes, which could facilitate cellular and/or organismal adaptation to changing environments.
tRNA methylation affects processing into tRNA fragments (tRFs)
The absence of strong Dnmt2 developmental phenotypes in most organisms suggested biological roles under non-standard laboratory conditions. Based on detailed analyses of Dnmt2 mutants in Drosophila reduced viability under heat and oxidative stress conditions was detected.5 In addition, tRNA fragmentation, which is a conserved response to various stresses, was increased in Dnmt2 mutant Drosophila. Using biochemical assays, it was shown that the absence of m5C at position 38 of 3 tRNAs (tRNAAspGUC, tRNAGlyGCC and tRNAValAAC) affected their fragmentation by stress-induced tRNA endonucleases5 (Fig. 5B). These findings uncovered a previously unknown molecular function for Dnmt2-mediated methylation, which influences the production of specific stress-induced tRNA-derived small RNAs through methylation of the anticodon loop. It is presently unclear whether the missing modification affects the folding of the tRNA, which in turn might allow increased access to the anticodon loop, or if m5C changes the sequence recognition of stress-induced endonucleases. Importantly, these results also sparked interest in studying the influence of m5C at other tRNA positions on stress-related tRNA stability.60 In addition, these observations were crucially important for the realization that certain aspects of Dnmt2 function can be uncovered when performing experiments under non-standard laboratory conditions.
It has been shown that tRFs inhibit translation without the need for complementary of tRF sequences to target sites in mRNA.61 In addition, tRFs could serve as long-distance signals. For instance, tRFs have been found in the roots of Arabidopsis thaliana , where their levels are increased upon phosphate starvation.62 Importantly, tRFs are present in various body fluids63 where they are thought to be constituents of exosomes. Interestingly, tRFs have also been found in mouse sperm and have been discussed as information carriers for diet-induced metabolic syndromes between generations.64,65 These observations indicate that tRFs are part of a repertoire of small RNAs that acts at a distance, thereby affecting other cells or tissues in response to stress such as changing nutrient availability, infection or other environmental stimuli. While stress-induced tRNA fragmentation is widely conserved, the scope of the biological impact of tRNA-derived small RNAs during stress but also in individuals with aberrant tRF production remains unclear. Most importantly, the mechanistic details of tRF interactions and activities are ill-defined and are a matter of active debate.66,67-69
Intriguingly, a Dicer-dependent class of human tRFs was identified, which exerted down-regulating activity on target genes in trans through differential Argonaute protein association, implying regulatory cross-talk of tRFs with siRNA pathways.4 Because tRNA fragments have repeatedly been found in association with small-interfering RNA (siRNA) components, it was tested whether increased tRNA fragmentation in Dnmt2 mutants affected the activity of siRNA pathways in Drosophila. Using RNA-immunoprecipitations it was found that Drosophila Dicer-2 (Dcr-2) and Argonaute-2 (Ago-2) protein complexes contained tRNA fragments from Dnmt2 substrate tRNAs that were not only halves (ca. 35 nucleotides) but also in the size range of siRNAs (18–21 nucleotides).70 Assays for Dcr-2 activity showed that Dcr-2 bound and degraded tRNA fragments into siRNA-sized RNAs. Importantly, tRNA fragments were able to inhibit Dcr-2 activity on exogenous long double-stranded RNAs (dsRNAs) in vitro experiments. Dcr-2 activity was also affected in vivo, causing not only the accumulation of endogenous dsRNAs but also reduced siRNA production and the mis-regulation of Dcr-2-dependent gene expression in Dnmt2.70 These findings revealed the impact of increased tRNA fragmentation in Dnmt2 mutants on small RNA pathways and indicated that Dnmt2/Trdmt1 function affects the regulation of small RNA homeostasis and gene expression, especially during stress conditions.
Part 3: Expression and regulation of Dnmt2 enzymes
Dnmt2 expression and sub-cellular localization
Although Dnmt2 enzymes are highly conserved, very little information exists about the expression patterns and sub-cellular localization of these proteins. Interestingly, all published or commercially available antibodies against Dnmt2 enzymes are only partially useful because they only detect Dnmt2 polypeptides in the denatured state (i.e. by Western blotting) but proved to be insufficient for in situ detection by, for instance, immunofluorescence approaches. Early experiments using Northern blotting and RNA in situ hybridization in flies indicated high expression of Dnmt2 in female germ line tissues and in embryos.71 Overexpression of an epitope-tagged human Dnmt2 in cell culture showed both cytoplasmic and nuclear localization.1 Biochemical fractionation experiments in Drosophila showed that Dnmt2 is mostly a cytoplasmic protein with a minor nuclear fraction tightly attached to the nuclear matrix.72 Furthermore, Drosophila Dnmt2 is ubiquitously expressed throughout embryonic development into adulthood, where the protein is enriched in female germ line tissues. Using Dnmt2-EGFP constructs that express a fusion protein under the minimal Dnmt2 promoter and immunofluorescence detection it was found that Dnmt2-EGFP is strongly expressed in highly proliferating or metabolically active cells/tissues such as the neuroblasts of the developing embryonic or larval central nervous systems as well as in specific regions of the adult germ line.72 Of note, using Dnmt2-EGFP and live cell imaging in synchronously dividing embryonic nuclei a cell cycle-dependent re-localization of the protein from cytoplasmic speckles during interphase to spindle-like structures during mitoses was detected.72 Importantly, larval mitotic cells allowed Dnmt2 access to nuclear sequences as evidenced by a trans-activating reporter system that involved Dnmt2-GAL4 fusion proteins mediating the activation of UAS-EGFP sequences in the nucleus.72 These results suggest a function of Dnmt2 enzymes during the cell cycle in actively dividing cells, which includes access of Dnmt2 to DNA sequences.
