Abstract
Hexamethylene bis-acetamide inducible protein 1 (HEXIM1) is identified as a novel inhibitor of estrogen stimulated breast cell growth, and it suppresses estrogen receptor-α transcriptional activity. HEXIM1 protein level has been found to be downregulated by estrogens. Recently, HEXIM1 has been found to inhibit androgen receptor transcriptional activity as well. Researchers have used Hexamethylene bis-acetamide (HMBA) for decades to stimulate HEXIM1 expression, which also inhibit estrogen stimulated breast cancer cell gene activation and androgen stimulated prostate cancer gene activation. However, the direct molecular targets of HMBA that modulate the induction of HEXIM1 expression in mammalian cells have not been identified. Based on HMBA and its more potent analog 4a1, we designed molecular probes to pull down the binding proteins of these compounds. Via proteomic approach and biological assays, we demonstrate that HMBA and 4a1 are actually heat shock protein 70 (HSP70) binders. The known HSP70 activator showed similar activity as HMBA and 4a1 to induce HEXIM1 expression, suggesting that HMBA and 4a1 might be putative HSP70 activators. Molecular target identification of HMBA and 4a1 could lead to further structural optimization of the parental compound to generate more potent derivatives to stimulate HEXIM1 expression, which could be a novel approach for hormone dependent breast cancer and prostate cancer treatment.
Keywords: HEXIM1, HMBA, Target identification, HSP70, Estrogen receptor
1. Introduction
Hexamethylene bis-acetamide inducible protein 1 (HEXIM1) was initially identified as an estrogen receptor transcription inhibitor and also down regulated by estrogen, therefore was named EDG1 (estrogen down regulated gene 1) (Fig. 1) [1]. It could be induced by a small molecule Hexamethylene bis-acetamide (HMBA), which leads to the current name HEXIM1 [2,3].
Fig. 1.
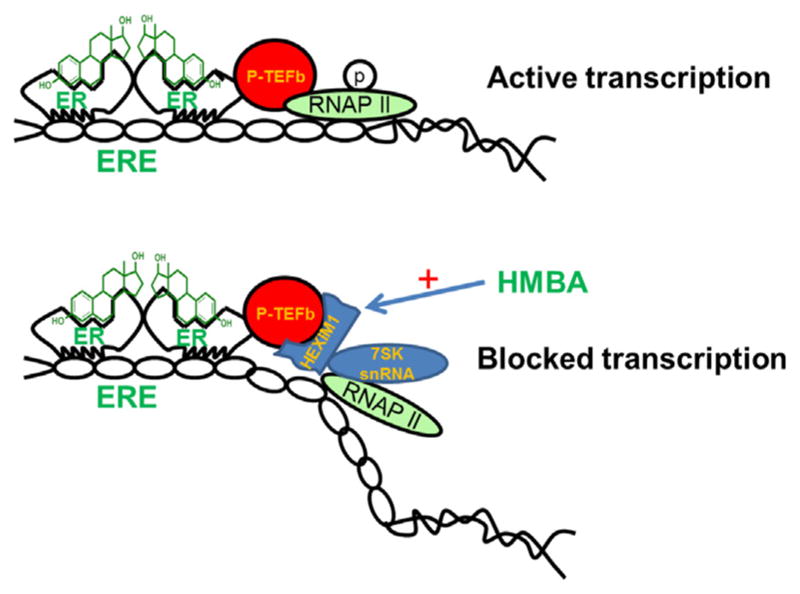
HMBA induces HEXIM1 expression and blocks estrogen receptor transcription.
HMBA is a small molecule that has been investigated by the National Cancer Institute due to its potent anti-cancer and cell differentiation activities [4]. HMBA induces cell differentiation via upregulation of HEXIM1 in breast cancer cells [1–3]. However, HMBA failed at the Phase II clinical trial because of a dose-dependent toxicity [4]. The main toxicity is the thrombocytopenia, which is due to the very short biological half-life of HMBA that necessitates administration of the agent through infusion at a high dosage [4–6].
HEXIM1 binds to 7SK snRNA, a highly abundant non-coding RNA [7]. This complex acts as a potent inhibitor of positive transcription elongation b (P-TEFb) and prevents elongation of RNA Pol II generated transcripts [2,8,9] (Fig. 1). The ratio of inactive to active P-TEFb in cells is regulated by HEXIM1/7SK snRNP, which plays a critical role in the regulation of expression of a wide range of genes such as estrogen and glucocorticoid receptor regulated genes [10–12]. We also reported the inhibitory role of HEXIM1 in prostate tumorigenesis [13,14]. HEXIM1 functions as an androgen receptor co-repressor as it physically interacts with the androgen receptor and is required for the ability of anti-androgens to inhibit androgen-induced target gene expression and cell proliferation [14]. Stimulating HEXIM1 expression is a novel strategy for the treatment of hormone dependent breast and prostate cancer.
The induction of HEXIM1 transcription by HMBA likely occurs through the recruitment of P-TEFb to HEXIM1 promoter. Peterlin and colleagues reported that AKT/PI3K is involved in the phosphorylation of the elongation factor inhibitory complex, which causes the complex to dissociate and HEXIM1 expression to be transiently induced before the recruitment of newly formed elongation factor inhibitory complexes [15,16]. However, the direct molecular targets of HMBA that transiently induce HEXIM1 are not well-defined.
In breast cells, HEXIM1 suppresses cancer metastasis by inhibiting cell invasion, angiogenesis, and the premetastatic niche [17]. However, HEXIM1 expression is lost during breast and prostate tumor progression [11,14]. To enhance HEXIM1 expression, we developed polymer-mediated delivery of HMBA to mammary tumors that resulted in increased HEXIM1 expression and inhibited tumor metastasis. This delivery strategy did not result in thrombocytopenia, the toxicity associated with HMBA in clinical trials. If the druggable features of HMBA could be improved through structural optimization, more promising drug candidates may be developed to increase HEXIM1 expression. These new drug candidates potentially have a broad application in hormone dependent breast and prostate cancer treatment.
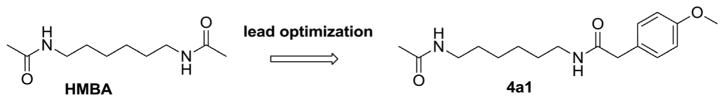
To enhance the potential translational impact of our studies, we conducted a study that focused on the lead optimization of HMBA, which resulted in the generation of more potent inducers of HEXIM1 expression such as compound 4a1(Fig. 2) [18]. The finding provides a unique unsymmetrical molecular scaffold that can induce HEXIM1 expression in prostate and breast cancer cells, and has opened a new lead optimization direction for HMBA [18,19]. Our results support the potential of both symmetrical and unsymmetrical HMBA derivatives as new drug candidates. However before we can conduct further lead optimization of HMBA to generate more potent HEXIM1 inducers, a critical step that needs to be taken is the identification of the molecular targets of HMBA and the derivatives.
