Abstract
Background
As part of a program to develop a Dengue virus vaccine which avoids the deleterious effects of antibody dependent enhancement (ADE) of infection mediated by antibodies to Dengue virus structural proteins, we have begun to investigate the possibility of designing Dengue vaccines based on non-structural proteins.
Results
Dengue constructs which lack major structural proteins replicate intracellularly in tissue culture. These replicons are capable of prolonged expression of Dengue virus non-structural proteins for at least seven days in culture.
Conclusions
Dengue virus genomes lacking major structural proteins can, like other flaviviruses, replicate intracellularly and express virus non-structural proteins with minimal toxicity to host cells. These findings pave the way for the development of dengue virus replicons as a form of live, attenuated virus vaccine.
Background
The mosquito-borne flavivirus, Dengue, is estimated to cause in each year 100 million cases of Dengue fever (DF), 500,000 cases of Dengue Hemorrhagic fever (DHF) and 25,000 deaths, with 2.5 billion people at risk [1]. Although a successful vaccine against the prototypical flavivirus, yellow fever (YF) virus, has been in use since the 1930s and vaccines to two other flaviviruses, Japanese encephalitis (JE) virus and tick-borne encephalitis (TBE) virus are currently available, there is as yet no Dengue vaccine approved for use [2].
Dengue virus has a typical flavivirus genome structure, as described in Figure 1A. The structural proteins, C, prM (M) and E, are involved in packaging, export and subsequent entry. The non-structural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5 include an RNA-directed RNA polymerase, and a protease function involved in cleaving certain positions of the long viral polyprotein which contains all the viral genes [3,4].
Figure 1.
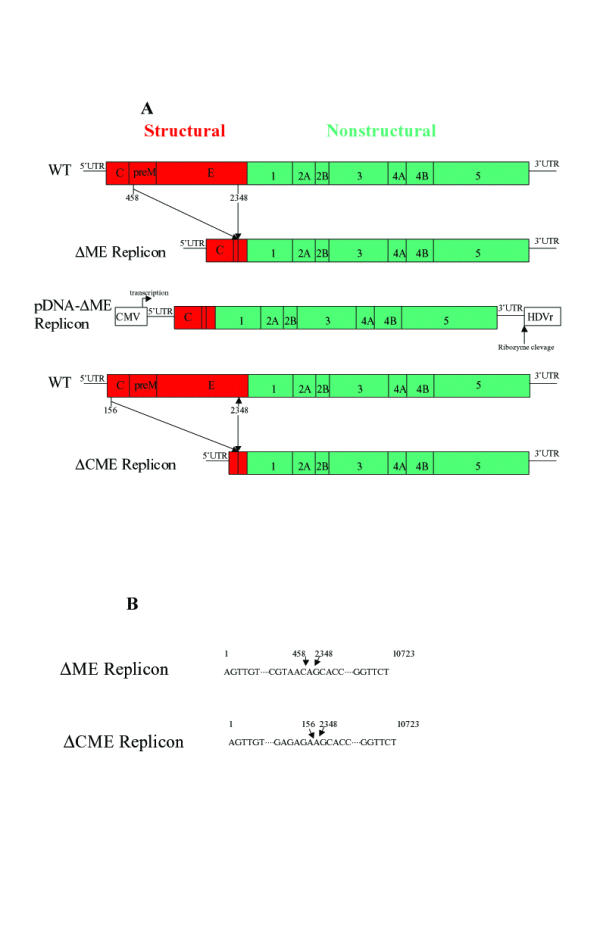
A. Diagram of Dengue virus genome and deletion mutations in this study. B. Sequences at the deletion points of mutants used in this study.
The four serotypes of Dengue virus ("1" through "4") share approximately 60%–74% amino acid residue identity with one another in the E gene [5] and induce cross-reacting antibodies [6]. However, neutralizing antibodies to the structural proteins of one serotype of Dengue typically not only fail to provide protection against other serotypes, but appear to cause the enhanced replication of virus seen in Dengue hemorrhagic fever, which is generally seen upon reinfection by Dengue virus of a different serotype. This antibody-dependent enhancement of infection (ADE), which is believed to be mediated by enhancement of viral uptake by macrophages [7] complicates Dengue vaccine development, since an inadequate or modified immunogen may contribute to disease, rather than prevent infection [8].