Support for a cell cycle-dependent function of Dnmt2 enzymes comes from studies in Dictyostelium. Of note, DnmA in the Dictyostelium laboratory strain AX2 is very low and not detectable by Northern blots. On the other hand, the parental wild type isolate, NC4, which is recalcitrant to routine molecular experimentation, shows a 5-fold higher DnmA expression in growing cells and a 30-fold increase of expression during specific stages of development as detected by quantitative PCR. DnmA mRNA expression in AX2 peaks at 16 hours of the 24 hour long developmental life cycle.19 Exactly at this time in the development of Dictyostelium mostly synchronous mitotic cell divisions occur. This peak of DnmA expression corresponds to a 5-fold increase over basal expression levels in vegetative growing cells. When examining DnmA expression levels during the cell cycle in experimentally synchronised cells, an approximately 6-fold increase in mRNA levels in late mitosis or early S-phase was found. Since no suitable antibodies against DnmA were available, DnmA localization was examined after overexpressing GFP-tagged DnmA, and it was found that the protein localized mostly to the nucleus. Time-lapse fluorescence microscopy showed that DnmA was rapidly lost from nuclei shortly before mitosis and re-accumulated after the completion of cell division during S-phase. Since Dnmt2-mediated RNA methylation impacts tRNA stability during stress conditions, DnmA expression after temperature shock or oxidative stress was examined. It was found that neither heat shock nor oxidative stress changed DnmA mRNA levels. However, during recovery from cold shock, DnmA mRNA levels increased transiently about 40-fold indicating that DnmA expression responds to particular stresses. Importantly, increased DnmA mRNA expression during NC4 development was accompanied by significantly higher tRNAAspGUC methylation at the DnmA target site C38.19 However, no increased enzymatic activity was observed for DnmA during the recovery from cold shock indicating that mRNA expression and enzyme activity are not necessarily strictly correlated. Taken together, the available information indicates that developmental timing, the cell cycle, the nature of the model organism (lab strain or wild type isolate) as well as stress conditions all affect the expression, sub-cellular localization and, ultimately, the activity of Dnmt2 enzymes. Especially, the involvement of Dnmt2 enzymes in stress-related cellular mechanisms raises the possibility that the high evolutionary conservation of Dnmt2 proteins might be explained by partaking in biological responses to environmental challenges (i.e., stress) that facilitate, for instance, nucleotide-based pathogens to enter cells, including varying RNA and DNA substrates, a situation that is not easily recapitulated under standard laboratory conditions.73
Regulation of Dnmt2 activity by the micronutrient queuine
Until recently, relatively little was known about how Dnmt2 activity is regulated in order to modulate the effect of C38 methylation on the diverse aspects of tRNA biology, both in different cell types and under varying environmental conditions. Insight into this question comes from work on the Dnmt2 homolog from fission yeast (Schizosaccharomyces pombe, Pmt1). A first surprising observation was that in vivo Dnmt2-dependent tRNAAspGUC methylation in S. pombe was induced by the presence of peptone in the growth medium.18 The realization that peptone contains the modified guanine base queuine led to the discovery that in vivo Dnmt2 activity shows a striking enhancement on tRNAAspGUC after prior incorporation of queuine into the tRNA59 (Fig. 5C). Queuosine (Q, which designates the nucleoside; the respective base is called queuine, q) is a hyper-modified 7-deaza-guanosine that replaces guanosine at the wobble position (position 34) of tRNAs with a GUN anticodon (tRNAAsnGUU, tRNAAspGUC, tRNAHisGUG and tRNATyrGUA74). Although queuosine was first reported not to alter the codon-recognizing properties of guanosine,75 its presence in tRNAs may yet alter decoding of mRNA. Interestingly, queuosine is present in prokaryotic as well as eukaryotic tRNAs. Remarkably, only bacteria can synthesize queuine de novo, whereas eukaryotes depend on scavenging the precursor queuine from the anticodons of bacterial tRNAs that are obtained from nutritional sources and/or are maintained in microbiomes. Queuine is incorporated into eukaryotic tRNAs by the tRNA-guanine transglycosylase (TGT).76
The initial observation was an increase in vivo of C38 methylation of tRNAAspGUC from 14% to 100% when S. pombe was cultivated in the presence of queuine.59 This stimulation depended on the incorporation of queuine into the tRNA, since it was abrogated when the genes encoding components of the TGT were deleted. However, S. pombe Dnmt2 was found to not be strictly dependent on the queuosine modification, because unmodified tRNAs that were obtained from in vitro transcription were good methylation substrates for Pmt1 in vitro. Yet, the in vitro activity was stimulated on a queuosine-containing substrate. This argues for a direct activation of Dnmt2 by the queuosinylated target, but also suggests additional regulatory mechanisms for Dnmt2 in vivo that were not recapitulated in vitro. It will be interesting to see whether queuosine in the tRNA increases Dnmt2 activity by increasing the affinity of the enzyme for its substrate. In this regard, structural information of Dnmt2 with the queuosinylated substrate may allow insights into the mechanism of activation.
Sequential modification of tRNAs has been previously described. The characterization of mitochondrial tRNAAspGUC species isolated from opossum revealed that queuosine modification (Q34) occurs subsequent to methylation at positions A9 and G10, pseudouridine conversions and editing of the anticodon (C to U). Interestingly, however, C38 methylation of tRNAAspGUC was not described in this study.77 Furthermore, the efficient formation of wybutosine at G37 of tRNAPhe in yeasts depends on prior 2′-O-methylation events imparted by Trm7.78 Also, the SPOUT methyltransferase TrmL from E. coli introduces a 2′-O-methyl group of tRNALeuCAA and tRNALeuUAA only after prior N6-isopentenyladenosine modification at position 37.79 An important future goal will be to determine whether the queuosine modification regulates other tRNA modifications apart from C38 methylation.
Queuosine incorporation and C38 methylation are both modifications of the tRNAAspGUC anticodon loop, raising the question of whether they affect translation. One scenario is that the 2 modifications co-ordinately regulate the interaction between codon and anticodon during translation. Queuosine is present at the wobble position (Q34) of tRNAAspGUC. Two synonymous codons for aspartate exist, GAC and GAU, and they are distributed with a ratio of 0.4 (GAC : GAU) in the S. pombe genome.80 However, since S. pombe tRNA genes only encode tRNAAspGUC, this tRNA must decode both Asp codons.
Conflicting data exist regarding the effect of Q on translation. Queuosinylation of tRNAHisQUN when injected in Xenopus oocytes improved translation of NAU codons more than that of NAC codons, thus providing a means of “equalization” between the 2 codons.81 Conversely, Q-modified tRNA bound C-ending codons more stably than U-ending codons.82 Also, Q-modified tRNA had a higher apparent affinity for ribosomes than the equivalent G-containing tRNA.81,83 The proximity of C38 to the anticodon suggests that C38 methylation may also be implicated in decoding. The combined modifications queuosine34 and m5C38 may modify the decoding of aspartate codons, and thus fine-tune the speed of translation, across the S. pombe genome, which is expected to be particularly relevant for proteins with a high aspartate content53 (see above). Furthermore, the modifications may coordinately suppress the misincorporation of amino acids into proteins and may thus improve the accuracy of translation.