Fig. 2.
Structures of HMBA and 4a1.
The detail mechanism of the induction of HEXIM1 expression by HMBA is poorly understood, with no any known molecular candidates for direct interaction with HMBA. The elucidation of the target of HMBA is necessary in order to further optimize its therapeutic potential and minimize the side effects. Target identification will be valuable in understanding how HMBA and derivatives act in the cells. Moreover, there may be existing chemotherapeutic approaches or drugs that have the same molecular target as HMBA, and thus may substitute for HMBA in the induction of HEXIM1 expression for the treatment of cancer. In addition, HMBA and 4a1 induce cell differentiation and is considered less cytotoxic than the existing cancer chemotherapeutic agents.
In the current study, we designed molecular probes based on the structures of HMBA and its more potent analog 4a1 to pull down the binding proteins of these compounds. Via proteomic approaches and biological assays, we found that HMBA and 4a1 bind to heat shock protein 70 kDa (HSP70) to induce HEXIM1 expression. To the best of our knowledge, HSP70 is the first potential molecular target of HMBA identified, which sheds light on the upstream signaling involved in the induction of HEXIM1 expression. This finding provides a solid foundation for further optimization to improve the therapeutic potential of HMBA, and minimize its side effects.
2. Materials and methods
2.1. Biotinylation of 4a1 and HMBA
Chemicals were commercially available and used as received without further purification unless otherwise noted. Moisture sensitive reactions were carried out under a dry argon atmosphere in pre-dried glassware. Thin-layer chromatography was performed on pre-coated silica gel F254 plates (Whatman). Silica gel column chromatography was performed using silica gel 60A (Merck, 230–400 Mesh), and hexane/ethyl acetate was used as the elution solvent. Mass spectra were obtained on the Micromass Quadrupole Time-of-Flight (QTOF) Electrospray mass spectrometer at Cleve-land State University MS facility Center. All the NMR spectra were recorded on a Bruker 400 MHz in either DMSO-d6 or CDCl3. Chemical shifts (δ) for 1H NMR spectra are reported in parts per million to residual solvent protons. The probe synthesis is illustrated in Schemes 1 and 2. The final product characterization is as follows.
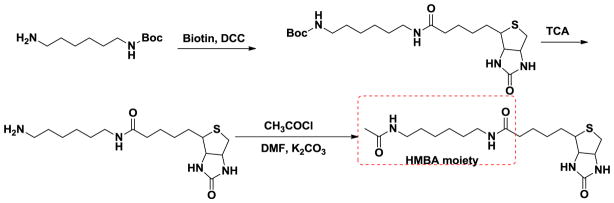
Scheme 1.
Biotin conjugated HMBA probe.
Scheme 2.
Biotin conjugated 4a1 probe.
4a1 probe: 1H NMR (400 MHz, DMSO-d6) δ 7.907 (1H, t, J = 5.2 Hz), 7.742 (2H, m), 7.137 (2H, d, J = 8.4 Hz), 6.834 (2H, d, J = 8.4 Hz), 6.408 (1H, s), 6.343 (1H, s), 4.297 (1H, m), 4.126 (1H, m), 3.915 (2H, t, J = 6.8 Hz), 3.293 (2H, s), 3.096 (1H, m), 3.0016H, m), 2.816 (1H, dd, J = 5.2, 12.4 Hz), 2.575 (1H, d, J = 12.4 Hz), 2.047 (2H, m), 1.779 (3H, s), 1.719–1.229 (22H, m); ESI–MS calculated for C32H50N5O5SNa [M+Na]+: 639.3, found: 640.1
HMBA probe: 1H NMR (400 MHz, DMSO-d6) δ 7.791 (1H, t, J = 5.2 Hz), 7.749 (1H, t, J = 5.6 Hz), 6.445 (1H, s), 6.374 (1H, s), 4.298 (1H, m), 4.115 (1H, m), 3.092 (1H, m), 2.997 (4H, m), 2.817 (1H, dd, J = 5.2, 12.4 Hz), 2.572 (1H, d, J = 12 Hz), 2.038 (2H, t, J = 7.2 Hz), 1.775 (3H, s), 1.593-1.223 (14H, m); ESI–MS calculated for C18H32N4O3SNa [M+Na]+: 407.2, found: 406.9
2.2. Biological methods and materials
2.2.1. Cell culture
LNCaP cells were obtained from ATCC (Rockville, MD). The cells were maintained in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mmol/L L-Glutamine, 1 mmol/L sodium pyruvate, 100 U/mL penicillin-streptomycin. FBS was heat inactivated for 30 min in a 56 °C water bath before use. Cell cultures were grown at 37 °C, in a humidified atmosphere of 5% CO2 in a Hereaus CO2 incubator.
2.2.2. Cell viability analysis
The effects of compounds on LNCaP cell viability were assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H- tetrazolium bromide assay in four replicates. Cells were grown in RPMI1640 medium in 96-well, flat-bottomed plates for 24 h, and were exposed to various concentrations of the compounds dissolved in DMSO (final concentration ≤0.1%) in media for 72 h. Controls received DMSO vehicle at a concentration equal to that in drug-treated cells. The medium was removed, replaced with 200 μL of 0.5 mg/ml of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide in fresh media, and cells were incubated in the CO2 incubator at 37 °C for 2 h. Supernatants were removed from the wells, and the reduced 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide dye was solubilized in 200 μL/well DMSO. Absorbance at 570 nm was determined on a plate reader. Statistical and graphical information was determined using GraphPad Prism software (GraphPad Software Incorporated) and Microsoft Excel (Microsoft Corporation).
2.2.3. Cell differentiation imagines
The effects of compounds on LNCaP cell differentiation were assessed using six well plates. Cells were grown in RPMI1640 medium in 6-well, flat-bottomed plates for 24 h, and were exposed to various concentrations of the compounds dissolved in DMSO (final concentration ≤0.1%) in media for 5 days. Controls received DMSO vehicle at a concentration equal to that in drug-treated cells. Cell morphological images in 6-well microplate were carried out manually on an inverted VWR VistaVision microscope (Bridgeport NJ) equipped with VWR VistaVision camera DV-2D (10 X objective, scan bar: 200 μm).