Two strategies suggest themselves for circumventing the problems caused by cross reacting antibodies against the major structural proteins, prM and E. One strategy is to immunize with multiple strains of Dengue virus to elicit high affinity, neutralizing antibodies against the multiple Dengue serotypes. At least one vaccine to do this (using dengue vaccine candidates DEN-1 PDK13, DEN-2 PDK53, DEN-3 PGMK 30/F3, and DEN-4 PDK48) has been in clinical trials [9,10]. A second strategy is to induce immunity only to viral proteins other than prM and E. Several studies have shown that the nonstructural glycoprotein NS1 can play an important role in protection against Dengue. Mice immunized with purified Dengue-2 NS1 protein injected intramuscularly and boosted after 3 days and two weeks were protected from developing lethal Dengue encephalitis upon subsequent challenge with Dengue 2 virus. [11]. Similarly, mice immunized with recombinant vaccinia virus expressing authentic NS1 [12] were protected against the development of Dengue-4 virus encephalitis when challenged by intracerebral injection. Inoculation of mice with specific combinations of MAbs directed against Dengue-2 NS1 [13] also protects against lethal virus encephalitis upon intracerebral Dengue-2 challenge. Other nonstructural proteins are also immunogenic and may participate in eliciting protection [14].
Towards the goal of devising a "live" vaccine based on only non-structural Dengue proteins, we have attempted to construct Dengue virus genomes from which the pre-M and E genes have been deleted. Upon introduction into a host's cells, these sub-genomic fragments should replicate intracellularly and support prolonged expression of Dengue non-structural proteins without producing the deleted structural proteins and without forming infectious virions. Sub-genomic replicons of several positive-strand RNA animal viruses have been reported, particularly yellow fever and Kunjin among the flaviviruses. These replicons, when introduced into host cells, replicate and make viral proteins for over 41 days [15], but cannot form infectious virions because they lack critical structural proteins. Effectively delivered to host cells in vivo, such replicons should efficiently induce immunologic reactions against the expressed proteins remaining in the sub-genomic construct. Here we describe the successful construction of two Dengue virus sub-genomic constructs which replicate in LLC-MK2 cells in tissue culture when transfected in as full length RNA. We also report that expression of Dengue virus proteins from at least one of these replicons can be supported by transfection of a DNA-based expression vector containing the replicon.
Results
Immunofluorescent analysis of cell cultures 48 hrs post transfection demonstrates efficient expression of Dengue virus proteins from wild type Dengue virus as well as from both the ΔprM-E and ΔC-prM-E mutants (see Figure 1 and Figure 2A,2B & 2C). By this time point, the wild type virus has had a limited opportunity to be transmitted in secondary rounds of infection. Interestingly, pairs of fluorescent cells were often seen, suggesting cells continued to replicate after being successfully transfected by replication competent Dengue replicons. This is particularly evident in Figure 2B & 2C.
Figure 2.
