Abstract
Many prokaryotic cells are encapsulated by a surface layer (S-layer) that consists of repeating units of S-layer proteins (SLPs). S-layers protect cells from the outside, provide mechanical stability, and also play roles in pathogenicity. In situ structural information on this highly abundant class of proteins is scarce, therefore atomic details of how S-layers are arranged on the surface of cells have remained elusive. Here, using purified C. crescentus’ sole SLP RsaA we obtained a 2.7 Å X-ray structure that shows the hexameric S-layer lattice. Next, we solved a 7.4 Å structure of the S-layer through electron cryotomography (cryo-ET) and sub-tomogram averaging of cell stalks. The X-ray structure was docked unambiguously into the cryo-ET map, resulting in a pseudo-atomic level description of the in vivo S-layer, which agrees completely with the atomic X-ray lattice model. This study spans different spatial scales from atoms to cells by combining X-ray crystallography with cryo-ET and sub-nanometre resolution sub-tomogram averaging. The cellular S-layer atomic structure shows that the S-layer is porous, with the largest gap dimension being 27 Å, and is stabilised by multiple Ca2+ ions bound near interfaces.
S-layer proteins (SLPs) are a diverse class of molecules found in many prokaryotes including Gram-positive and Gram-negative bacteria and archaea 1–5. SLPs assemble to form planar sheets called S-layers on the surface of cells, where they are anchored usually through non-covalent interactions with other surface molecules such as lipopolysaccharide (LPS) in Gram-negative bacteria 2,6. S-layers act as the outermost permeability barriers protecting prokaryotic cells from extracellular attack and provide mechanical support to membranes 7. S-layers also play a role in pathogenicity of some bacteria including Clostridium difficile and Bacillus anthracis 2. The regularity and symmetry of S-layers have fascinated cell biologists, microscopists and structural biologists for many decades 8–10. Low-resolution EM studies have been conducted on bacterial and archaeal S-layers 11,12 and X-ray structures of some SLPs have been elucidated 13–16. Thus far, no high-resolution structure of an intact S-layer has been resolved directly on a prokaryotic cell surface.
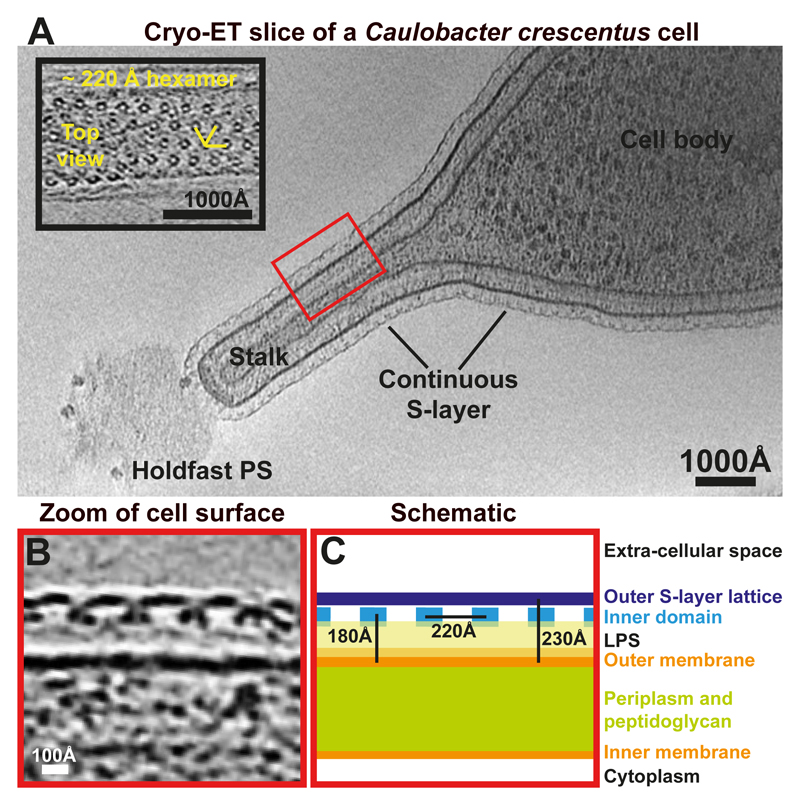
To address this problem, we turned to Caulobacter crescentus, a well-studied Gram-negative alphaproteobacterium with a characteristic ultrastructure and a complex life cycle 17. A ~120 nm thick extension of the cell envelope called the stalk emanates from one pole of the C. crescentus cell body 18. Cryo-ET analysis of C. crescentus CB15 cells showed that the cells are covered with a S-layer that is continuous between the cell body and the stalk (Figure 1A, Movie S1). The density corresponding to the S-layer of C. crescentus CB15 is almost perfectly hexameric (Figure 1A, inset) with a ~220 Å repeat distance seen in tomographic top views of the cell surface, confirming previous electron microscopy and tomography studies on the C. crescentus S-layer 19,20. Tomographic side views showed that the S-layer density is located roughly 180 Å away from the outer membrane (Figure 1B-C). Two discrete densities were observed in the S-layer, the outer, highly-connected S-layer lattice and the discrete inner domains located around the centres of the hexamers. Weak, fuzzy density could be seen between the outer membrane and the inner domain of the S-layer, presumably corresponding to LPS molecules in which the S-layer is most likely anchored 21.
Figure 1. Arrangement of the Caulobacter crescentus S-layer on cells and stalks.
(A) A tomographic slice of a CB15 C. crescentus cell. The S-layer is continuous between the cell body and the stalk. Inset – a magnified tomographic slice through a S-layer of a stalk. A hexameric lattice with a ~220 Å spacing is seen (see also Movie S1). (B) A magnified tomographic slice of a side view of the cell surface. The S-layer is arranged in two layers and is seen ~180 Å away from the outer membrane of the cell. The outer S-layer lattice is highly inter-connected while the inner domains are ~220 Å apart from each other. (C) A schematic representation of the cell surface.