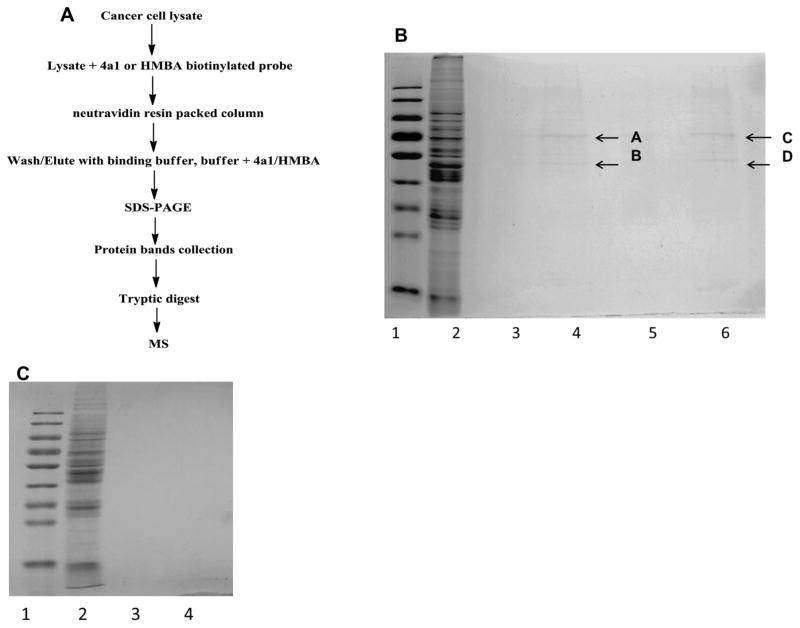
2.2.4. Biotin-neutravidin pull-down assay
The pull down assay was performed according to the protocol of neutravidin (Thermofisher). LNCaP cells (1.0 × 107) were disrupted in NP-40 lysis buffer and sonicated, with freshly added protease inhibitor cocktail (Roche). The cell lysate was incubated with the probes (final probe concentration in the lysate was 200 μM) for 1 h at room temperature, and then incubated with neutravidin resin for 30 mins. The mixture was loaded onto a separation column and centrifuged. The resin was washed with binding buffer for 5 times. The last wash elution solution was kept. The resin was further washed with buffer containing compound 4a1 (200 μM) or HMBA (1 mM). The last buffer without drug wash solution (lane 3 and 5) (Fig. 5), wash solution with compound 4a1 and HMBA (lane 4 and 6), were denatured in SDS sample buffer, and separated by SDS-PAGE. Lane 1 represents the molecular weight markers and lane 2 is the whole cell lysate. Visualization of the separated proteins with coomassie blue stain showed the proteins bound to 4a1 or HMBA. In addition, to the probes of HMBA and 4a1, LNCaP prostate cancer cell lysate was also exposed to a negative probe for 1 h at room temperature, and then incubated with neutravidin resin for 30 mins. The mixture was loaded on a separation column and centrifuged. The resin was washed with binding buffer for 5 times. The last wash elution solution was kept. The resin was further washed with buffer containing HMBA (1 mM). The last buffer without drug wash solution (lane 3), wash solution with HMBA (lane 4), were denatured in SDS sample buffer, and separated by SDS-PAGE. Lane 1 represents the molecular weight markers and lane 2 is the whole cell lysate. Visualization of the separated proteins was completed with coomassie blue staining.
Fig. 5.
Affinity isolation of 4a1 and HMBA bound proteins. (A) Procedure for the purification of 4a1 and HMBA bound proteins from LNCaP cell lysate. (B) The protein bands pullout by the probes. Visualization of the separated proteins with coomassie blue stain showed two major the proteins bound to 4a1 or HMBA. (C) Cell lysate exposed to a negative probe and the visualization of the separated proteins with coomassie blue stain showed no visible protein bands.
2.2.5. Peptide analysis of the binding protein via mass spectrometry
Bands visualized by coomassie blue staining were cut and transferred to 0.65 mL siliconized tubes (National Scientific Supply, Claremont, CA, USA). Proteins were in-gel digested by trypsin (Sequencing Grade, Modified; Promega, Madison, WI, USA). The protein digest was reconstituted in 20 μL of 0.1% (v/v) trifluoroacetic acid prior to LC-QTOF/MS analysis. Peptide separation was carried out using 10-μL sample injection at 50 μL/min flow rate on a Vydac® Protein & Peptide C18 (5 μm, 300 Å, 1 mm × 150 mm) column (Grace Discovery Sciences, Deerfield, IL, USA) proceeded by an inline filter (0.5 μm pore) (Upchurch Scientific, Oak Harbor, WA, USA). The gradient elution profile consisted of 1% of mobile phase A for 5 min, then brought to 60% of mobile phase B over 90 min, and followed by 90% of mobile phase B for 8 min. The total run time was 105 min. Mobile phase A was 0.1% (v/v) formic acid in HPLC-grade ddH2O, and the mobile phase B was 0.1% (v/v) formic acid in HPLC-grade acetonitrile. Peptide detection was done using the positive information-dependent-acquisition (IDA) mode of AB Sciex QStar™ Elite Q-TOF mass spectrometer (AB Sciex, Foster City, CA, USA). Data acquisition was performed using AB Sciex Analyst QS (v. 2.0). Protein identification through peptides matching was accomplished using Mascot MS/MS Ions Search (http://www.matrixscience.com).
2.2.6. Western blot
To determine the effect of HMBA, 4a1, SW02, Pifitrin-μ, synthesized probes on the HEXIM1 expression, LNCaP cells were treated with these chemicals or their combinations at indicated concentrations respectively for 24 h. Total cell lysates were extracted using M-PER reagent (Pierce). Equal amounts of proteins (50 μg) samples were separated on 12% SDS-polyacrylmide gel and transferred onto a polyvinylidene diflouride (PVDF) membrane (Pall Cooperation, FL.). After blocking for 1 h, the membrane was incubated in PBST containing 5% BSA and primary antibody specific to HEXIM1, p21 or actin (Cell signaling, MA) overnight at 4 °C. HRP-conjugated anti-rabbit IgG or anti-mouse IgG (Cell signaling, MA) were used as secondary antibody and incubated at room temperature for 1 h. The membrane was incubated in ECL plus reagent (GE health) and then exposed to hyper film. The densitometry measurement of the band intensities of HEXIM1, p21 and β-actin were quantified using ImageJ (NIH https://imagej.nih.gov/ij/) and the data was used to generate the corresponding bar figures. The normalization results are based on three independent experiments (±SD), and only one of the representative bands is presented.
2.2.7. Statistical analysis
Statistical and graphical information were determined using GraphPad Prism software (GraphPad Software Incorporated) and Microsoft Excel (Microsoft Corporation). Statistically significant differences were calculated with the two-tailed unpaired Student’s t-test and P values reported at 95% confidence intervals.