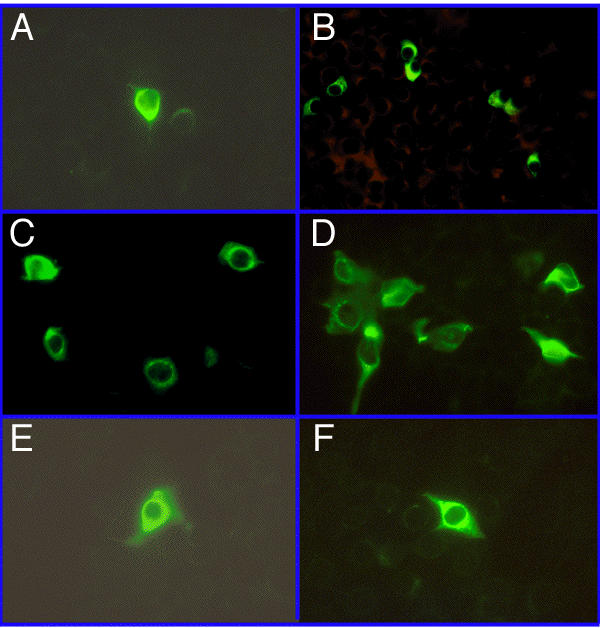
Expression of Dengue virus as determined by immunofluorescent staining with mouse anti-Dengue virus-2. Cells were counterstained with Evans Blue, but different filtration systems available on the different microscopes variably blocked visualization of the background of cells fluorescing red. Transfection efficiencies were generally in the range of 0.01% to 1% and in the background of each photograph in this figure are numerous non-fluorescent cells, best visualized on an Apple Macintosh. In the experiments here, cells were photographed variously with either 40X or 60X objectives. A, Dengue 2 wild type virus (48 hrs); B, ΔprM-E (48 hrs);C, ΔC-prM-E (48 hrs); D, Dengue 2 wild type (8 days); E, ΔprM-E (8 days); F, ΔC-prM-E (8 days).
By 8 days post transfection, wild type Dengue virus expression is more widespread throughout the culture than it was 48 hrs post transfection, presumably because it was able to undergo multiple rounds of replication and transmission (Figure 2D). Cell cultures transfected with ΔprM-E (Figure 2E) or ΔC-prM-E (Figure 2F) still have cells efficiently expressing Dengue proteins at 8 days, but they are more rare than they were at 48 hrs post transfection. This is consistent with the inability of these mutants to make infectious virions and suggests that the viral proteins remaining in the Dengue replicons may moderately retard cell growth and replication. However, in the experiments presented here, cells were trypsinized and replated on day 7 post transfection, so they may not have had sufficient time to recover and replicate prior to harvest for immunofluorescence on day 8.
No fluorescent cells were ever seen at either 48 hrs or 8 days post transfection with Dengue deletion mutant ΔE, from which most of the E gene has been deleted. However, negative results are hard to interpret in this system.
The prolonged expression of Dengue virus proteins by the ΔprM-E and ΔC-prM-E replicons presumably is dependent on the ability of these sub-genomic fragments to replicate. Consistent with this presumption, ΔprM-E RNA was seen to sequentially increase in cultures over at least the first 48 hrs post transfection (Figure 3), whereas no Dengue virus RNA was seen over this time period after transfection with ΔE RNA. RNA from both of these constructs is undetectable at 6 hrs post transfection, suggesting that most of the transfecting RNA is rapidly degraded.
Figure 3.
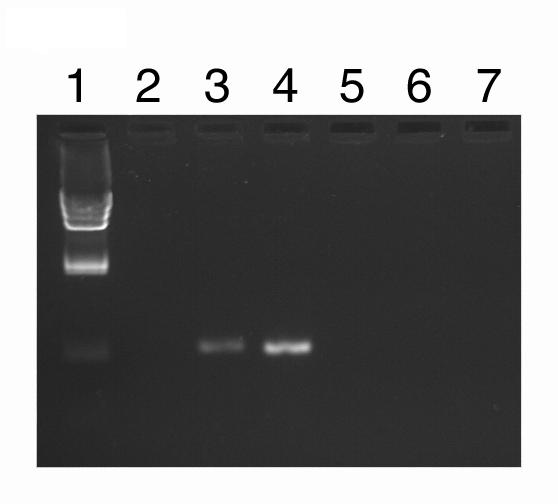
Dengue virus RNA in transfected cells as determined by RT-PCR, normalized to total RNA: Lane 1, λ Hind-Ill molecular weight markers; Lanes 2–4, ΔprM-E; 5–7, ΔE; 2 & 5, 6 hrs; 3 & 6, 24 hrs; 4 & 7, 48 hrs.
When the Δ-prM-E replicon is placed under the control of a cytomegalovirus (CMVB) promoter, it is still capable of active Dengue virus protein expression when transfected into cells in the form of plasmid DNA (see Figure 4).