An alternative hypothesis for the biological function of queuosine and C38 methylation is that either one, or both in cooperation, protect tRNAAspGUC from cleavage in the anticodon loop by ribonucleases, which generate tRNA halves that may have regulatory roles outside of translation (see other parts of this review). Such a protection from cleavage and concomitant stabilization of tRNAAspGUC by Dnmt2-dependent methylation has been documented in flies and mice.5,39,55 Interestingly, the bacterial ribonuclease colicin E5 was reported to selectively cleave all tRNAs with a Q34,84 though it is not clear whether the presence of queuosine would affect cleavage. We speculate that Q34 and C38 methylation might cooperate to provide protection of tRNAAspGUC from cleavage, and that they thus limit the production of tRNA fragments.
Evolutionary conservation of queuine-mediated Dnmt2 stimulation
An important question that was immediately raised by the discovery of a queuosine dependence of Dnmt2 in S. pombe is how universal this activation is across evolution. Has the queuosine dependence in other organisms not been observed previously, because they are routinely cultivated in high queuine conditions? Or are other Dnmt2 homologs refractory to queuosine? Intriguingly, a stimulation of Dnmt2 in D. discoideum upon cultivation in the presence of queuine was observed,59 arguing for at least some level of evolutionary conservation of this mode of activation.
A further promising candidate for Q-dependent stimulation is Ehmeth, the Dnmt2 homolog from E. histolytica. The fact that queuine in mammals is taken up in the gut from microbiome sources,74 lends support to the idea that the pathogenicity of this parasite might be linked to queuine, since it has become evident over the last few decades that its pathogenicity is related to its interaction with the gut microbiota. The composition of the gut flora in patients suffering from amebiasis showed a significant decrease in the population size of bacteroides, Clostridium coccoides, Clostridium leptum, Lactobacillus and Campylobacter and an increase in Bifidobacterium, while there was no change in Ruminococcus compared to healthy patients.85 These findings suggest that the pathogenesis of amebiasis might be driven by a dysregulated microbiome or crosstalk between enteropathic bacteria, the parasite, and the intestinal immune system. This crosstalk may be modulated by chemical signaling molecules, such as short-chain fatty acids (SCFAs), which are released by the bacteria.86 The discovery of a queuosine dependence of Dnmt2 in S. pombe opens up the possibility that the biology of the parasite may be affected by queuine.
Significantly, preliminary work indicates that queuine metabolism and methylation of C38 in tRNAAspGUC by Ehmeth are in fact linked: the methylation level of C38 in tRNAAspGUC of queuine-cultivated trophozoites is significantly higher than the level in control trophozoites (trophozoites cultivated in absence of queuine). This may alter the physiology of the parasite, since the trophozoites that were cultivated in the presence of queuine were more resistant to OS and NS than those that were cultivated in its absence (S. Ankri, unpublished).
Given the 3 documented cases of S. pombe, D. discoideum and A. histolytica, how conserved is the queuine dependence of Dnmt2 in metazoans, including mammals? The involvement of the micronutrient queuine in Dnmt2 regulation potentially has far-reaching implications. Since eukaryotes, including humans, incorporate queuine from bacterial sources into their tRNA,74 this raises the question whether mammalian Dnmt2 is also stimulated by the presence of queuosine in the target tRNA. If true, this would be a possibility for the environment in the broadest sense, and in particular nutrition and the microbiome in the mammalian gut, to manipulate translation and proteome composition of the host. Thus, differences in the composition of the microbiome, which have been associated with obesity, cancer and inflammatory diseases,87 could cause altered translation and thus affect disease states both positively and negatively due to differences in queuosine production by the microbiota.
An interesting further question will be whether the queuosine content is equal in all tissues and at all developmental stages, or whether differences in queuosine levels differentially affect translation in different cell types. In Drosophila, the queuosine content of tRNAs varies across development, being lowest in fast growing third-instar larvae and highest in adult flies.88,89 Interestingly, these queuine levels may explain codon preference in genes that are differentially expressed during development as well as across different Drosophilid species.90 It will thus be important in the future to assess the in vivo effect of queuosine in mammals and the sensitivity of mammalian Dnmt2 to queuosinylated tRNAs.
Between nutrition and stress: Extrinsic stimuli affect activity of Dnmt2
While tRNA methylation in dependence on glucose may arguably be perceived in a nutritional context, glucose starvation (GS) is one of the most studied metabolic stresses and particularly relevant to E. histolytica. This pathogenic single cell eukaryote lives in the colon, a niche where the amount of available glucose for fermentation is usually small due to the high absorptive capacity of the glucose transporters in the small intestine.91,92,93 On rare occasions, it has been reported that E. histolytica trophozoites leave the colon, possibly as a reaction to low glucose levels, and migrate to the liver, where the concentration of glucose was estimated to be twice that of perfusing blood.94 It was found that E. histolytica is capable of responding to changes in its surrounding glucose concentration: short term glucose starvation (12 hours) led to the accumulation of enolase, a glycolytic enzyme, in the nucleus. Enolase interacted with the catalytic site of Ehmeth, subsequently inhibiting its tRNA methyltransferase activity.95 Up to this day, enolase is the only identified protein partner for Ehmeth. Extending the condition of glucose starvation beyond 12 hours led to the progressive death of most of the parasite's population. Surprisingly, some individual clones survived and adapted to this absence of glucose in the media. Adaptations included a number of metabolic changes including expression changes of various catabolic enzymes involved in amino acid regulation. The Ankri and Ficner groups recently solved the crystal structures of both E. histolytica enolase96 and of Ehmeth.16 Concerning E. histolytica enolase, it displays a disulfide bond formed between Cys147 and Cys169 in each monomer despite the presence of 1 mM dithiothreitol during the purification. This disulfide bond has not been observed in any other enolase structures solved to date. Therefore, the relevance of this disulfide bond in the redox-regulation of E. histolytica enolase needs still to be determined. Despite these interesting molecular details on Ehmeth and enolase, the structure of the Ehmeth-enolase hybrid remained elusive.
The above insights on nutritional stimulation of Ehmeth in E. histolytica are reminiscent of observations in S. pombe that indicate stimulation of Dnmt2 activity by other factors than queuosine. Specifically, growth of S. pombe cells in minimal rather than full medium stimulated tRNAAspGUC methylation to approx. 23% in the complete absence of queuosine, thus revealing a queuine-independent mode of Dnmt2 stimulation. Furthermore, nitrogen depletion also increased tRNA methylation in vivo. Interestingly, Dnmt2-dependent tRNA methylation in S. pombe is regulated by the nutrient signaling kinase Sck2, a homolog of S. cerevisiae Sch9, which is a major target of the Tor pathway.18 Nutrient sensing thus leads to the activation of a signaling pathway that also activates Dnmt2-dependent tRNA methylation, possibly by direct phosphorylation of Dnmt2. Alternatively, some of the induction may be due to transcriptional regulation of the Dnmt2 gene in S. pombe. As for any Dnmt2-related observations, it will be important to determine how evolutionarily conserved the regulation of Dnmt2 by nutritional cues is.