3. Results and discussions
3.1. Construction of the biotinylated probes based on compound 4a1 and HMBA
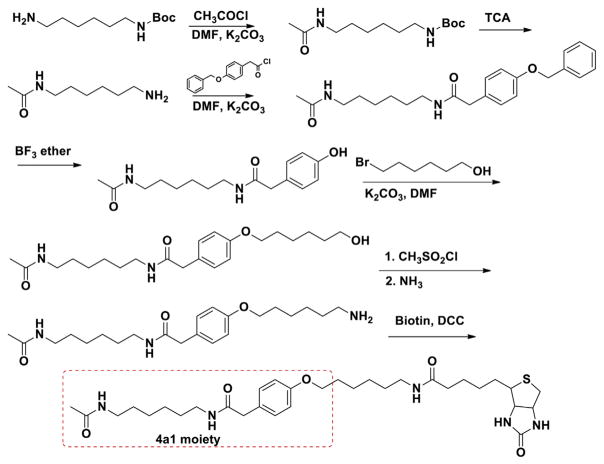
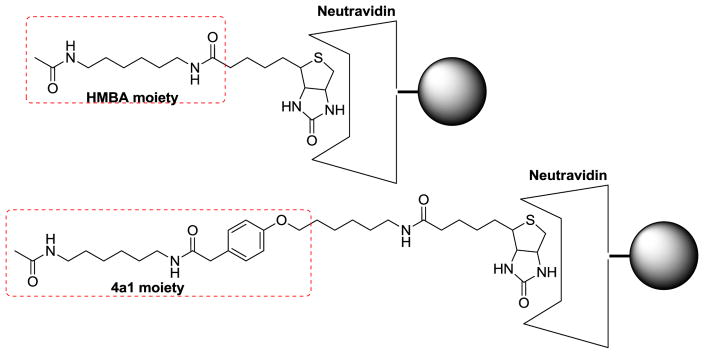
Numerous studies have demonstrated the induction of cell differentiation and HEXIM1 expression by HMBA [3,20]. However, the HMBA concentrations used in these studies are in the millimolar range. We speculate that HMBA binds to its molecular targets with low affinity, and therefore, using HMBA to design molecular probes to pull down its targets is not optimal, and the potential for false positive could be high. On the other hand, compound 4a1, a new HMBA analog developed recently, showed increased potency in the induction of HEXIM1 expression when compared to HMBA, perhaps due to enhanced affinity of 4a1 for its target [18]. Thus, 4a1 may be a better molecular probe for the identification of binding proteins of HEXIM1 inducers. Target identification represents a major challenge in small molecule drug development. A method used for target identification involves immobilizing the compound via a chemical linker on a solid-phase support, followed by affinity purification of the cellular targets. Therefore, we designed and synthesized a biotin-linked 4a1 as the probe for the target identification (Fig. 3).
Fig. 3.
Structures of 4a1 and HMBA probes.
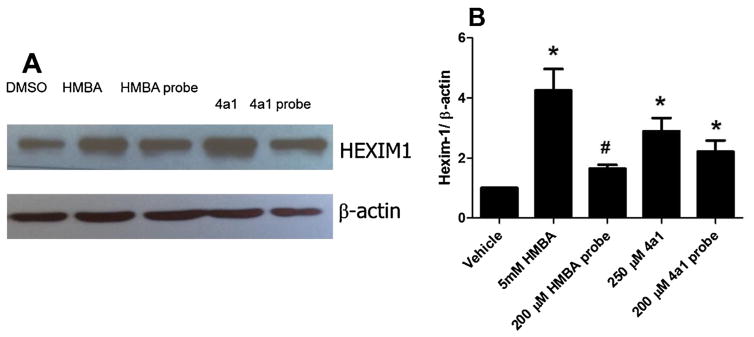
In order to determine if the HMBA binding targets are the same as 4a1, we synthesized an HMBA biotin-linked probe as well (Schemes 1 and 2) to examine if the two probes would pull down the same proteins. To test if the probes retained the biological activity of the parental compounds HMBA and 4a1, HEXIM1 expression was examined using western blots after treatment of cells with the probes. Treatment of LNCaP prostate cancer cells with either biotinylated-4a1 or –HMBA resulted in the induction of HEXIM1 expression, comparable to what was observed after treatment with parental compounds HMBA or 4a1 (Fig. 4). It should be noted that HMBA probe concentration used was much lower than that of HMBA due to the relatively lower solubility of the probe. These results suggest that the linkage and biotin moiety did not attenuate the activity of the parental compounds much, and the compound moiety can still bind to their targets and induce HEXIM1 expression.
Fig. 4.
Probes stimulate HEXIM1 expression in LNCaP cells. The cells were treated with DMSO, HMBA (5 mM), HMBA probe (200 μM), 4a1 (250 μM), 4a1 probe (200 μM) for 24 h. The results represent three independent experiments and the representative one or the mean is presented (±SD). (A) A representative western blot result of HEXIM1 expression. (B) The densitometry measurement of the band intensities of three western blots. *P <0.01 vs. DMSO by unpaired t test, #p <0.05 vs DMSO by unpaired t test.
3.2. Affinity purification of 4a1 and HMBA bound proteins
We hypothesize that 4a1 and HMBA bind to certain cellular factors in order to stimulate HEXIM1 expression. Therefore, we incubated 4a1 or HMBA probes with LNCaP cell lysates in order to allow binding of cellular proteins to the 4a1 or HMBA moiety of the probes, respectively. For protein isolation, the biotin moiety of the probe was bound to the neutravidin resin to immobilize the probe. After extensive washing with binding buffer, the non-specific binding proteins were eluted. Then, 4a1 and HMBA were used as competing agents to elute proteins bound to the probes. The major steps are shown in Fig. 5A. We next examined the molecular weight of the binding proteins using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) and stained the gel with coomassie blue reagent (Fig. 5B). Lane 2 is the whole cell lysate, lane 3 is the final elution solution of 4a1 probe, lane 4 contains the proteins eluted with 4a1, lane 5 is the final elution of probe HMBA, and lane 6 contains the proteins eluted with HMBA. There are two protein bands visible in lanes 4 and 6 with molecular weights around 70 kDa and 55 kDa respectively. The results reveal that both probes pulled down proteins with similar molecular weights, suggesting that HMBA and 4a1 have the same molecular targets. While these two protein bands are the most visible ones using our current methodology, there are likely less abundant proteins that bind to the probes as well. We will identify the visible proteins first and verify if they are the molecular targets of HMBA and 4a1. In the future, we will use a more sensitive staining method to identify the less abundant bound proteins. To rule out that the neutravidin resin might retain non-specific and highly abundant proteins, we also used biotin as a negative control to repeat the same experiment (Fig. 5C). The results revealed that the negative control probe did not pull out any visible proteins with our current methodology. The proteins collected from the pull down assay of the two probes are specific to HMBA and 4a1.