Figure 4.
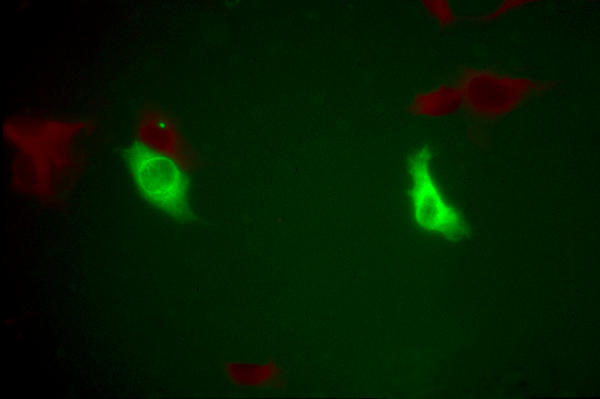
Expression of Dengue virus proteins by DNA-ΔprM-E transfected into cells in the form of plasmid DNA.
Discussion
The expression of Dengue virus RNA and proteins in cultures transfected with the Δ-prM-E and ΔC-prM-E mutants is consistent with replication of the viral sub-genomes in these host cells. This is consistent with similar replicons constructed from Kunjin virus [15]. Most encouraging, however, is the finding of viral protein expression at long time points (8 days) subsequent to transfection with the ΔprM-E and ΔC-prM-E replicons in the absence of selection. Previous experiments with other flavivirus replicons have demonstrated expression for as long as 41 days post transfection [15], but these replicons expressed neomycin and were grown under selective pressure. The longest, previously reported time for pure replicon expression in the absence of selection was just 72 hours [15]. We have not yet searched for continued expression beyond 8 days post transfection, but we have no reason to believe that is not readily achievable.
In order for the Dengue replicons reported here to be of immunologic value, they need to be expressible in a convenient form and (we anticipate) they need to produce NS1 protein. Formally, the data presented here do not directly prove NS1 production, because the antisera used detect multiple viral structural and nonstructural proteins. Attempts to visulize Dengue replicon protein synthesis by Western blotting were unsuccessful in our hands (data not shown), presumably because of the comparatively low transfection efficiencies achieved. However, the previously demonstrated dependence of Dengue virus replication on NS1 production [16] and the fact that Dengue virus RNA levels increase with time after transfection in the cultures used here (implying active Dengue RNA replication) together strongly imply the production of significant quantities of NS1 protein by these replicons. As for a suitable form of delivery, the ability of pDNA-ΔprM-E to make high levels of Dengue proteins after transfection into cells in the form of DNA suggests the possibility of using DNA transfection to achieve immunization [17].
Conclusions
We have demonstrated the prolonged expression of Dengue virus proteins from sub-genomic Dengue RNA fragments, lacking major structural genes, transfected into tissue culture cells. This prolonged expression is associated with detectable increases in Dengue RNA in the transfected tissue cultures, implying that the sub-genomic fragments are replicating, and implying the synthesis of NS1 protein and other viral non-structural proteins known to be required for viral genomic replication. The ability to express one of these sub-genomic Dengue replicons from transfected DNA offers the possibility of using DNA-based, Dengue replicon vaccines. Experiments are currently in progress to develop other delivery methods, including the development of packaging cell lines.
Materials & methods
Culturing of Dengue virus
Dengue virus strains DEN1/WP and DEN2/NGC, kindly provided by Dr. Lewis Markoff, [18,19] were passaged in monkey LLC-MK2 cells at 37° C in a humidified incubator under 5% CO2, using Medium 199 plus 10% fetal bovine serum (FBS) and 50 ug of Gentamicin per ml. The cells were trypsinized a day before virus infection and plated to reach approximately 80% confluence on the day of infection. Infections were typically at an MOI of 0.01 PFU/cell in Medium 199 plus 2% FBS.