Involvement of Ehmeth in resistance to oxidative and nitrosative stress
Passage of E. histolytica from the anoxic luminal colon into the tissues/bloodstream of the human host brings about a dramatic change in environmental pO2. Moreover, the parasite must now withstand the assaults of the human immune system, including oxidative bursts of superoxide anions. The role of Dnmt2 in the resistance to oxidative stress (OS) was first investigated in D. melanogaster. Dnmt2 overexpression induced small Heat Shock Protein (HSP) expression in D. melanogaster.97 This facilitated the stabilization/sequestration of damaged or misfolded proteins that result from oxidative damage.98 Similarly, it was demonstrated that Ehmeth overexpressing trophozoites exhibited significantly greater resistance/survivability to H2O2 (an inducer of OS) exposure. This resistance is correlated with an upregulation of HSP70 expression.99 Ehmeth expression did not directly induce HSP70 expression via methylation of its promoter, implying that there might be other players or mediators involved in the process.99 Furthermore, it was recently reported that overexpression of Ehmeth confers resistance to nitrosative stress (NS).100 Ehmeth-mediated resistance to NS was found to be associated with (i) high level of tRNAAspGUC methylation, (ii) persistence of protein synthesis under NS and (iii) specific expression of proteins like ADH2 and peroxyredoxin, which are involved in protein translation, protein transport, vacuolar sorting protein signaling and resistance to OS and NS.100 Since hypermethylation of C38 in tRNAAspGUC was associated with OS and NS resistance,99,100 it was posited that C38 methylation in tRNAAspGUC might lead to selective translation of proteins that are encoded by GAC-enriched genes and that these proteins might be involved in the parasite's resistance to OS and NS. In support of this postulate, it was found that, of the 2 codons for Asp (GAT, GAC) in E. histolytica, GAC is rarely used (see http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=5759). It was also found that 21 genes exclusively use GAC to code for Asp and 5 of these genes encode ribosomal proteins. Three of these ribosomal proteins were upregulated in Ehmeth-overexpressing trophozoites which were exposed to NS.100 These responses are similar to those found when Saccharomyces cerevisiae was exposed to OS, where Trm4-dependent increases in tRNALeuCAA methylation and a consequent increased translation of ribosomal proteins from TTG-enriched genes were reported.101 These findings suggest that C38 methylation in tRNAAspGUC might lead to selective translation of proteins from GAC-enriched genes, and that these proteins might be involved in the response of oxidatively- and nitrosatively-stressed E. histolytica trophozoites.
Part 4: Potential implications of Dnmt2 enzymes in cancer, retrotransposon silencing, and epigenetic heredity
Functional role of somatic cancer mutations in Dnmt2
Recent data indicate that the expression levels of tRNA methyltransferases are frequently altered in cancer cells.102 In correlation with this finding, Dnmt2 was found up-regulated in hundreds of tumor samples listed in the COSMIC data base103 and more than 60 somatic mutations in Dnmt2 were found in tumors originating from various tissues. Based on this data, Dnmt2 is not among the most frequently mutated genes, but mutations occur frequently and regularly. The distribution of mutations found in the dnmt2 gene were compared and recurrent exchanges at some specific residues were observed, which suggests potential significance of functional effects resulting from the mutations.104 To investigate this further, 13 mutations (out of the 48 missense mutations at 37 amino acid residues reported in the database in spring 2014 when that study was initiated) were selected, and the mutant genes were generated, followed by expression and purification of the mutated Dnmt2 proteins.104 All proteins were properly folded as determined by circular dichroism spectroscopy. Their RNA methylation activity was tested using in vitro generated tRNAAspGUC. Two exchanges led to a strong decrease in activity (G155S and L257V) and 2 more mutant proteins were almost inactive (R371H and G155V). On the other hand, one of the somatic cancer mutations (E63K) caused a twofold increase in activity. The observed effects of the somatic mutations can be interpreted with respect to structure-function relationship in some cases, indicating a good level of understanding in this respect. R371 is located next to the active center and part of the conserved motif X, which has a role in the substrate interaction. A similar decrease of methylation activity was previously observed after an exchange of R371 to Ala which was shown to be caused by reduced tRNA binding.48 G155 is a hotspot of somatic cancer mutations in Dnmt2. This residue is located on the back-side of the protein, but it is located right in front of motif VIII, which has an important catalytic role in DNA-(cytosine C5)-methyltransferases and Dnmt2.14 L257 is located on the back-side of Dnmt2 in a non-conserved part of the amino acid sequence and it is unclear why a relatively conservative exchange of this residue by Val caused a strong reduction of activity. The E63 residue, exchange of which to K caused an increase in activity, is also located on the back-side of Dnmt2. It is interesting that Dnmt2 enzymes from other eukaroytic species like S. pombe, E. histolytica or D. melanogaster carry a Lys at the position corresponding to E63. It has been shown previously that mutations changing a position to the phylogenetic consensus residue at this place often increase protein stability and activity105 which may also explain the stimulatory effect of the E63K mutation.
Potential involvement of Dnmt2 in the control of silencing processes
Lower expression levels of Dnmt2 in “domesticated” laboratory strains compared to wild type populations, as discussed above, might also depend on possible differences in content and organization of specific sequences including intact retrotransposons or tandem and dispersed repetitive elements. Consequently it was important to identify putative DNA target sequences of Dnmt2. In Drosophila, Dnmt2 loss-of-function mutations were isolated either by gene knockout1 or after re-mobilization of a P-element transposon inserted upstream of the Dnmt2 gene.38,39 Homozygotes mutants isolated after imprecise P-element excision do not express Dnmt2 protein. Like Dnmt2 knockouts or gene disruptions in other organisms, also in Drosophila the Dnmt2 mutations are viable, fertile and without obvious phenotypic defects.34,37 Only upon closer inspection, subtle molecular differences were detected in Dnmt2 mutant animals.5 Importantly, utilizing genetic systems monitoring heterochromatic silencing processes identified an essential role for Dnmt2 in the control of retrotransposons and repeat- dependent gene silencing. Most silencing monitoring systems in Drosophila depended on juxtaposition of the white marker gene to genomic regions of heterochromatic chromatin conformation resulting in a white variegated phenotype.106 In about 40 white variegated transgenes inserted into LTR regions of Invader4, HeT-A, Doc and Copia retrotransposons, the Dnmt2 mutation caused dominant suppression of gene silencing at the white gene.38 In addition, RT-PCR analysis revealed significant over-expression of retrotransposons in Dnmt2 mutant embryos suggesting a genome-wide role of Dnmt2 in retrotransposon silencing. Dnmt2 mutations also strongly suppressed silencing at a tandemly repeated white transgene.107 However, Dnmt2 mutations showed no effect on gene silencing associated with pericentric heterochromatin. Two different Dnmt2-dependent silencing pathways could be identified. Retrotransposon silencing includes, beside Dnmt2, the H4K20 methyltransferase SUV4-20 and the JIL1 H3S10 phosphatase whereas repeat-dependent gene silencing was controlled by Dnmt2, the histone H3K9 methyltransferase SETDB1, the heterochromatin protein HP1a and the JIL1 H3S10 phosphatase.38 Currently it still remains to be resolved whether site-specific or transient DNA methylation or other molecular processes that include RNA methylation are associated with the epigenetic function of Dnmt2.