3.3. Identification of the bound proteins
The eluted proteins were separated by SDS/PAGE and collected by excising visible protein bands (A–D). Several very faint bands that are above bands A and C were also collected for analysis. The samples were subjected to tryptic digestion in situ. The resulting peptide mixture was identified with mass spectrometry. The molecular weight of the peptide mass fingerprint was used to identify the protein via Mascot database (Matrix Science Mascot, Boston, MA). The proteins with the best score (highest possibility) are listed in Table 1. Bands A and C were identified to be heat shock protein 70 kDa (HSP70), and B and D were identified to be tubulin. The signals from the faint bands were not adequate for mass spectrometry analyses. These proteins may either have weak binding affinity to the probes or are expressed at low levels in LNCaP cells. Our current probe based pull-down strategy will be optimized to identify these weaker binding proteins in the future. We decided to pursue the identified bands as potential targets of HMBA and 4a1.
Table 1.
Peptides were sequenced by electrospray ionization quadrupole time-of-fiight tandem mass spectrometry, and internal sequences were searched using the Mascot Database.
| Protein band | Molecular weight (estimated based on the marker) | Peptides (bold and red) matching with potential protein | Percentage of the matching | Protein identity |
|---|---|---|---|---|
| A/C | 70KD | MMKFTVVAAA LLLLGAVRAE EEDKKEDVGT VVGIDLGTTY SCVGVFKNGR VEIIANDQGN RITPSYVAFT PEGERLIGDA AKNQLTSNPE NTVFDAKRLI GRTWNDPSVQ QDIKFLPFKV VEKKTKPYIQ VDIGGGQTKT FAPEEISAMV LTKMKETAEA YLGKKVTHAV VTVPAYFNDA QRQATKDAGT IAGLNVMRII | 24% | Human heat shock protein 70 |
| NEPTAAAIAY GLDKREGEKN ILVFDLGGGT FDVSLLTIDN GVFEVVATNG DTHLGGEDFD QRVMEHFIKL YKKKTGKDVR KDNRAVQKLR REVEKAKRAL SSQHQARIEI ESFFEGEDFS ETLTRAKFEE LNMDLFRSTM KPVQKVLEDS DLKKSDIDEI VLVGGSTRIP KIQQLVKEFF NGKEPSRGIN PDEAVAYGAA | ||||
| VQAGVLSGDQ DTGDLVLLDV CPLTLGIETV GGVMTKLIPR NTVVPTKKSQ IFSTASDNQP TVTIKVYEGE RPLTKDNHLL GTFDLTGIPP APRGVPQIEV TFEIDVNGIL RVTAEDKGTG NKNKITITND QNRLTPEEIE RMVNDAEKFA EEDKKLKERI DTRNELESYA YSLKNQIGDK EKLGGKLSSE DKETMEKAVE | ||||
| EKIEWLESHQ DADIEDFKAK KKELEEIVQP IISKLYGSGG PPPTGEEDTS EKDEL | ||||
| B/D | 55KD | MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLDRISVY YNEATGGKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDSVLDVV RKEAESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEYP DRIMNTFSVV PSPKVSDTVV EPYNATLSVH QLVENTDETY | 27% | Bovin Tubulin beta chain |
| CIDNEALYDI CFRTLKLTTP TYGDLNHLVS ATMSGVTTCL RFPGQLNADL RKLAVNMVPF PRLHFFMPGF APLTSRGSQQ YRALTVPELT QQVFDAKNMM AACDPRHGRY LTVAAVFRGR MSMKEVDEQM LNVQNKNSSY FVEWIPNNVK TAVCDIPPRG LKMAVTFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG | ||||
| EGMDEMEFTE AESNMNDLVS EYQQYQDATA EEEEDFGEEA EEEA |
3.4. Biological activity of HSP70 ligands to HEXIM1 expression
We determined whether the binding of HMBA and 4a1 to HSP70 and tubulin could interfere with the functions of these two proteins. Tubulin can be affected in two manners: Taxanes and epothilones stabilize tubulin polymerization, whereas vinca alkaloids, halichondrins and colchicine inhibit tubulin polymerization [21,22]. Both tubulin stabilizers and destabilizers cause cell cycle arrest and subsequent cell apoptosis and death. HMBA and 4a1 induce cell differentiation, but do not cause cell apoptosis and cell death. Therefore, they are less likely to be a tubulin stabilizer or destabilizer.
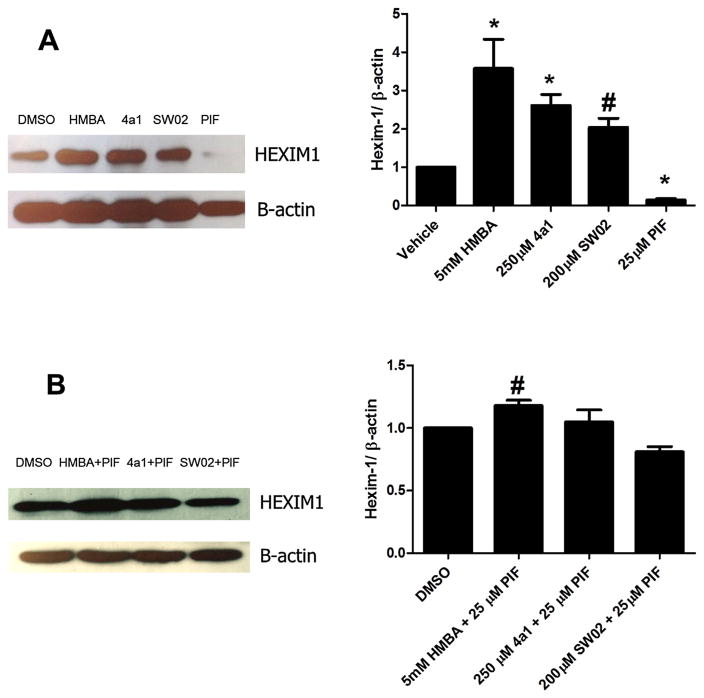
HSP70 can be affected in two ways, inhibition or activation. SW02 {4-(4-Bromophenyl)- 5-(ethoxycarbonyl)-3,4-dihydro-6-methyl-2-oxo-1(2H)-pyrimidinebutanoic acid methyl ester} has been demonstrated to be an HSP70 activator and Pifitrin-μ(2-phenylethynesulfonamide, PIF) has been used as an HSP70 inhibitor [23–25]. We used these two compounds as tools to examine if HSP70 ligands could regulate HEXIM1 expression in LNCaP cells. The results of the study could provide us indirect evidence of whether HSP70 is involved in HEXIM1 regulation. After 24 h treatment with compound SW02 and Pifitrin-μ, we observed that SW02 increased HEXIM1 expression whereas Pifitrin-μ decreased HEXIM1 expression (Fig. 6A). The results suggest that activation of HSP70 enhances HEXIM1 expression, while inhibition attenuates basal HEXIM1 expression in prostate cancer cells. To determine if the activator and inhibitor of HSP70 could antagonize each other, we performed combination treatments with SW02 and Pifitrin-μ. We observed that the induction of HEXIM1 expression by SW02 is attenuated by the Pifitrin-μ (Fig. 6B), and similar phenomenon happens to 4a1 and HMBA as well. Our results suggest that HSP70 plays a role in the regulation of HEXIM1 expression, HSP70 is a potential target of HEXIM1 inducers, and HMBA and 4a1 are potential HSP70 activators.