In vitro mutagenesis
DNA fragments used for spanning the desired deletions (see Figure 1) were synthesized by polymerase chain reaction (PCR) from two short overlapping primers. For the ACME replicon, the 5' primer was 5ATCATTATGCTGATTCCAACAGTGATGGCGTTCCATTTAACCACACGTAA CAGCACCTCACTGTCTGTG3. For the ΔprME replicon, the 5' primer was 5ACAGCTGTCGCTCCTTCAATGACAATGCGTTGCATAGGAATATCAAATA GAAGCACCTCACTGTCTGTG3. For both mutants, the 3' PRIMER was 5ATACAGCGTCACGACTCCCACCAATACTAGTGACACAGACAGTGAGGTG CT3
PDNA-ΔprM-E was constructed by placing the ΔprM-E replicon RNA under the transcriptional control of a CMV promoter and placing it upstream of a Hepatitis Delta Virus (HDV) self cleaving ribozyme.
Dengue-2 virus cDNA cloned in the yeast shuttle vector pBR424, linearized by excision of a short Bam HI fragment was transfected into competent [20] S. cerevisiae YPH857 (kindly provided by Barry Falgout (CBER/FDA), along with the appropriate PCR fragment spanning the desired deletion. Yeast colonies which grew on tryptophan minus plates represented vectors which had recircularized by homologous recombination with these PCR fragments [20]. DNA from these colonies was transformed into E. coli Stbl 2 cells (Life Technologies, Inc.) to make sufficient quantities of Dengue recombinant, genomic-length DNAs for characterization and analysis.
Expression of virus & replicons in cells
The full length virus and replicon cDNA plasmids isolated from Stbl 2 cells were linearized with Sac I, purified by Qiagen chromatography, and eluted by RNase-free water in preparation for transcription. The transcription reaction mixtures contained lug of linearized DNA; 0.5 mM (each) ATP, CTP, and UTP; 0.1 mM GTP; 0.5 mM cap analog (NEBL); 10 mM DTT; 40 U of RNasin (Promega); 30 U of SP6 RNA polymerase; and 1 × SP6 RNA polymerase buffer (Promega) in a volume of 30 ul. The reaction mixtures were incubated at 40°C for 2 hr. Aliquots (12.5 ul) of the reaction mixtures, containing full length viral RNA, were used to transfect approximately 2 × 106 monkey LLC-MK2 cells in phosphate-buffered saline (PBS) by electroporation in a 0.4 cm gap electroporation cuvette. Each cuvette was pulsed at 200 V, 950 uF using a BioRad Genepuls electroporator. The cells were then resuspended in growth medium and plated on the appropriate tissue culture dish. Plasmid DNA-ΔprM-E was transfected into cells by electroporation using identical conditions.
After electroporation, cells were either plated directly on multiwell plates for harvest at short time periods (typically 48 hrs or less) or on tissue culture dishes for trypsinization and seeding onto multiwell plates on the day before final harvest for longer time periods (typically 8 days post transfection).
Immuno-histochemical methods
Cells growing on chamber slides were rinsed in room-temperature PBS and then fixed in cold acetone for 10 min at -20°C. After being air dried, each chamber was covered with 50 ul of a 1:50 dilution of DEN2-specific hyperimmune mouse ascitic fluid (HMAF, American Type Culture Collection) in PBS plus 2% normal goat serum. Samples were incubated at room temperature for 1 h in a humidified atmosphere and then rinsed twice in PBS. Samples were similarly incubated with a 1:100 dilution of fluorescein isothiocyanate-labeld goat anti-mouse antibodies (Life Technologies) and rinsed twice in PBS. Cells in some experiments were counterstained with 0.02% Evans Blue.
Acknowledgments
Acknowledgements
We thank Dr. Owen Wood for help with immunofluorescence photomicrography.
Contributor Information
Xiaowu Pang, Email: dayton@cber.fda.gov.
Mingjie Zhang, Email: dayton@cber.fda.gov.
Andrew I Dayton, Email: dayton@cber.fda.gov.