Dnmt2 is involved in RNA-mediated epigenetic heredity
Evidence for a role of Dnmt2 in the transgenerational inheritance of epigenetic signals was provided in mice. It was reported that Dnmt2 is required for both the KittmAlf-induced and miR-124-induced Sox9 paramutations,108,109 suggesting that RNA methylation is an essential step for paramutation establishment and/or transmission. Although details of the mechanism are not known, some evidence connects the effect to RNA present in sperm. Microinjection of such RNA into oocytes can emulate the observed epigenetic heredity, and the presence of m5C in microinjected RNA was reported to be crucial for such an effect. A recent report of epigenetic heredity mediated by tRNA derived small RNAs, akin to tRFs, also mentions an altered modification status of the latter.65 Finally, a report on transgenerational inheritance of altered mouse phenotypes specifically implies fragments of the Dnmt2 substrate tRNAGlyGCC.64,65 Although these latter reports do not directly concern Dnmt2, their association with tRFs, their modification status, and epigenetic heredity clearly may suggest a role of Dnmt2, particularly considering that the generation of tRFs has been shown to be impeded by Dnmt2-mediated tRNA methylation.5
Conclusions
Since the initial discovery of Dnmt2 as a tRNA methyltransferase, knowledge on the functional consequences of tRNA methylation has significantly increased. Although our understanding is far from complete, Dnmt2 has advanced to be one of the most studied and best understood tRNA methyltransferases. This in part, is due to the fact that few modification enzymes have been examined in a comparative fashion in several organisms. The effects of extrinsic stimuli, including nutrition and stress, on tRNA methylation and its downstream effects are a new field, and the same is true for tRFs, in particular where the impact of tRNA modification on tRNA cleavage is concerned. Next to these new and exciting findings, some old questions remain open. Dnmt2 mediated methylation of RNA other than tRNA still awaits verification and functional analysis, as does DNA methylation. Certainly, regulation of Dnmt2 activity has not been exhaustively analyzed yet, and only an advanced understanding of these features will allow an assessment of its function in epigenetic heredity.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work has been supported by the DFG research group FOR1082. The PIs like to thank numerous Ph.D. students and postdocs involved in the projects for their enthusiastic work, including: Maria Becker, Zeljko Durdevic, Winfried Elhardt, Mark Hartmann, Steffen Kaiser, Stefanie Kellner, Reinhardt Liebers, Madeleine Meusburger, Martin Müller, Sara Müller, Olaf Nickel, Sameer Phalke, Katharina Schmid, Isabelle Schuster, Raghuvaran Shanmugam, Kathrin Thüring, Ayala Tovy, Francesca Tuorto.
References
- 1.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006; 311:395-8; PMID:16424344; http://dx.doi.org/ 10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- 2.Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett 2014; 588:4287-96; PMID:25304425; http://dx.doi.org/ 10.1016/j.febslet.2014.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol 2015; 12:603-14; PMID:25892531; http://dx.doi.org/ 10.1080/15476286.2015.1031947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010; 16:673-95; PMID:20181738; http://dx.doi.org/ 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24:1590-5; PMID:20679393; http://dx.doi.org/ 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saikia M, Hatzoglou M. The Many Virtues of tRNA-derived Stress-induced RNAs (tiRNAs): Discovering Novel Mechanisms of Stress Response and Effect on Human Health. J Biol Chem 2015; 290:29761-8; PMID:26463210; http://dx.doi.org/ 10.1074/jbc.R115.694661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Wyngaert I, Sprengel J, Kass SU, Luyten WH. Cloning and analysis of a novel human putative DNA methyltransferase. FEBS Lett 1998; 426:283-9; PMID:9599025; http://dx.doi.org/ 10.1016/S0014-5793(98)00362-7 [DOI] [PubMed] [Google Scholar]
- 8.Yoder JA, Bestor TH. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum Mol Genet 1998; 7:279-84; PMID:9425235; http://dx.doi.org/ 10.1093/hmg/7.2.279 [DOI] [PubMed] [Google Scholar]
- 9.Jurkowski TP, Jeltsch A. On the evolutionary origin of eukaryotic DNA methyltransferases and Dnmt2. PLoS One 2011; 6:e28104; PMID:22140515; http://dx.doi.org/ 10.1371/journal.pone.0028104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patalano S, Vlasova A, Wyatt C, Ewels P, Camara F, Ferreira PG, Asher CL, Jurkowski TP, Segonds-Pichon A, Bachman M, et al.. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc Natl Acad Sci U S A 2015; 112:13970-5; PMID:26483466; http://dx.doi.org/ 10.1073/pnas.1515937112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shanmugam R, Aklujkar M, Schafer M, Reinhardt R, Nickel O, Reuter G, Lovley DR, Ehrenhofer-Murray A, Nellen W, Ankri S, et al.. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res 2014; 42:6487-96; PMID:24711368; http://dx.doi.org/ 10.1093/nar/gku256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res 2001; 29:439-48; PMID:11139614; http://dx.doi.org/ 10.1093/nar/29.2.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeltsch A, Nellen W, Lyko F. Two substrates are better than one: dual specificities for Dnmt2 methyltransferases. Trends Biochem Sci 2006; 31:306-8; PMID:16679017; http://dx.doi.org/ 10.1016/j.tibs.2006.04.005 [DOI] [PubMed] [Google Scholar]
- 14.Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA 2008; 14:1663-70; PMID:18567810; http://dx.doi.org/ 10.1261/rna.970408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Du J, Yang H, Yin J, Ding J, Zhong J. Functional and structural characterization of DNMT2 from Spodoptera frugiperda. J Mol Cell Biol 2013; 5:64-6; PMID:23103599; http://dx.doi.org/ 10.1093/jmcb/mjs057 [DOI] [PubMed] [Google Scholar]
- 16.Schulz EC, Roth HM, Ankri S, Ficner R. Structure analysis of Entamoeba histolytica DNMT2 (EhMeth). PLoS One 2012; 7:e38728; PMID:22737219; http://dx.doi.org/ 10.1371/journal.pone.0038728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell 1994; 76:357-69; PMID:8293469; http://dx.doi.org/ 10.1016/0092-8674(94)90342-5 [DOI] [PubMed] [Google Scholar]