Fig. 6.
HSP70 ligands regulate HEXIM1 expression in LNCaP prostate cancer cells. (A) A HSP70 inhibitor and activator show opposing effects to HEXIM1 expression; (B) A HSP70 inhibitor and an activator were antagonistic to each other to stimulate HEXIM1expression. Cells were treated with DMSO, 5 mM HMBA, 200 μM 4a1, 250 μM SW02, 25 μM PIF or their combinations with the indicated concentrations for 24 h. The results are representative of three independent experiments and the representative one or the mean are presented (±SD). The densitometry measurement of the band intensities of three western blots were listed on the right. *P <0.01 vs. DMSO by unpaired t test, #p <0.05 vs DMSO by unpaired t test.
HSP70 is a well-documented molecular chaperone protein. It can maintain the stability of its client proteins under normal conditions and prevent stress-induced cellular damage, which is accomplished by maintaining the appropriate protein folding, prevention of protein aggregation, increase or decrease of proteasomal degradation of selected proteins. HSP70 is also known to play an important and complex role in apoptosis via interacting with components of apoptosis pathway or activating anti-apoptotic mediators [24,25]. HSP70 inhibitors have been well documented to induce apoptosis in cancer cells, while the role of HSP70 activators in cancer cells has not yet been well investigated. Our results indicate that HSP70 activators might play a role in regulating HEXIM1 expression, therefore affect cell differentiation.
3.5. HSP70 activators induce cell differentiation
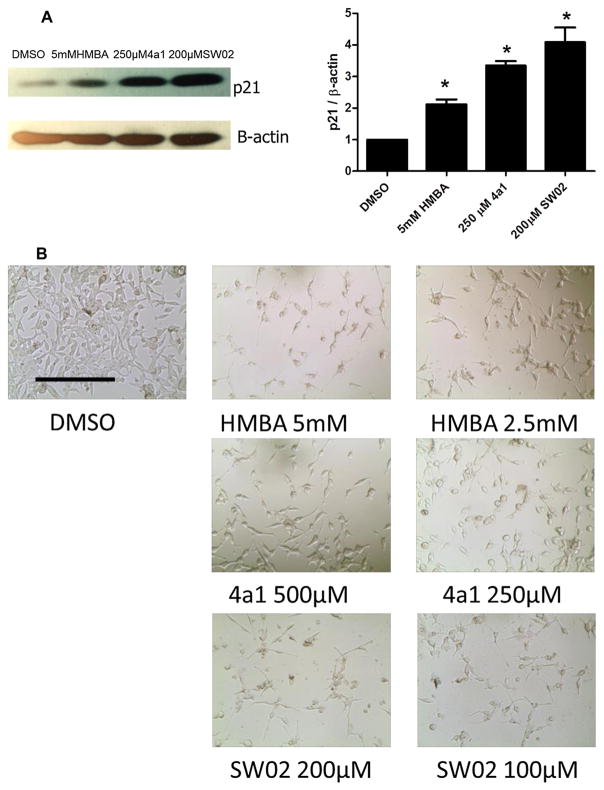
HMBA is well known for its ability to induce cell differentiation [26]. We hypothesized that since HMBA and its analog 4a1 are potential HSP70 activators, other HSP70 activators could be potential inducers of cell differentiation as well. LNCaP cells were treated with HMBA, 4a1 or HSP70 activator SW02. The HSP70 activator did not show acute cell toxicity compared to HSP70 inhibitors, and they caused changes in cell morphology after long term treatment (5 days), resulting in a more elongated shape compared to the control cells (Fig. 7B) [27]. P21 protein expression is a biomarker for cell differentiation, and all three compounds significantly increased p21 expression compared to the control (Fig. 7A), which is consistent with the study of HMBA and 4a1 in breast cancer cells [19]. The results suggest that an HSP70 activator could induce cell differentiation, which is different from what has been reported for HSP70 inhibitors.
Fig. 7.
HSP70 activators induce cancer cell differentiation. (A) LNCaP cells were treated with DMSO, HMBA, 4a1 or SW02 using the indicated concentrations for 24 h. P21 expression was examined with western blot. The results are representative of three independent experiments and the representative one or the mean is presented (±SD). The densitometry measurement of the band intensities of three western blots were listed on the right. *P <0.01 vs. DMSO by unpaired t test. (B) LNCaP cells were treated with DMSO, HMBA, 4a1 or SW02 with indicated concentrations for 5 days, and images of cell morphology were taken (10 X objective, scan bar: 200 μm).
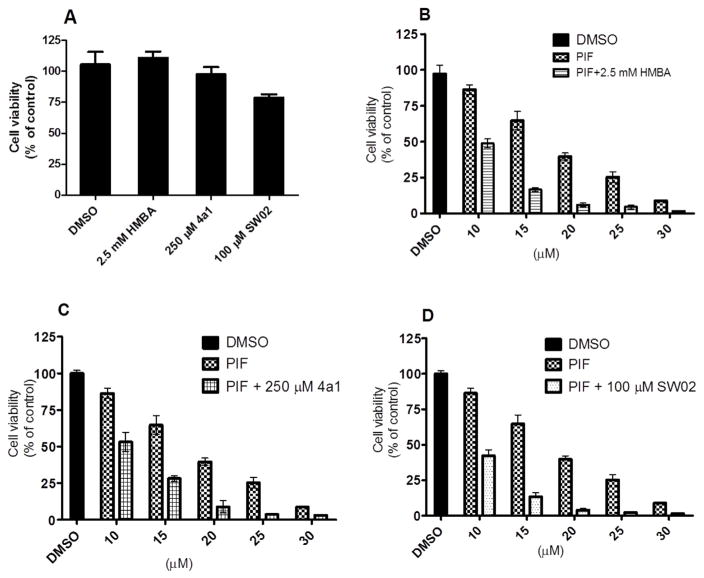
3.6. HSP70 activator and inhibitor show additive effect to inhibit cancer cell growth
HSP70 inhibitors were investigated as agents that induce cell apoptosis and therefore have potential as cancer therapeutics. However, HSP70 activators did not show acute cytotoxicity and have not been as well investigated as cancer therapeutic agent. Based on our results, HSP70 activators induce cell differentiation. Since the activators and inhibitors of HSP70 were antagonistic with regards to the induction of HEXIM1 expression, we determined if the same is true with regards to regulation of cancer cell proliferation. After 72 h treatment, HMBA and 4a1 did not cause significant decreases in cell number, and SW02 slightly decreased cell number (Fig. 8). HSP70 inhibitor Pifitrin-μ significantly inhibited cell proliferation in a dose dependent manner. Contrary to what we expected, when we combined Pifitrin-μ with either HSP70 activator SW02, HMBA or 4a1, we found that the activator augmented growth inhibition by Pifitrin-μ. The combination treatment resulted in an additive effect, and is different from the effect of the combination treatment on HEXIM1 expression. These findings suggest that regulation of cell growth by HSP70 activators and inhibitors may occur independently of HEXIM1 induction. It is likely that HSP70 has other client proteins that can regulate cell proliferation and account for the different effects of combination treatment on HEXIM1 expression and cell proliferation.