We purified the sole component of the C. crescentus CB15N (NA1000) S-layer, the ~98 kDa RsaA protein (Figure 2A) directly from cells using a previously described S-layer extraction procedure that employs low pH 22. Large quantities of pure RsaA protein could be obtained from cells using this procedure (Figure S1A). We confirmed that the purified protein retained its characteristic polymerisation function by reconstituting S-layers in solution at physiological pH (Figure 2B). Incubation with divalent alkaline earth cations Ca2+ or Sr2+ resulted in the formation of two-dimensional sheets showing the characteristic 220 Å hexagonal lattice (Figure S1B-C, Movie S2). The reconstituted RsaA sheets showed only short-range order and were not perfectly planar (Movie S2), indicating that monomers of RsaA in the two-dimensional lattice and lattice contacts possessed significant conformational flexibility. Although purified RsaA had a tendency to degrade in solution even at 4 ºC, addition of divalent cations such as Ca2+ or Sr2+ moderately protected the protein from cleavage (Figure S1D), presumably because monomeric RsaA polymerised into sheets.
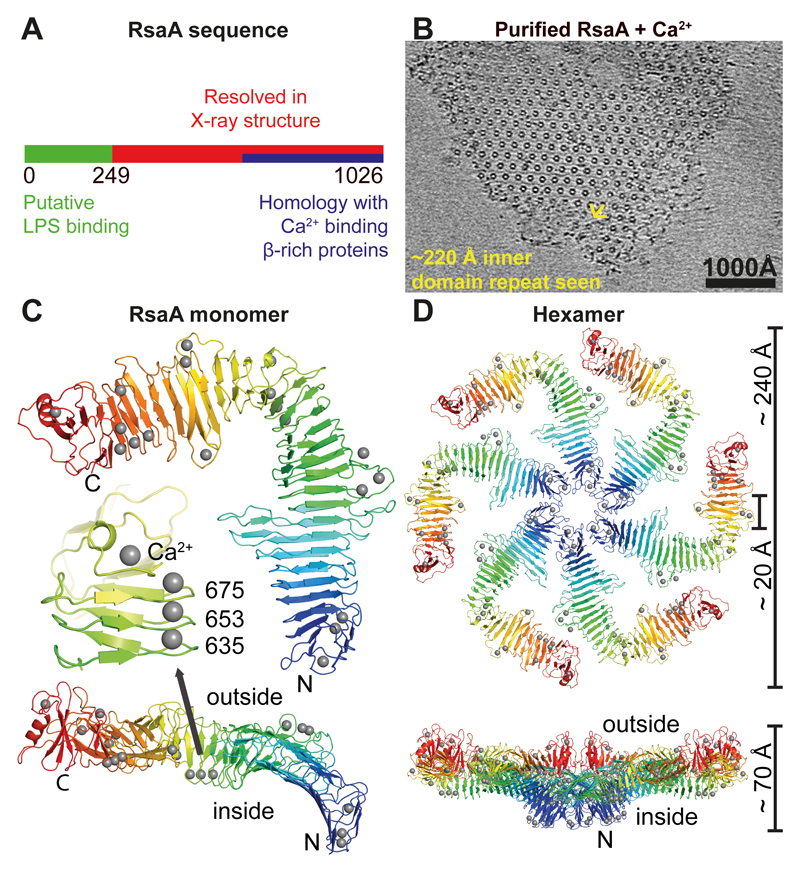
Figure 2. 2.7 Å X-ray structure of the outer S-layer lattice.
(A) Schematic diagram of the RsaA protein sequence. The N-terminal region of RsaA is a putative LPS binding region (see Figure S2), residues 249-1026 are resolved in the X-ray structure (below) and residues 618-1026 have predicted sequence similarity with Ca2+-dependent β-sheet rich proteins (HHPred server 50). (B) On addition of Ca2+ ions to RsaA, 2D sheets were obtained with the same appearance and repeat as S-layers on cells. A cryo-ET slice through a reconstituted sheet is shown (see also Movie S2). (C) Crystal structure of RsaA: one monomer of RsaA (see Table S1) shown as ribbon diagram, coloured as a rainbow, N- to C-terminus from blue to red. Two orthogonal orientations are shown with the N- and C-termini of the protein marked. Ca2+ ions are shown as grey spheres. The inset shows a region around amino acid 653 where three tightly bound structural Ca2+ ions stabilise the fold. (D) Non-crystallographic hexamer of RsaA (forming the asymmetric unit of the crystals) resolved in the X-ray structure, shown in two orthogonal orientations. The central pore is roughly 20 Å wide while the lattice is about 70 Å thick. The N-termini of the six monomers of RsaA point towards the cell surface.
To resolve the structure of RsaA, we screened a variety of crystallisation conditions and obtained diffraction quality co-crystals of RsaA with Ca2+ or Sr2+. Using the obtained crystals we determined a 2.7 Å X-ray structure by holmium SAD (single-wavelength anomalous dispersion) coupled with six-fold NCS (non-crystallographic symmetry) averaging and phase extension (Figure 2C-D, Table S1). RsaA amino acid residues 249-1026 were resolved in the resulting electron density map (Movie S3). Peptide mapping of protein obtained from dissolved crystals indicated that N-terminal residues of RsaA had been cleaved during crystallisation. At least amino acid residues 256-1026 were still present in the crystallised protein as determined by mass spectrometry after protease cleavage into peptides, a method that does not reveal the exact N- and C-termini of the analysed polypeptide (Figure S2A, C).