- 18.Becker M, Muller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res 2012; 40:11648-58; PMID:23074192; http://dx.doi.org/ 10.1093/nar/gks956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller S, Windhof IM, Maximov V, Jurkowski T, Jeltsch A, Forstner KU, Sharma CM, Gräf R, Nellen W. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res 2013; 41:8615-27; PMID:23877245; http://dx.doi.org/ 10.1093/nar/gkt634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengesbach M, Meusburger M, Lyko F, Helm M. Use of DNAzymes for site-specific analysis of ribonucleotide modifications. RNA 2008; 14:180-7; PMID:17998290; http://dx.doi.org/ 10.1261/rna.742708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmid K, Thuring K, Keller P, Ochel A, Kellner S, Helm M. Variable presence of 5-methylcytosine in commercial RNA and DNA. RNA Biol 2015; 12:1152-8; PMID:26274337; http://dx.doi.org/ 10.1080/15476286.2015.1076612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 2009; 37:e12; PMID:19059995; http://dx.doi.org/ 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al.. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530:441-6; PMID:26863196; http://dx.doi.org/ 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauenschild R, Tserovski L, Schmid K, Thuring K, Winz ML, Sharma S, Entian KD, Wacheul L, Lafontaine DL, Anderson J, et al.. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res 2015; 43:9950-64; PMID:26365242; http://dx.doi.org/ 10.1093/nar/gkv895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, et al.. Transcriptome-wide mapping reveals reversible and dynamic N-methyladenosine methylome. Nat Chem Biol 2016; 12:311-6, PMID: 26863410, http://dx.doi.org/26075521 10.1038/nchembio.2040 [DOI] [PubMed] [Google Scholar]
- 26.Tserovski L, Marchand V, Hauenschild R, Blanloeil-Oillo F, Helm M, Motorin Y. High-throughput sequencing for 1-methyladenosine (mA) mapping in RNA. Methods 2016. S1046-2023(16)30029-9; PMID: 26922842, http://dx.doi.org/26075521 10.1016/j.ymeth.2016.02.012 [DOI] [PubMed] [Google Scholar]
- 27.Li X, Zhu P, Ma S, Song J, Bai J, Sun F, et al.. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 2015; 11:592-7; PMID:26075521; http://dx.doi.org/ 10.1038/nchembio.1836 [DOI] [PubMed] [Google Scholar]
- 28.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012; 40:5023-33; PMID:22344696; http://dx.doi.org/ 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess AL, David R, Searle IR. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol 2015; 15:199; PMID:26268215; http://dx.doi.org/ 10.1186/s12870-015-0580-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS genetics 2013; 9:e1003602; PMID:23825970; http://dx.doi.org/ 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amort T, Souliere MF, Wille A, Jia XY, Fiegl H, Worle H, Micura R, Lusser A. Long non-coding RNAs as targets for cytosine methylation. RNA Biol 2013; 10:1003-8; PMID:23595112; http://dx.doi.org/ 10.4161/rna.24454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 2013; 31:458-64; PMID:23604283; http://dx.doi.org/ 10.1038/nbt.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller S. Biologische Funktionsanalyse und Identifizierung neuer Substrate der Methyltransferase DNMT2. Universität Kassel. Kassel: Kassel, 2012. [Google Scholar]
- 34.Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol 2006; 301:203-25; PMID:16570849; http://dx.doi.org/ 10.1007/3-540-31390-7 [DOI] [PubMed] [Google Scholar]
- 35.Raddatz G, Guzzardo PM, Olova N, Fantappie MR, Rampp M, Schaefer M, Reik W, Hannon GJ, Lyko F. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A 2013; 110:8627-31; PMID:23641003; http://dx.doi.org/ 10.1073/pnas.1306723110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh M, Curk T, Xu Q, Zupan B, Kuspa A, Shaulsky G. Developmentally regulated DNA methylation in Dictyostelium discoideum. Eukaryot Cell 2006; 5:18-25; PMID:16400165; http://dx.doi.org/ 10.1128/EC.5.1.18-25.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Söderbom F, et al.. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res 2005; 33:6405-17; PMID:16282589; http://dx.doi.org/ 10.1093/nar/gki952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat Genet 2009; 41:696-702; PMID:19412177; http://dx.doi.org/ 10.1038/ng.360 [DOI] [PubMed] [Google Scholar]
- 39.Schaefer M, Lyko F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat Genet 2010; 42:920-1; author reply 1; PMID:20980983; http://dx.doi.org/ 10.1038/ng1110-920 [DOI] [PubMed] [Google Scholar]
- 40.Capuano F, Mulleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem 2014; 86:3697-702; PMID:24640988; http://dx.doi.org/ 10.1021/ac500447w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takayama S, Dhahbi J, Roberts A, Mao G, Heo SJ, Pachter L, Martin DI, Boffelli D. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res 2014; 24:821-30; PMID:24558263; http://dx.doi.org/ 10.1101/gr.162412.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev 2007; 21:261-6; PMID:17289917; http://dx.doi.org/ 10.1101/gad.1472907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng X. Structure and function of DNA methyltransferases. Annu Rev Biophys Biomol Struct 1995; 24:293-318; PMID:7663118; http://dx.doi.org/ 10.1146/annurev.bb.24.060195.001453 [DOI] [PubMed] [Google Scholar]
- 44.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem 2002; 3:274-93; PMID:11933228; http://dx.doi.org/ 10.1002/1439-7633(20020402)3:4%3c274::AID-CBIC274%3e3.0.CO;2-S [DOI] [PubMed] [Google Scholar]
- 45.Gowher H, Loutchanwoot P, Vorobjeva O, Handa V, Jurkowska RZ, Jurkowski TP, Jeltsch A. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J Mol Biol 2006; 357:928-41; PMID:16472822; http://dx.doi.org/ 10.1016/j.jmb.2006.01.035 [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Santi DV. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc Natl Acad Sci U S A 2000; 97:8263-5; PMID:10899996; http://dx.doi.org/ 10.1073/pnas.97.15.8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res 2004; 32:2453-63; PMID:15121902; http://dx.doi.org/ 10.1093/nar/gkh564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jurkowski TP, Shanmugam R, Helm M, Jeltsch A. Mapping the tRNA binding site on the surface of human DNMT2 methyltransferase. Biochemistry 2012; 51:4438-44; PMID:22591353; http://dx.doi.org/ 10.1021/bi3002659 [DOI] [PubMed] [Google Scholar]
- 49.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol 2007; 366:1-13; PMID:17187822; http://dx.doi.org/ 10.1016/j.jmb.2006.11.046 [DOI] [PubMed] [Google Scholar]
- 50.El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 2012; 46:69-95; PMID:22905870; http://dx.doi.org/ 10.1146/annurev-genet-110711-155641 [DOI] [PubMed] [Google Scholar]