Fig. 8.
HSP70 activators and inhibitor show additive effects to inhibit cancer cell growth. LNCaP cancer cells were treated with DMSO, HMBA, 4a1, SW02, Pifitrin-μ or their combinations at the indicated concentrations for 72 h in four replicate experiments. The cell viability was evaluated with MTT assay. The results are a representative one of three independent experiments and the mean (±SD) is represented.
4. Conclusion
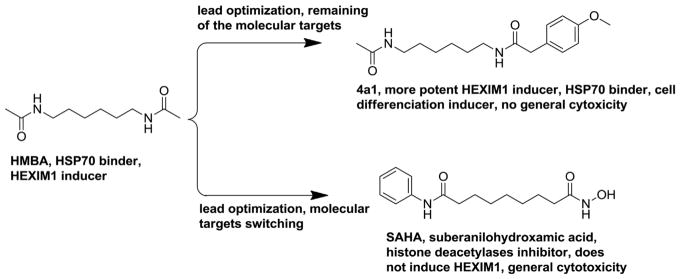
Tamoxifen and aromatase inhibitors are the standard treatments of estrogen dependent breast cancer. However, patients eventually develop resistance to these endocrine disrupting agents. At this stage, estrogen receptor still plays a critical role in cancer cell proliferation, but does not respond to these agents [28,29]. HEXIM1 interferes with estrogen receptor in a different mechanism compared to tamoxifen and aromatase inhibitors, and may be able to solve the endocrine therapy resistant breast cancer [19]. HMBA is a well-used tool drug to induce HEXIM1 expression, but its toxicity limits the clinical application. In addition, the direct molecular target of HMBA is unclear, which also decreases the further drug development efforts based on this compound as the lead.
In fact, based on the anti-cancer activity and the bipolar moieties of HMBA, SAHA (suberanilohydroxamic acid) was developed and identified as a HDAC (histone deacetylases) inhibitor (Fig. 9) [26]. However, SAHA does not induce HEXIM1 expression, which suggests that the structural modification from HMBA to SAHA altered its mechanism of action. To focus on HEXIM1 induction as the outcome, we performed lead optimization and evaluated new analogs using HEXIM1 expression change as the readout. The more potent HEXIM1 inducer 4a1 with asymmetric structure was developed based on HMBA with symmetric moieties. To identify the molecular targets of the agents and elucidate the mechanism of the HEXIM1 induction, we designed and synthesized biotinylated 4a1 and HMBA as probes. Proteins bound to the compounds were isolated, subjected to mass spectrometric identification, and the most prevalent binding proteins were determined to be tubulin and HSP70. Since HMBA and 4a1 did not induce significant cell growth inhibition or apoptosis inducing activity, it is less likely that they are tubulin stabilizers or de-stabilizers. Further investigation revealed that the HSP70 activator SW02 showed similar activity as HMBA and 4a1, including induction of HEXIM1 expression and induction of cell differentiation. In addition, HSP70 inhibitor Pifitrin-μ could antagonize the induction of HEXIM1 expression by HMBA and 4a1. These results provide indirect evidence that HMBA and 4a1 are likely to be HSP70 activators. Although the HSP70 activation and inhibition antagonize each other in the induction of HEXIM1 expression, HSP70 activation and inhibition are additive with regards to cancer cell growth inhibition. It is possible that HSP70 has other targets besides HEXIM1 that can regulate cell proliferation and account for the different effects of combination treatment on HEXIM1 expression and cell proliferation.
Fig. 9.
Drug development based on HMBA as a lead compound.
Conversely, HSP70 may not be the only molecular target of HMBA and 4a1, since other proteins appear to bind to the biotinylated compounds. Due to the low binding affinity of these proteins to the probes or their low abundance, we are unable to identify these other putative binding proteins currently. Studies are now underway to further optimize 4a1 in order to generate more potent derivatives to induce HEXIM1 expression. In addition, we will identify the binding domain of HMBA and 4a1 on HSP70. The binding pocket will be the basis for structure activity relationship studies of HMBA analogs as potential HSP70 activators.
Acknowledgments
This research was supported Center for Gene Regulation in Health and Disease (GRHD) of Cleveland State University and Ohio Department of Development (ODOD). Chunfang Gan was supported by National Natural Science Foundation of China (No: 21562007).