RsaA249-1026 adopts a predominantly β-helical fold with ~30 right-handed turns (Figure 2C-D), forming an overall L-shape (Figure 2C). A domain formed by the C-terminal residues 933-1026 is not β-helical and also contains the only α-helix in the entire structure. In the crystals, the L-shaped monomers were arranged into hexamers that have a central pore with a size of ~20 Å (Figure 2D). The N-termini of all six RsaA monomers point towards the central pore, while the C-termini are located on the outside of the hexamer, as dictated by the linear β-helical fold. Although the space group of the crystal was P21, the unit cell is almost hexameric (Table S1) with an inter-hexameric spacing of between ~215 and ~220 Å. Intriguingly, this pseudo-hexameric repeat is close to the repeat observed in cellular S-layers in tomograms (Figure 1). Hence we hypothesised that the structure of RsaA249-1026 corresponds to the outer S-layer lattice (Figure 1B), and that the N-terminal residues missing in the crystal structure contain the membrane-proximal LPS-associated domain. We tested whether removal of the N-terminus would remove LPS association to RsaA. In protein gels of full-length RsaA stained for LPS a significant band was observed corresponding to LPS, which was not seen when Coomassie stained (Figure S2B). We compared full-length RsaA with a gently trypsin-cleaved version missing the first 184 N-terminal residues and found that on removal of the N-terminal domain, the LPS band was also lost (Figure S2B). This indicates that indeed, the X-ray structure contains only the outer S-layer lattice and not the inner LPS-associated domain of the S-layer.
All residues in the RsaA249-1026 X-ray structure were well ordered, reflecting a requirement that the S-layer lattice must avoid flexible loops, as they would be easy targets for hostile extra-cellular proteases. The fully ordered C-terminus of RsaA up to residue 1026 is almost completely buried in the C-terminal domain that deviates significantly from a canonical β-helical fold (Figure 2C, residues 933-1026). Amino acid side chains buried inside the β-helix domain of RsaA are either hydrophobic, or are negatively charged and co-ordinated tightly to structural Ca2+ ions (Figure 2C-D, grey spheres), with very few exceptions. Some Ca2+ ions are in close proximity to the central pore of the hexamer (Figure 2D, Movie S4), where they appear to stabilise the hexameric interface. Inspection of the crystal packing revealed that RsaA hexamers were packed into regular, near-hexagonal, two-dimensional lattices that show repeat distances closely related to S-layers observed on cells and those seen reconstituted in vitro as described above.
In order to verify that the lattice found in the crystals is the same as the in vivo S-layer structure, we attempted to resolve the structure of the C. crescentus S-layer using electron cryotomography (cryo-ET) of cells and subsequent sub-tomogram averaging. Because the quality of tomograms is affected by the thickness of the sample, we used a mutant of C. crescentus, strain YB2811 23, which efficiently sheds cell stalks for cryo-ET imaging (Figure S3, Movie S5). Using a regularised likelihood algorithm for refinement 24, we obtained an ab initio sub-tomogram averaging model of the S-layer at ~30 Å resolution.
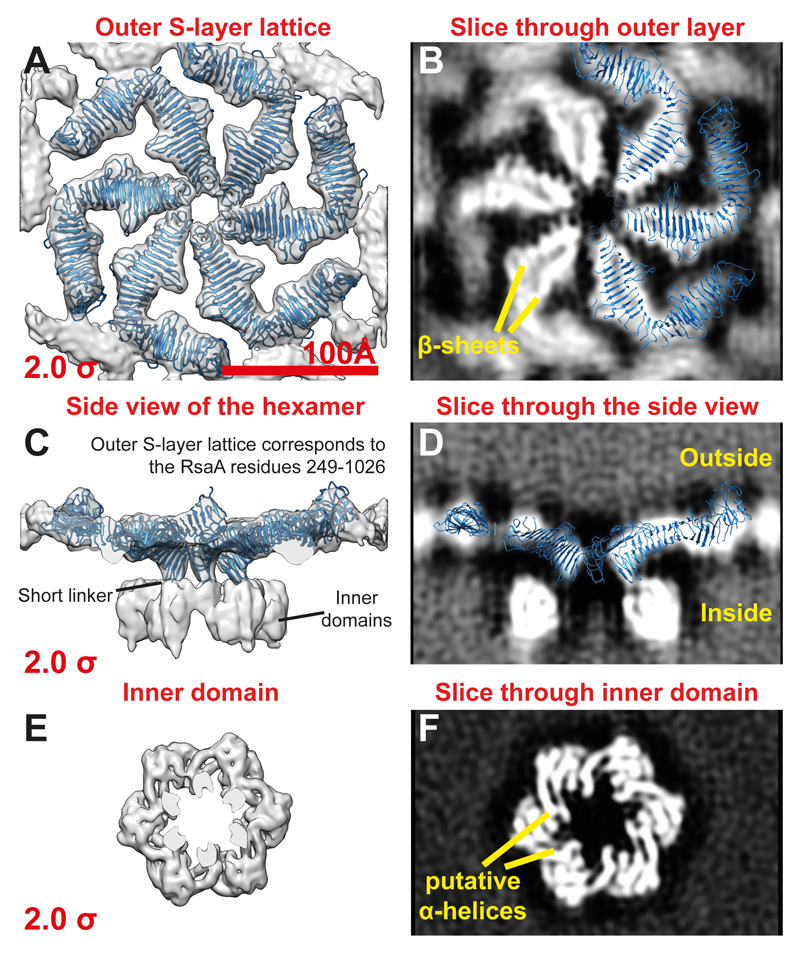
Next, to resolve a higher resolution structure of the hexameric repeating asymmetric unit, we utilised a pipeline for data collection and image processing that has recently allowed near-atomic resolution sub-tomogram averaging refinement 25. The initial model obtained above was used as input for high-resolution sub-tomogram refinement. The final refined 7.4 Å structure (Figure S4A) allowed us to locate different domains of the S-layer in the map (Figures S4B-C) and identify secondary structure in the S-layer (Figures 3A-F and S4D-F). As expected from inspection of tomograms (Figure 1B), the outer S-layer lattice is highly inter-connected and consists mainly of β-sheets (Figures 3A-B) while the inner domain is discrete and appears to contain cylindrical densities that are most likely α-helical (Figures 3E-F).
Figure 3. 7.4 Å cryo-ET and sub-tomogram averaging map of the C. crescentus S-layer.