References
- Monath TP. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardosa MJ. Dengue vaccine design: issues and challenges: British Medical Bulletin. 1998;54 (No.2):395–405. doi: 10.1093/oxfordjournals.bmb.a011696. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Chang HS, Galler R, Rice CM. Flavivirus Genome Organization, Expression and Replication. Ann Rev Microbiol. 1990;44:649–6888. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Rice CM. Flaviviridae: the virus and their replication: Fields Virology 3rd ed. Philadelphia, Pa. Lippincott-Raven Publishers, 1996. pp. p931–996.
- Thomas CJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Ann Rev Microbiol. 1990;44:649–88. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Heinz FX. Epitope mapping of flavivirus glycoproteins. Adv Vims Res. 1986;31:103–168. doi: 10.1016/s0065-3527(08)60263-8. [DOI] [PubMed] [Google Scholar]
- Morens DM. Antibody-dependent enhancement of infection and the pathogenesis of viral disease: Clinical Infectious Diseases. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Pathogenesis of dengue: Challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- Bhamarapravati N, Sutee Y. Live attenuated tetravalent dengue vaccine. Vaccine. 2000;18 Suppl 2:44–47. doi: 10.1016/s0264-410x(00)00040-2. [DOI] [PubMed] [Google Scholar]
- Kanesa-thasan N, Sun W, Kim-Ahn G, Van Albert S, Putnak JR, King A, Raengsakulsrach B, Christ-Schmidt H, Gilson K, Zahradnik JM, Vaughn I, Innis BL, Saluzzo J, Hoke CH., Jr Safety and immunogenicity of attenuated dengue virus vaccines (Aventis Pasteur) in human volunteers. Vaccine. 2001;19(23-24):3179–3188. doi: 10.1016/s0264-410x(01)00020-2. [DOI] [PubMed] [Google Scholar]
- Schlesinger JJ, Brandriss MW, Walsh EE. Immunization with the dengue 2 virus nonstructural glycoprotein NS1. J Gen Virol. 1987;68:853–857. doi: 10.1099/0022-1317-68-3-853. [DOI] [PubMed] [Google Scholar]
- Falgout B, Bray M, Schlesinger JJ, Lai C-J. Immunization of Mice with recombinant vaccinia virus expressing authentic dengue virus nonstructural protein NS1 protects against lethal dengue virus encephalitis. J Virol. 1990;64(9):4356–4363. doi: 10.1128/jvi.64.9.4356-4363.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchal EA, Henchal LS, Schlesinger JJ. Synergistic interactions of anti-NS1 monoclonal antibodies protect passively immunized mice from lethal challenge with dengue 2 virus. J Gen Virol. 1988;69 (Pt 8):2101–2107. doi: 10.1099/0022-1317-69-8-2101. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Kurane I, Mathew A, Zeng L, Shi PY, Rothman A, Ennis FA. Immune mediated and inherited defenses against flaviviruses. Clin Diagn Virol. 1998;10(2-3):129–39. doi: 10.1016/s0928-0197(98)00039-7. [DOI] [PubMed] [Google Scholar]
- Khromykn AA, Westaway ED. Sub-genomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71(2):1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Ward G, Rieder E, Chinsangaram J, Grubman MJ, Mason PW. Development of DNA vaccines for foot-and-mouth disease, evaluation of vaccines encoding replicating and non-replicating nucleic acids in swine. J Biot. 1999;73:243–249. doi: 10.1016/s0168-1656(99)00142-x. [DOI] [PubMed] [Google Scholar]
- Polo S, Ketner G, Levis R, Falgout B. Infectious RNA Transcripts from full-length dengue virus type 2 cDNA clones made in Yeast. J Virol. 1997;71(7):5366–74. doi: 10.1128/jvi.71.7.5366-5374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pur B, Polo S, Hayes CG, Falgout B. Construction of a full length infection clone for dengue-1 virus western pacific 74 strain. Virus Genes. 2000;20(1):57–63. doi: 10.1023/a:1008160123754. [DOI] [PubMed] [Google Scholar]
- Spencer F, Ketner G, Connelly C, Hieter P. Targeted recombination-based cloning and manipulation of large DNA segments in yeast. Methods Companion Methods Enzymol. 1993;5:161–175. [Google Scholar]