Abbreviations
- HEXIM1
hexamethylene bis-acetamide inducible protein 1
- HMBA
hexamethylene bis-acetamide
- HSP70
heat shock protein 70
- SAHA
suberanilohydroxamic acid
- HDAC
histone deacetylases
- SDS/PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- FBS
fetal bovine serum
- PVDF
polyvinylidene diflouride
References
- 1.Wittmann BM, Wang N, Montano MM. Identification of a novel inhibitor of breast cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 2003;63(16):5151–5158. [PubMed] [Google Scholar]
- 2.He N, Pezda AC, Zhou Q. Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol Cell Biol. 2006;26(19):7068–7076. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turano M, Napolitano G, Dulac C, Majello B, Bensaude O, Lania L. Increased HEXIM1 expression during erythroleukemia and neuroblastoma cell differentiation. J Cell Physiol. 2006;206(3):603–610. doi: 10.1002/jcp.20502. [DOI] [PubMed] [Google Scholar]
- 4.Andreeff M, Stone R, Michaeli J, Young CW, Tong WP, Sogoloff H, Ervin T, Kufe D, Rifkind RA, Marks PA. Hexamethylene bisacetamide in myelodysplastic syndrome and acute myelogenous leukemia: a phase II clinical trial with a differentiation-inducing agent. Blood. 1992;80(10):2604–2609. [PubMed] [Google Scholar]
- 5.Young CW, Fanucchi MP, Declan Walsh T, Baltzer L, Yaldaei S, Stevens YW, Gordon C, Tong W, Rifkind RA, Marks PA. Phase I trial and clinical pharmacological evaluation of hexamethylene bisacetamide administration by ten-day continuous intravenous infusion at twenty-eight-day intervals. Cancer Res. 1988;48(24 Pt 1):7304–7309. [PubMed] [Google Scholar]
- 6.Conley BA, Egorin MJ, Sinibaldi V, Sewack G, Kloc C, Roberts L, Zuhowski EG, Forrest A, Van Echo DA. Approaches to optimal dosing of hexamethylene bisacetamide. Cancer Chemother Pharmacol. 1992;31(1):37–45. doi: 10.1007/BF00695992. [DOI] [PubMed] [Google Scholar]
- 7.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12(4):971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 8.Espinoza-Derout J, Wagner M, Salciccioli L, Lazar JM, Bhaduri S, Mascareno E, Chaqour B, Siddiqui MA. Positive transcription elongation factor b activity in compensatory myocardial hypertrophy is regulated by cardiac lineage protein-1. Circ Res. 2009;104(12):1347–1354. doi: 10.1161/CIRCRESAHA.108.191726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23(13):2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montano MM, Doughman YQ, Deng H, Chaplin L, Yang J, Wang N, Zhou Q, Ward NL, Watanabe M. Mutation of the HEXIM1 gene results in defects during heart and vascular development partly through downregulation of vascular endothelial growth factor. Circ Res. 2008;102(4):415–422. doi: 10.1161/CIRCRESAHA.107.157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogba N, Chaplin LJ, Doughman YQ, Fujinaga K, Montano MM. HEXIM1 regulates 17beta-estradiol/estrogen receptor-alpha-mediated expression of cyclin D1 in mammary cells via modulation of P-TEFb. Cancer Res. 2008;68(17):7015–7024. doi: 10.1158/0008-5472.CAN-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu N, Ouchida R, Yoshikawa N, Hisada T, Watanabe H, Okamoto K, Kusuhara M, Handa H, Morimoto C, Tanaka H. HEXIM1 forms a transcriptionally abortive complex with glucocorticoid receptor without involving 7SK RNA and positive transcription elongation factor b. Proc Natl Acad Sci U S A. 2005;102(24):8555–8560. doi: 10.1073/pnas.0409863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascareno EJ, Belashov I, Siddiqui MA, Liu F, Dhar-Mascareno M. Hexim-1 modulates androgen receptor and the TGF-beta signaling during the progression of prostate cancer. Prostate. 2012;72(9):1035–1044. doi: 10.1002/pros.21510. [DOI] [PubMed] [Google Scholar]
- 14.Yeh IJ, Song K, Wittmann BM, Bai X, Danielpour D, Montano MM. HEXIM1 plays a critical role in the inhibition of the androgen receptor by anti-androgens. Biochem J. 2014;462(2):315–327. doi: 10.1042/BJ20140174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contreras X, Schweneker M, Chen CS, McCune JM, Deeks SG, Martin J, Peterlin BM. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284(11):6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3(10):1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ketchart W, Smith KM, Krupka T, Wittmann BM, Hu Y, Rayman PA, Doughman YQ, Albert JM, Bai X, Finke JH, Xu Y, Exner AA, Montano MM. Inhibition of metastasis by HEXIM1 through effects on cell invasion and angiogenesis. Oncogene. 2013;32(33):3829–3839. doi: 10.1038/onc.2012.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong B, Lama R, Ketchart W, Montano MM, Su B. Lead optimization of HMBA to develop potent HEXIM1 inducers. Bioorg Med Chem Lett. 2014;24(5):1410–1413. doi: 10.1016/j.bmcl.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ketchart W, Yeh IJ, Zhou H, Thiagarajan PS, Lathia J, Reizes O, Exner A, Su B, Montano MM. Induction of HEXIM1 activities by HMBA derivative 4a1: functional consequences and mechanism. Cancer Lett. 2016;379(1):60–69. doi: 10.1016/j.canlet.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richon VM, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind RA, Marks PA. Second generation hybrid polar compounds are potent inducers of transformed cell differentiation. Proc Natl Acad Sci U S A. 1996;93(12):5705–5708. doi: 10.1073/pnas.93.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai R, Nguyen TL, Burnett JC, Atasoylu O, Munro MH, Pettit GR, Smith AB, 3rd, Gussio R, Hamel E. Interactions of halichondrin B and eribulin with tubulin. J Chem Inf Model. 2011;51(6):1393–1404. doi: 10.1021/ci200077t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingston DG. Tubulin-interactive natural products as anticancer agents. J Nat Prod. 2009;72(3):507–515. doi: 10.1021/np800568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chafekar SM, Wisen S, Thompson AD, Echeverria A, Walter GM, Evans CG, Makley LN, Gestwicki JE, Duennwald ML. Pharmacological tuning of heat shock protein 70 modulates polyglutamine toxicity and aggregation. ACS Chem Biol. 2012;7(9):1556–1564. doi: 10.1021/cb300166p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granato M, Lacconi V, Peddis M, Lotti LV, Di Renzo L, Gonnella R, Santarelli R, Trivedi P, Frati L, D’Orazi G, Faggioni A, Cirone M. HSP70 inhibition by 2-phenylethynesulfonamide induces lysosomal cathepsin D release and immunogenic cell death in primary effusion lymphoma. Cell Death Dis. 2013;4:e730. doi: 10.1038/cddis.2013.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leu JI, Pimkina J, Pandey P, Murphy ME, George DL. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol Cancer Res. 2011;9(7):936–947. doi: 10.1158/1541-7786.MCR-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci U S A. 1998;95(6):3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan TC, Veeramani S, Lin FF, Kondrikou D, Zelivianski S, Igawa T, Karan D, Batra SK, Lin MF. Androgen deprivation induces human prostate epithelial neuroendocrine differentiation of androgen-sensitive LNCaP cells. Endocr Relat Cancer. 2006;13(1):151–167. doi: 10.1677/erc.1.01043. [DOI] [PubMed] [Google Scholar]
- 28.Guest SK, Ribas R, Pancholi S, Nikitorowicz-Buniak J, Simigdala N, Dowsett M, Johnston SR, Martin LA. Src is a potential therapeutic target in endocrine-resistant breast cancer exhibiting low estrogen receptor-mediated transactivation. PLoS One. 2016;11(6):e0157397. doi: 10.1371/journal.pone.0157397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan VC, Fan P, Abderrahman B, Maximov PY, Hawsawi YM, Bhattacharya P, Pokharel N. Sex steroid induced apoptosis as a rational strategy to treat anti-hormone resistant breast and prostate cancer. Discov Med. 2016;21(117):411–427. [PubMed] [Google Scholar]