(A) A top view (from outside the cell) of the outer layer of the sub-tomogram averaging map (grey density) with the X-ray structure (blue ribbon) fitted into the density (see also Figure S4). The cryo-EM density is shown at an isosurface threshold level of 2.0 σ away from the mean (red number on the bottom left). The entire hexamer as determined by X-ray crystallography is fitted into the outer S-layer lattice as a rigid body without further adjustments, demonstrating very strong agreement between the X-ray structure determination and cryo-ET. (B) A slice through the outer S-layer lattice with three of the six RsaA molecules from the X-ray hexamer overlaid (see also Figures S4-5 and Movie S6). (C) Side view of the sub-tomogram averaging map shows that the N-terminal RsaA residue 249 in the X-ray structure is located in the short linker between the two domains of the S-layer (see Figures S4E). (D) A single slice through the cryo-ET map, with the fitted RsaA hexamer from the X-ray model overlaid (a clipping plane has been applied at the front and the back). (E) A top view of the hexamer of inner domains of the S-layer. (F) A slice through the inner domain density suggests that it might contain some α-helical secondary structure elements.
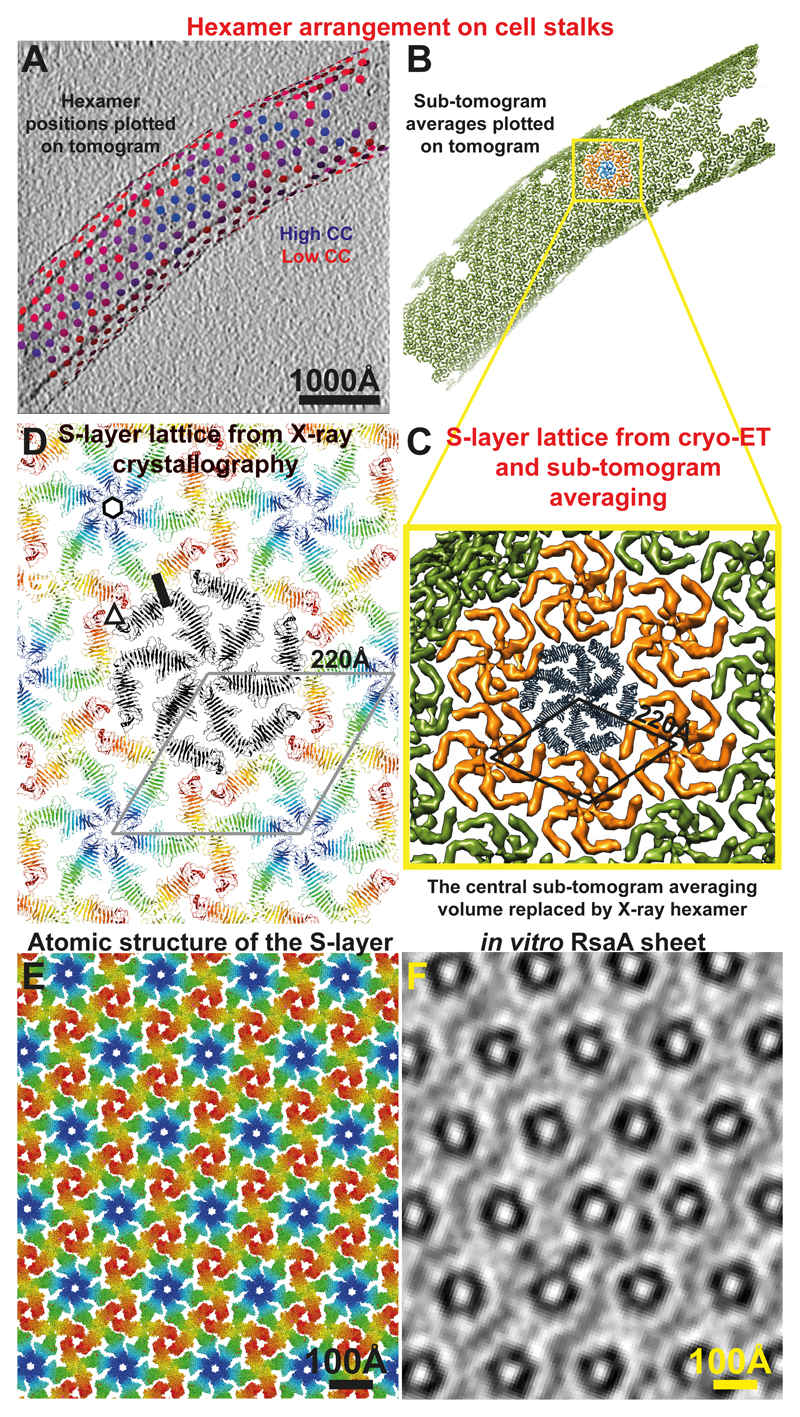
We then docked the hexamer from the RsaA249-1026 X-ray structure into the cryo-ET density map of the outer S-layer lattice as a rigid body (Figures 3A-D, S5A-F, Movie S6). The fit shows the precise position of the outer S-layer lattice in the cryo-ET map and shows that the inner domains are separated from the outer lattice by a short linker region between the two domains (Figure 3C and S4E). The size of the resolved inner domain density suggests that it likely contains all 248 N-terminal residues of RsaA missing from the X-ray structure. Extended densities emanating from the inner domains towards the outer membrane of the cell could be seen at low threshold levels (Figure S5C-D). Importantly, the docked atomic model very convincingly shows that the lattices formed by RsaA in the crystals and on cells are extremely similar (Figure 4A-D) and allows us to describe the arrangement of the S-layer lattice on cells at the atomic level (Figure 4A-D, Movie S7).
Figure 4. The S-layer lattices observed in the X-ray structure and the cryo-ET map are exceptionally similar.