- 51.Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet 2011; 7:e1002403; PMID:22144916; http://dx.doi.org/ 10.1371/journal.pgen.1002403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. et al.. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 2012; 19:900-5; PMID:22885326; http://dx.doi.org/ 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- 53.Shanmugam R, Fierer J, Kaiser S, Helm M, Jurkowski TP, Jeltsch A. Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discovery 2015; 1:15010; http://dx.doi.org/ 10.1038/celldisc.2015.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 1998; 26:5017-35; PMID:9801296; http://dx.doi.org/ 10.1093/nar/26.22.5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Gröne HJ, et al.. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 2015; 34:2350-62; PMID:26271101; http://dx.doi.org/ 10.15252/embj.201591382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci U S A 2013; 110:12289-94; PMID:23836657; http://dx.doi.org/ 10.1073/pnas.1300781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zinshteyn B, Gilbert WV. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 2013; 9:e1003675; PMID:23935536; http://dx.doi.org/ 10.1371/journal.pgen.1003675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nedialkova DD, Leidel SA. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015; 161:1606-18; PMID:26052047; http://dx.doi.org/ 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller M, Hartmann M, Schuster I, Bender S, Thuring KL, Helm M, Katze JR, Nellen W, Lyko F, Ehrenhofer-Murray AE. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res 2015; 43:10952-62; PMID:26424849; http://dx.doi.org/ 10.1093/nar/gkv980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al.. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 2014; 33:2020-39; PMID:25063673; http://dx.doi.org/ 10.15252/embj.201489282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 2013; 10:553-63; PMID:23563448; http://dx.doi.org/ 10.4161/rna.24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsieh LC, Lin SI, Kuo HF, Chiou TJ. Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots. Plant Signal Behav 2010; 5:537-9; PMID:20404547; http://dx.doi.org/ 10.4161/psb.11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Y, Hu Q, Deng Z, Hang Y, Wang J, Wang K. MicroRNAs in body fluids as biomarkers for non-small cell lung cancer: a systematic review. Technol Cancer Res Treat 2014; 13:277-87; PMID:24066954; http://dx.doi.org/ 10.7785/tcrt.2012.500377 [DOI] [PubMed] [Google Scholar]
- 64.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al.. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016; 351:391-6; PMID:26721685; http://dx.doi.org/ 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al.. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016; 351:397-400; PMID:26721680; http://dx.doi.org/ 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- 66.Pederson T. Regulatory RNAs derived from transfer RNA? RNA 2010; 16:1865-9; PMID:20719919; http://dx.doi.org/ 10.1261/rna.2266510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hurto RL. Unexpected functions of tRNA and tRNA processing enzymes. Adv Exp Med Biol 2011; 722:137-55; PMID:21915787; http://dx.doi.org/ 10.1007/978-1-4614-0332-6_9 [DOI] [PubMed] [Google Scholar]
- 68.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013; 10:1798-806; PMID:24351723; http://dx.doi.org/ 10.4161/rna.27177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Durdevic Z, Schaefer M. tRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress? Bioessays 2013; 35:323-7; PMID:23315679; http://dx.doi.org/ 10.1002/bies.201200158 [DOI] [PubMed] [Google Scholar]
- 70.Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep 2013; 4:931-7; PMID:24012760; http://dx.doi.org/ 10.1016/j.celrep.2013.07.046 [DOI] [PubMed] [Google Scholar]
- 71.Lyko F, Whittaker AJ, Orr-Weaver TL, Jaenisch R. The putative Drosophila methyltransferase gene dDnmt2 is contained in a transposon-like element and is expressed specifically in ovaries. Mech Dev 2000; 95:215-7; PMID:10906465; http://dx.doi.org/ 10.1016/S0925-4773(00)00325-7 [DOI] [PubMed] [Google Scholar]
- 72.Schaefer M, Steringer JP, Lyko F. The Drosophila cytosine-5 methyltransferase Dnmt2 is associated with the nuclear matrix and can access DNA during mitosis. PloS one 2008; 3:e1414; PMID:18183295; http://dx.doi.org/ 10.1371/journal.pone.0001414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, Schaefer M. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep 2013; 14:269-75; PMID:23370384; http://dx.doi.org/ 10.1038/embor.2013.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fergus C, Barnes D, Alqasem MA, Kelly VP. The queuine micronutrient: charting a course from microbe to man. Nutrients 2015; 7:2897-929; PMID:25884661; http://dx.doi.org/ 10.3390/nu7042897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yokoyama S, Miyazawa T, Iitaka Y, Yamaizumi Z, Kasai H, Nishimura S. Three-dimensional structure of hyper-modified nucleoside Q located in the wobbling position of tRNA. Nature 1979; 282:107-9; PMID:388227; http://dx.doi.org/ 10.1038/282107a0 [DOI] [PubMed] [Google Scholar]
- 76.Garcia GA, Kittendorf JD. Transglycosylation: a mechanism for RNA modification (and editing?). Bioorg Chem 2005; 33:229-51; PMID:15888313; http://dx.doi.org/ 10.1016/j.bioorg.2005.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morl M, Dorner M, Paabo S. C to U editing and modifications during the maturation of the mitochondrial tRNA(Asp) in marsupials. Nucleic Acids Res 1995; 23:3380-4; PMID:7567446; http://dx.doi.org/ 10.1093/nar/23.17.3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guy MP, Phizicky EM. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 2015; 21:61-74; PMID:25404562; http://dx.doi.org/ 10.1261/rna.047639.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou M, Long T, Fang ZP, Zhou XL, Liu RJ, Wang ED. Identification of determinants for tRNA substrate recognition by Escherichia coli C/U34 2′-O-methyltransferase. RNA Biol 2015; 12:900-11; PMID:26106808; http://dx.doi.org/ 10.1080/15476286.2015.1050576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res 2000; 28:292; PMID:10592250; http://dx.doi.org/ 10.1093/nar/28.1.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meier F, Suter B, Grosjean H, Keith G, Kubli E. Queuosine modification of the wobble base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J 1985; 4:823-7; PMID:2988936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grosjean HJ, de Henau S, Crothers DM. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A 1978; 75:610-4; PMID:273223; http://dx.doi.org/ 10.1073/pnas.75.2.610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noguchi S, Nishimura Y, Hirota Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J Biol Chem 1982; 257:6544-50; PMID:6804468 [PubMed] [Google Scholar]