(A) Final refined positions of sub-tomograms are shown plotted back onto a tomogram of a cell stalk with the corresponding refined orientations (see Movie S7). Positions have been coloured from blue (high cross-correlation of alignment) to red (low cross-correlation). One slice of the tomogram is shown with protein density black. (B) The same plot as panel A, except each hexamer position is illustrated with the sub-tomogram average (green volumes). One hexamer is highlighted in blue, and the hexamers directly contacting it are shown in orange. (C) A zoomed view of the hexameric lattice revealed by cryo-ET and sub-tomogram averaging. The 220 Å hexamer:hexamer spacing is highlighted. The central blue hexamer from panel B is replaced by one copy of the X-ray hexamer. (D) The RsaA lattice formed in the crystals through crystal packing is the same as the S-layer lattice observed on cells through cryo-ET. The 220 Å hexamer:hexamer repeat is shown. One hexameric, trimeric and dimeric interface, each have been highlighted with a black hexagon, a triangle and a line, respectively. In conclusion, both X-ray crystallography and cryo-ET used in this study show essentially the same lattice and arrangement of RsaA in C. crescentus S-layers. Also see Movies S4, 7. (E) Atomic structure of the S-layer. (F) A slice through an in vitro assembled RsaA sheet (see Movie S2) for comparison at the same scale as panel E.
In the 2D lattice formed by RsaA in the crystals, shown above by cryo-ET to be equivalent to the lattice formed in intact S-layers at 7.4 Å resolution, the hexameric interface is mediated mainly by amino acid residues between positions 256-260, making contact with the loop between residues 271-275 in the neighbouring subunit (Figures S6A-B, Movie S4, Table S2). Conspicuously, Ca2+ ions are bound near this interface (Figure S6B) and could play a role in stabilising the RsaA hexamer and the S-layer lattice. The hexameric interface is the most extensive protein:protein interface in the S-layer lattice (Table S2). Formation of this interface requires six RsaA molecules to assemble into a hexameric structure, and it is likely highly energetically favourable due to tight packing of the monomers around the six-fold axis.
In addition to the hexameric interface, there are connections between residues 667-668 and 688-693 on one RsaA molecule with residues 713-715 and 756-758 on another RsaA molecule in a dimeric interface between adjoining hexamers (Figures S6C-D, Table S2). This dimeric interface is built up of two spatially distinct smaller interfaces (brown circles in Figure S6D), thus making it another strong interface stabilising the S-layer lattice (Figure S6D). Again, like the hexameric interface described above, Ca2+ ions appear to be tightly bound to some interface residues. A less extensive trimeric contact is also observed in the S-layer lattice involving residues in the loop between positions 952-956 and residues 854-857 (Figures S6E-F and Movie S4). This interface is smaller than the hexameric and dimeric interfaces, and Ca2+ ions are located near some of the interface residues (Figures S6E-F).
Thus, a fascinating overall feature of the C. crescentus S-layer lattice is the presence of multiple Ca2+ ions bound tightly to RsaA, either tightly co-ordinated within the RsaA fold or bound near the hexameric, trimeric, and dimeric interfaces (Figure S6). Without a divalent alkaline earth ion like Ca2+ or Sr2+, no sheet formation was observed in our cryo-EM experiments nor were crystals of RsaA sheets obtained, indicating that Ca2+ ions could play a role in lattice stabilisation and assembly on cells. Strong Ca2+-dependence of RsaA polymerisation and S-layer lattice formation has been observed previously 22. Our results here are also in line with previous analysis of a Geobacillus stearothermophilus SLP SbsB 13 that also showed strong dependence on Ca2+ for oligomerisation.
The atomic structure of the S-layer lattice (Figures 4E-F and S7) suggests how the surface layer acts as a tough yet flexible permeability barrier, protecting cells from extra-cellular attack. The central circular pore is only ~20 Å wide (Figure 2D), and other gaps between RsaA monomers in the hexamer are no larger than ~27 Å along the largest dimensions, smaller than previous estimates of ~35 Å from low-resolution EM studies of purified RsaA sheets 20. This pore size is larger than that observed in purified S-layer sheets of the archaeal species Methanosarcina acetivorans 14. A 20 Å wide pore would not only occlude harmful entities like phages, but also prevent most macromolecules and even thin filaments (those larger than ~27 Å) from reaching the bacterial cell surface through the S-layer. Despite forming a continuous sheet, there is a requirement for flexibility in the S-layer lattice. RsaA monomers in the lattice must encase membranes with widely differing membrane curvatures from 550 nm wide C. crescentus cell bodies (Figure S7A-B) to 120 nm wide cell stalks (Figure S3) requiring RsaA to adopt slightly different conformations. Furthermore, gaps in the lattice will be needed for insertion of new RsaA molecules on growing cells.
The cryo-ET and X-ray structures together explain the modular architecture and function of RsaA, which was suggested by previous low-resolution cryo-ET studies 19. Such a modular organization in p6 S-layers has been suggested previously for the bacterium Acetogenium kivui 12. Having more than one domain can allow SLPs to perform the dual functions of S-layer lattice formation and membrane anchoring. This is an elegant solution to the problem of placing a rigid, lattice-forming protein in the correct position in a cell with only one part of the protein mediating anchoring (Figure S7A, inset).
The S-layer lattice structure on the surface of cells revealed in this study immediately suggests a number of experiments and potential applications. Residues exposed to the extracellular environment could be fluorescently tagged to investigate S-layer biogenesis and its morphological changes during the cellular life cycle of C. crescentus at the molecular and atomic levels. Also guided by the S-layer structure reported here, C. crescentus cells carrying mutated or fused versions of the rsaA gene may be used for surface display of molecules for nanotechnological or antigen display applications 26,27.
Recent improvements in cryo-ET imaging have revealed the architecture of protein complexes (e.g. type IV pili) on bacterial cells 28, and novel image processing techniques have allowed high-resolution refinement from purified virus (HIV-1) particles 25. Here we combine X-ray crystallography with cryo-ET imaging from cell stalks with sub-nanometre resolution refinement to bridge different spatial scales from atoms to cells (Figure S7). The integrated approach (Figure S7) used in this study to obtain the atomic S-layer structure on the surface of bacterial cells illustrates convincingly how in vitro reconstitution may be combined with high-resolution in vivo imaging to shed light on an important biological problem. We believe that we are now entering an era of cellular structural biology where it will be increasingly possible to determine structures of molecules in situ in their natural environments.
Methods
Purification of RsaA
Caulobacter crescentus CB15N (NA1000) cells were grown in peptone yeast extract (PYE) medium 29 for 48 hours at 30 ºC. Two litres of the resulting culture were centrifuged (4000 rcf, 4 ºC, 30 minutes). The pelleted cells were frozen in liquid nitrogen and stored at -80 ºC. On the day of the purification pellets were thawed and the cells were re-suspended in 50 mM HEPES buffer at pH 2.0 and incubated at 37 ºC for 10 minutes with vigorous shaking. Next, the suspension was centrifuged (16000 rcf, 4 ºC, 30 minutes). The pellet was discarded and the pH of the supernatant was adjusted to 7.0 with 5 M NaOH. The resulting liquid was filtered and from here on kept at 4 ºC. The protein solution was loaded onto a 5 mL HiTrap Q HP column (GE Healthcare), washed with 50 mM HEPES pH 7.0 and then eluted with the same buffer containing increasing concentrations of NaCl. Fractions containing pure RsaA were collected and concentrated to ~30 mg/mL. Aliquots were frozen in liquid nitrogen and stored at -80 ºC.
Crystallisation of Caulobacter crescentus RsaA
The RsaA protein solution was supplemented with 5 mM CaCl2 prior to crystallisation. Initial screens of full-length RsaA were setup using MRC Laboratory of Molecular Biology’s in-house robotic nano-litre crystallisation facility 30. After optimisation, the native RsaA crystals were grown at 19 ºC by sitting-drop vapour-diffusion in a drop composed of 100 nL of reservoir solution (0.07 M KSCN, 24 % (w/v) PEG 8000, 0.075 M TAPS pH 8.5) and 100 nL of protein solution at 30 mg/mL. Plate-like crystals appeared in 3-10 days and continued growing to a final size of 300 x 300 x 30 µm3. Crystals were flash frozen in liquid nitrogen for data collection using an additional 25% (v/v) PEG 200 as cryo-protectant. Heavy-atom crystal derivatives, selected based on their similarity to calcium, were obtained by using similar technique. Again, 100 nL of the reservoir solution (0.43 M KSCN, 18.5 % (w/v) PEG 8000, 0.02 M Sr(CH3COO)2, 0.075 M TAPS pH 8.5) was mixed with 100 nL of 26 mg/mL RsaA solution supplemented with 5 mM SrCl2. The resulting crystals were soaked for 7 min in the reservoir solution additionally containing 0.04 M HoCl3, 25% (v/v) PEG 200 and flash frozen in liquid nitrogen. Holmium was used for phasing as described below.
Trypsin cleavage and LPS staining
Full-length RsaA (~2 mg/mL) was incubated at a ratio of 1000:1 with trypsin at room temperature (22 ºC) for 24 hours with gentle shaking. The pH of the resulting solution was lowered to 2.0 with HCl to inactivate trypsin and then restored to pH 7.0 with NaOH. The solution was centrifuged, filtered and then subjected to gel fitration chromatography on a Superose 6 10/300 column (GE Healthcare) using 100 mM NaCl, 1 mM EGTA, 50 mM HEPES pH 7.0 buffer. Fractions containing cleaved RsaA were pooled and concentrated. Staining of full-length and trypsin-cleaved RsaA was performed using the Pro-Q Emerald 300 Lipopolysaccharide Gel Stain Kit (Thermo Fisher Scientific).
RsaA structure determination by X-ray crystallography
The structure was solved by holmium SAD (single-wavelength anomalous dispersion), phase transfer to a high-resolution native dataset by molecular replacement and by phase extension using six-fold, non-crystallographic (NCS) symmetry. A SAD dataset obtained from a holmium-soaked RsaA crystal was collected on beamline I04 at Diamond Light Source (Harwell, UK) at the holmium edge to 3.3 Å resolution (Table S1). Diffraction images were indexed and integrated with XDS 31 and scaled and merged with SCALA 32. Forty-two holmium sites were located with SHELXD 33. PHASER 34 completed this to 49 sites and an interpretable electron density map was obtained in one of the two possible hands after SAD phasing in PHASER, coupled with density modification in DM 35. BUCCANEER 36 was used to build a rough initial model that indicated 6-fold NCS symmetry but was not sufficient to complete atomic interpretation. Because crystals were not isomorphous to each other, density around one of the six monomers was cut out using CCP4 methods and used for molecular replacement into the native dataset, that went to 2.7 Å resolution in two directions, using PHASER (Table S1). The native dataset was collected on beamline I04 at Diamond Light Source and was first modified for severe anisotropy in the B* direction using the UCLA Diffraction Anisotropy Server (https://services.mbi.ucla.edu/anisoscale). Phases for the native dataset were slowly extended from 8 to 2.7 Å using DM and the six-fold NCS symmetry over 2000 cycles. The resulting electron density map was readily interpretable and was traced for all six monomers almost completely and automatically with BUCCANEER. The model was improved manually using MAIN 37 and refined with PHENIX and REFMAC 38,39 keeping the 5% randomly selected test set of reflections separate at all times. 19 calcium ions per RsaA chain were added in positions of otherwise unexplained strong electron density peaks located in chemically plausible environments such as being close to several carbonyl groups and aspartate and glutamate side chains. Structure factors and coordinates of the final model have been deposited in the Protein Data Bank (PDB) with accession code 5N8P.