- 84.Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 1999; 283:2097-100; PMID:10092236; http://dx.doi.org/ 10.1126/science.283.5410.2097 [DOI] [PubMed] [Google Scholar]
- 85.Verma AK, Verma R, Ahuja V, Paul J. Real-time analysis of gut flora in Entamoeba histolytica infected patients of Northern India. BMC Microbiol 2012; 12:183; PMID:22913622; http://dx.doi.org/ 10.1186/1471-2180-12-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Byers J, Faigle W, Eichinger D. Colonic short-chain fatty acids inhibit encystation of Entamoeba invadens. Cell Microbiol 2005; 7:269-79; PMID:15659070; http://dx.doi.org/ 10.1111/j.1462-5822.2004.00457.x [DOI] [PubMed] [Google Scholar]
- 87.Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annu Rev Pathol 2012; 7:99-122; PMID:21910623; http://dx.doi.org/ 10.1146/annurev-pathol-011811-132421 [DOI] [PubMed] [Google Scholar]
- 88.Jacobson KB, Farkas WR, Katze JR. Presence of queuine in Drosophila melanogaster: correlation of free pool with queuosine content of tRNA and effect of mutations in pteridine metabolism. Nucleic Acids Res 1981; 9:2351-66; PMID:6789305; http://dx.doi.org/ 10.1093/nar/9.10.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White BN, Tener GM. Activity of a transfer RNA modifying enzyme during the development of Drosophila and its relationship to the su(s) locus. J Mol Biol 1973; 74:635-51; PMID:4199662; http://dx.doi.org/ 10.1016/0022-2836(73)90054-5 [DOI] [PubMed] [Google Scholar]
- 90.Zaborske JM, Bauer DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA. Correction: A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol 2015; 13:e1002150; PMID:25923688; http://dx.doi.org/ 10.1371/journal.pbio.1002150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cummings J, Macfarlane G. Role of intestinal bacteria in nutrient metabolism. J Parenter Enteral Nutr 1997; 21:357-65; PMID:9406136; http://dx.doi.org/ 10.1177/0148607197021006357 [DOI] [PubMed] [Google Scholar]
- 92.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al.. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res 2009; 69:4918-25; PMID:19458066; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-4806 [DOI] [PubMed] [Google Scholar]
- 93.Kellett GL, Brot-Laroche E, Mace OJ, Leturque A. Sugar absorption in the intestine: the role of GLUT2. Annu Rev Nutr 2008; 28:35-54; PMID:18393659; http://dx.doi.org/ 10.1146/annurev.nutr.28.061807.155518 [DOI] [PubMed] [Google Scholar]
- 94.Appelboom JW, Brodsky WA, Rehm WS. The concentration of glucose in mammalian liver. J Gen Physiol 1959; 43:467-79; PMID:13793964; http://dx.doi.org/ 10.1085/jgp.43.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tovy A, Siman Tov R, Gaentzsch R, Helm M, Ankri S. A new nuclear function of the Entamoeba histolytica glycolytic enzyme enolase: the metabolic regulation of cytosine-5 methyltransferase 2 (Dnmt2) activity. PLoS Pathog 2010; 6:e1000775; PMID:20174608; http://dx.doi.org/ 10.1371/journal.ppat.1000775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schulz EC, Tietzel M, Tovy A, Ankri S, Ficner R. Structure analysis of Entamoeba histolytica enolase. Acta crystallographica Section D, Biological crystallography 2011; 67:619-27; PMID:21697600; http://dx.doi.org/ 10.1107/S0907444911016544 [DOI] [PubMed] [Google Scholar]
- 97.Lin MJ, Tang LY, Reddy MN, Shen CK. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J Biol Chem 2005; 280:861-4; PMID:15533947; http://dx.doi.org/ 10.1074/jbc.C400477200 [DOI] [PubMed] [Google Scholar]
- 98.Kaul G, Thippeswamy H. Role of heat shock proteins in diseases and their therapeutic potential. Indian J Microbiol 2011; 51:124-31; PMID:22654152; http://dx.doi.org/ 10.1007/s12088-011-0147-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fisher O, Siman-Tov R, Ankri S. Pleiotropic phenotype in Entamoeba histolytica overexpressing DNA methyltransferase (Ehmeth). Mol Biochem Parasitol 2006; 147:48-54; PMID:16497397; http://dx.doi.org/ 10.1016/j.molbiopara.2006.01.007 [DOI] [PubMed] [Google Scholar]
- 100.Hertz R, Tovy A, Kirschenbaum M, Geffen M, Nozaki T, Adir N, Ankri S. The Entamoeba histolytica Dnmt2 homolog (Ehmeth) confers resistance to nitrosative stress. Eukaryot Cell 2014; 13:494-503; PMID:24562908; http://dx.doi.org/ 10.1128/EC.00031-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 2012; 3:937; PMID:22760636; http://dx.doi.org/ 10.1038/ncomms1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Towns WL, Begley TJ. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: activities, predications, and potential roles in human health. DNA Cell Biol 2012; 31:434-54; PMID:22191691; http://dx.doi.org/ 10.1089/dna.2011.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al.. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2014; PMID:25355519; http://dx.doi.org/ 10.1093/nar/gku1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Elhardt W, Shanmugam R, Jurkowski TP, Jeltsch A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie 2015; 112:66-72; PMID:25747896; http://dx.doi.org/ 10.1016/j.biochi.2015.02.022 [DOI] [PubMed] [Google Scholar]
- 105.Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr Opin Chem Biol 2008; 12:151-8; PMID:18284924; http://dx.doi.org/ 10.1016/j.cbpa.2008.01.027 [DOI] [PubMed] [Google Scholar]
- 106.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harbor perspectives in biology 2013; 5:a017780; PMID:23906716; http://dx.doi.org/ 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 1994; 77:993-1002; PMID:8020105; http://dx.doi.org/ 10.1016/0092-8674(94)90439-1 [DOI] [PubMed] [Google Scholar]
- 108.Kiani J, Grandjean V, Liebers R, Tuorto F, Ghanbarian H, Lyko F, Cuzin F, Rassoulzadegan M. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet 2013; 9:e1003498; PMID:23717211; http://dx.doi.org/ 10.1371/journal.pgen.1003498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liebers R, Rassoulzadegan M, Lyko F. Epigenetic regulation by heritable RNA. PLoS genetics 2014; 10:e1004296; PMID:24743450; http://dx.doi.org/ 10.1371/journal.pgen.1004296 [DOI] [PMC free article] [PubMed] [Google Scholar]