Purification of C. crescentus stalks
Purification of stalks from cells was performed using a stalk-shedding C. crescentus strain (YB2811) with a differential centrifugation procedure described previously, but slightly modified here 23. Briefly, a 5 mL of starter culture of strain YB2811 in PYE medium was used to inoculate a 250 mL flask containing PYE medium. The new culture was grown at 30 ºC for 2-3 days. The culture was imaged using a wide-field light microscope to confirm efficient stalk shedding. The culture was then centrifuged to pellet whole C. crescentus cells (17000 rcf, 4 ºC, 30 minutes). The pellet was discarded, and the supernatant was centrifuged again to pellet stalks (41000 rcf, 4 ºC, 1 hour). The supernatant was carefully decanted, and the pellet was re-suspended in 100 µL of PYE and stored at 4 ºC or used immediately for cryo-EM sample preparation.
Cryo-EM sample preparation
Whole cells, purified stalks or reconstituted sheets were mixed with 10 nm protein-A gold (CMC Utrecht) and pipetted onto a freshly glow discharged Quantifoil R2/2 Cu/Rh 200 mesh grids in a Vitrobot Mark IV (FEI Company) maintained at 10 ºC at 100 % relative humidity. Grids were plunge-frozen into liquid ethane and stored in liquid nitrogen until the day of microscopy.
Cryo-EM and cryo-ET data collection
Initial cryo-EM assessment of samples was conducted on a Spirit microscope (FEI Company) with a Gatan 626 side-entry holder. Data collection for initial sub-tomogram averaging model generation was performed on a Titan Krios microscope (FEI) fitted with a Quantum energy filter (slit width 20 eV) and a K2 direct electron detector running in counting mode with a dose rate of ~ 10 e-/pixel/second (Gatan). Tilt series (22 in total) of purified stalks were collected between ±60º in two directions at a 2º increment using SerialEM 40. A total dose of 70 e-/Å2 was applied, the data was sampled at a pixel size of 4.45 Å. For obtaining a sub-nanometre resolution cryo-ET structure of the S-layer on cell stalks, we adopted a data collection strategy that has recently allowed high-resolution refinement from cryo-ET 25. Briefly, a dose-symmetric data collection scheme was used 41 and 110 tomograms were collected at a pixel size of 1.3 Å. A total dose of 140 e-/Å2 was applied over the tilt series collected between ±60º with 3º spacing between the tilts.
Sub-tomogram averaging image processing
Tomogram reconstructions were performed in IMOD 42. Sub-tomogram extraction from tomograms of tubular stalks was performed using previously described procedures implemented in MATLAB 43. Initial model generation was performed using a regularised likelihood refinement algorithm implemented in the RELION software 44. First, heavily binned sub-tomograms were subjected to RELION 3D classification (with a 3D contrast transfer function (CTF) model used) with wide angular and translational search range to locate all the hexamers of the S-layer in 3D. Once all the hexamers had been located, then sub-tomograms were re-extracted at the refined positions and a 3D auto-refinement sub-tomogram averaging was run in RELION. The final volume obtained had a resolution of 30 Å and this was used as input for high-resolution refinement. High-resolution sub-tomogram averaging was performed using a recently described image-processing scheme 25,41. CTF estimation of the input tilt series images was performed using CTFFIND 45. The estimated defocus values were used for CTF correction within IMOD to produce a CTF corrected tilt series stack. Next, dose filtration was performed on the stacks to down-weight the higher resolution Fourier components in the high-tilt images 25. The resulting dose-weighted and CTF corrected stack were used for tomogram generation in IMOD using weighted back projection. The centres of all stalks were picked manually and these central positions were used to generate the sub-tomogram extraction locations assuming a cylindrical arrangement of the RsaA hexamers around the stalk. Sub-tomograms were extracted from 8 times binned tomograms, and as before, and sub-tomogram alignment and averaging was performed to locate the positions of each hexamer in the tomogram 46,47. After locating the hexamer positions all poorly aligned sub-tomograms were removed, as judged from cross-correlation scores, as well from their position in lattice maps. Finally, re-extraction was performed with no binning on the data and with the extracted sub-tomograms centred on the 6-fold axis of each hexamer. C6 symmetry was applied during the final refinement of the total of 51,866 hexamers of S-layer from 110 tomograms leading to a 7.4 Å structure. Resolution estimation, FSC weighting and map sharpening was conducted in RELION 48. The final map has not been explicitly symmetrised 6-fold and has been deposited in the Electron Microscopy Data Bank (EMDB) with the accession code EMD-3604. Rigid body docking of the hexamer of RsaA from X-ray crystallography into the cryo-ET map was performed with UCSF Chimera 49.
Supplementary Material
Acknowledgements
The authors would like to thank Mark Skehel and Farida Begum for mass-spectrometric identification of proteins, Minmin Yu for help with X-ray data collection, Christos Savva for help with cryo-EM data collection, Sjors Scheres for help with RELION software, Florian Schur and William Wan for providing high-resolution image-processing code and scripts prior to publication and for advice on their implementation, Fusinita van den Ent for advice on protein purification and Toby Darling and Jake Grimmett for help with high-performance computing. Part of this work was funded by the European Molecular Biology Organization (aALTF 778-2015 to TAMB), the Medical Research Council (U105184326 to JL), the Wellcome Trust (095514/Z/11/Z to JL), and the National Institutes of Health (GM51986 to YVB). This work has been supported by iNEXT, project number 1482, funded by the Horizon 2020 program of the European Union.
Footnotes
Author contributions:
TAMB, YVB, and JL designed research. TAMB, DKC, GGH, EWY, JMD, WJHH and JL performed experiments. WJHH and JAGB supported high-resolution cryo-ET and image processing. TAMB, DKC, JMD, EWY and JL analysed data. TAMB and JL wrote the manuscript with support from all the authors.
Competing financial interests:
The authors declare no competing financial interests.
Materials and Correspondence:
Structure factors and coordinates of the final model have been deposited in the Protein Data Bank (PDB) with accession code 5N8P. The final cryo-ET map has been deposited in the Electron Microscopy Data Bank (EMDB) with the accession code EMD-3604. Please contact the corresponding authors Tanmay A.M. Bharat or Jan Löwe for further information.
References
- 1.Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- 2.Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014;12:211–222. doi: 10.1038/nrmicro3213. [DOI] [PubMed] [Google Scholar]
- 3.Glauert AM. The fine structure of bacteria. Br Med Bull. 1962;18:245–250. doi: 10.1093/oxfordjournals.bmb.a069988. [DOI] [PubMed] [Google Scholar]
- 4.Sara M, Sleytr UB. S-Layer proteins. J Bacteriol. 2000;182:859–868. doi: 10.1128/jb.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu C, et al. Diversity in S-layers. Progress in biophysics and molecular biology. 2016 doi: 10.1016/j.pbiomolbio.2016.08.002. [DOI] [Google Scholar]
- 6.Kirk JA, Banerji O, Fagan RP. Characteristics of the Clostridium difficile cell envelope and its importance in therapeutics. Microbial biotechnology. 2016 doi: 10.1111/1751-7915.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sleytr UB, Beveridge TJ. Bacterial S-layers. Trends Microbiol. 1999;7:253–260. doi: 10.1016/s0966-842x(99)01513-9. [DOI] [PubMed] [Google Scholar]
- 8.Houwink AL. A macromolecular mono-layer in the cell wall of Spirillum spec. Biochim Biophys Acta. 1953;10:360–366. doi: 10.1016/0006-3002(53)90266-2. [DOI] [PubMed] [Google Scholar]
- 9.Sleytr UB, Glauert AM. Ultrastructure of the cell walls of two closely related clostridia that possess different regular arrays of surface subunits. J Bacteriol. 1976;126:869–882. doi: 10.1128/jb.126.2.869-882.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumeister W, Wildhaber I, Phipps BM. Principles of organization in eubacterial and archaebacterial surface proteins. Canadian journal of microbiology. 1989;35:215–227. doi: 10.1139/m89-034. [DOI] [PubMed] [Google Scholar]
- 11.Kessel M, Wildhaber I, Cohen S, Baumeister W. Three-dimensional structure of the regular surface glycoprotein layer of Halobacterium volcanii from the Dead Sea. EMBO J. 1988;7:1549–1554. doi: 10.1002/j.1460-2075.1988.tb02974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupas A, et al. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J Bacteriol. 1994;176:1224–1233. doi: 10.1128/jb.176.5.1224-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranova E, et al. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature. 2012;487:119–122. doi: 10.1038/nature11155. [DOI] [PubMed] [Google Scholar]
- 14.Arbing MA, et al. Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans. Proc Natl Acad Sci U S A. 2012;109:11812–11817. doi: 10.1073/pnas.1120595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing H, et al. Archaeal surface layer proteins contain beta propeller, PKD, and beta helix domains and are related to metazoan cell surface proteins. Structure. 2002;10:1453–1464. doi: 10.1016/s0969-2126(02)00840-7. [DOI] [PubMed] [Google Scholar]
- 16.Kern J, et al. Structure of surface layer homology (SLH) domains from Bacillus anthracis surface array protein. J Biol Chem. 2011;286:26042–26049. doi: 10.1074/jbc.M111.248070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang C, Brown PJ, Ducret A, Brun YV. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature. 2014;506:489–493. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol Microbiol. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- 19.Amat F, et al. Analysis of the intact surface layer of Caulobacter crescentus by cryo-electron tomography. J Bacteriol. 2010;192:5855–5865. doi: 10.1128/JB.00747-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smit J, Engelhardt H, Volker S, Smith SH, Baumeister W. The S-layer of Caulobacter crescentus: three-dimensional image reconstruction and structure analysis by electron microscopy. J Bacteriol. 1992;174:6527–6538. doi: 10.1128/jb.174.20.6527-6538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ford MJ, Nomellini JF, Smit J. S-layer anchoring and localization of an S-layer-associated protease in Caulobacter crescentus. J Bacteriol. 2007;189:2226–2237. doi: 10.1128/JB.01690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nomellini JF, Kupcu S, Sleytr UB, Smit J. Factors controlling in vitro recrystallization of the Caulobacter crescentus paracrystalline S-layer. J Bacteriol. 1997;179:6349–6354. doi: 10.1128/jb.179.20.6349-6354.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ireland MM, Karty JA, Quardokus EM, Reilly JP, Brun YV. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol Microbiol. 2002;45:1029–1041. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- 24.Bharat TAM, Scheres SHW. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nature protocols. 2016;11:9–20. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schur FK, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 26.Howorka S. Rationally engineering natural protein assemblies in nanobiotechnology. Curr Opin Biotech. 2011;22:485–491. doi: 10.1016/j.copbio.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Mark SS, et al. Bionanofabrication of metallic and semiconductor nanoparticle arrays using S-layer protein lattices with different lateral spacings and geometries. Langmuir. 2006;22:3763–3774. doi: 10.1021/la053115v. [DOI] [PubMed] [Google Scholar]
- 28.Chang YW, et al. Architecture of the type IVa pilus machine. Science. 2016;351 doi: 10.1126/science.aad2001. aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriological reviews. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stock D, Perisic O, Löwe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Progress in biophysics and molecular biology. 2005;88:311–327. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheldrick GM. SHELX applications to macromolecules. Nato Adv Sci I C-Mat. 1998;507:401–411. [Google Scholar]
- 34.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 37.Turk D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr D Biol Crystallogr. 2013;69:1342–1357. doi: 10.1107/S0907444913008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. S1047-8477(05)00152-8 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Hagen WJ, Wan W, Briggs JA. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J Struct Biol. 2016 doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. S1047-8477(96)90013-1 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Bharat TA, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001196. e1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bharat TA, Russo CJ, Löwe J, Passmore LA, Scheres SH. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure. 2015;23:1743–1753. doi: 10.1016/j.str.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. doi:S1047847703000698 [pii] [DOI] [PubMed] [Google Scholar]
- 46.Briggs JA, et al. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. 0903535106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Förster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci U S A. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. 0409178102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.