Abstract
RNA molecules are flexible yet foldable. Proteins must cope with this structural duality when forming biologically active complexes with RNA. Recent studies of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs)-mediated RNA immunity illustrate some remarkable mechanisms with which proteins interact with RNA. Currently known sstructure of CRISPR-Cas6 endoribonucleases bound with RNA suggest a conserved protein recognition mechanism mediated by RNA stem-loops. However, a survey of CRISPR RNA reveals that many repeats either lack a productive stem-loop(Relaxed) or possess stable but inhibitory structures (Tight), which raises the question of how the enzyme processes structurally diverse RNA. In reviewing recent literature, we propose a bivalent trapping and an unwinding mechanism for CRISPR-Cas6 to interact with the Relaxed and the Tight repeat RNA, respectively. Both mechanisms aim to create an identical RNA conformation at the cleavage site for accurate processing.
Keywords: Cas6, CRISPR, endoribonuclease, protein-RNA interactions, RNA binding proteins, stem-loop
Introduction
As a major class of biopolymers, RNA fulfills both informational and functional roles in cells, and is associated with many diseases including neurodegenerative diseases and cancer. Of the physicochemical properties RNA have, the most distinct is their structural flexibility [1]. RNA can fold into complex structures similar to proteins that are catalytic or versatile binders for small molecules and proteins [2]. However, RNA must also stay unfolded as templates for information transfer [2]. This biophysical dichotomy creates challenges in structure characterization of RNA, whether as isolated molecules or in complex with proteins. To gain insight into how structural flexibility in RNA impacts their structure and function, large sets of experimentally obtained structures of RNA complexes should be analyzed.
The recently discovered small RNA-guided CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated) immunity in prokaryotes offers a rare opportunity to achieve a deeper understanding of protein-RNA interactions [3–6]. In particular, the evolutionarily conserved Cas6 family proteins recognize and excise the CRISPR repeat RNA that vary widely in primary and secondary structures and have been the subject of extensive structural and biochemical studies [7–10]. The outcome of these studies has provided a set of useful principles applicable to other RNA binding proteins. Chief among them is the ability of Cas6 in preparing CRISPR RNA, regardless of their starting structure, into a conserved form cleavable by Cas6. Though there are well over thousands of Cas6 proteins, only ten have been characterized structurally and eight with their binding substrate RNA [7, 8, 11]. Thus the principles learned so far remain limited and biased toward those known. Continued studies of structure and function of Cas6-RNA pairs are needed. We analyzed a set of repeat RNA associated with Cas6 and identified, besides the canonical recognition motif, many that are either of no structure (Relaxed) orhyperstabilized (Tight). This analysis suggests potentially novel mechanisms of Cas6 processing. We present a hypothesis of how Cas6 is able to recognize and process these types of repeat RNA.
Cas6 processes CRISPR RNA required for the CRISPR-Cas immumity
CRISPR-Cas loci are found in nearly half of bacterial and all archaeal genomes that have been sequenced to date [5, 6, 12]. A CRISPR-Cas locus is typically comprised of contiguous repeat sequences interrupted by distinct spacer sequences and the adjacent Cas protein encoding genes (cas genes). To elicit defense activity, a CRISPR-Cas system acquires spacer sequences from the invading genetic elements and then employs the RNA transcripts of the acquired spacers as guides in degrading the same invaders during subsequent infections. Cas family proteins perform each of these functional steps. Among them, Cas6 is responsible for processing the precursor RNA transcript of the repeat-spacer-repeat array into small CRISPR RNAs (crRNA) associated with the Types I and III CRISPR-Cas effector complexes. Cas6 binds and excises within and near the 3′ end of the repeat, releasing short spacer RNAs flanked by both 3′ and 5′ halves of the repeat (Fig. 1). In some cases, the flanking repeat 3′ to the spacer is further processed by currently unknown mechanisms unrelated to Cas6 [7, 8].
Figure 1.
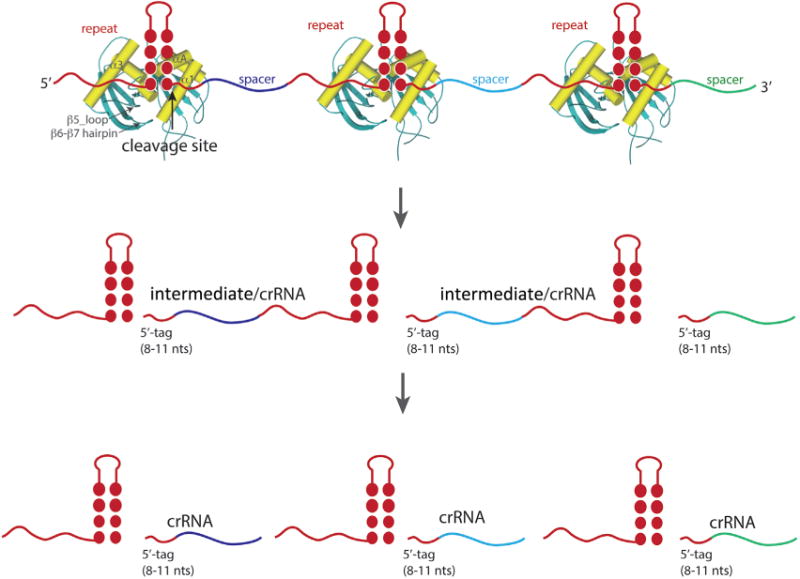
Schematic recognition and processing of CRISPR repeat-spacer array by Cas6. Repeats are colored in red and spacers are in various colors. In general, the palindromic feature in CRISPR repeats leads to an RNA stem-loop structure recognized by Cas6 (represented by the crystal structure of Pyrococcus furiosus Cas6 PDB id 3I4M). The secondary elements of Cas6 involved in binding RNA are labeled for one of the Cas6 models. Cleavage at the base of the stem releases the intermediate for the Type III or mature crRNA for the Type I CRISPR systems. The intermediate is further processed by uncharacterized activities to the final crRNA that contains the 5′-tag and the spacers.
How Cas6 interacts with structurally different CRISPR RNA
In order to gain a full description of structural features of repeat RNA processed by Cas6, we examined repeat RNA sequences found in CRISPRdb [13] that are associated with Types I & III CRISPR-Cas systems and confirmed their orientations by CRISPRmap [14]. We then computationally predicted their secondary structures by a thermodynamics-based method as implemented in mFold [15]. Select structures of the analyzed repeats are shown in Fig. 2. Many repeats display a clear palindromic feature that would result in a stable stem-loop structure immediately preceding the cleavage site (Canonical). Note that the 3′ cleavage product, which forms the 5′ handle of a mature crRNA, is primarily 8nt in length but seems to vary from 7 to 11nts. The second class of repeats, which we term Relaxed, lack the characteristic stem-loop structure and form multiple, and short stem-loops at variable locations relative to the predicted site of cleavage (Relaxed) (Fig. 2). The third class of repeats, which we term Tight, form surprisingly hyperstabilized long stems (Tight).If we assume that the Tight repeats are also processed to leave a similar5′-handleof7–11 ntsinlength, the cleavage site would fall within the stem, rather than at the base of the stem as in the Canonical stem-loop class (Fig. 2C). Even if one takes into account the fact that mFold does not necessarily provide an accurate prediction of RNA secondary structures, it is clear that the substrates for Cas6 vary widely in structure, which raisesthequestionofhowCas6processes such range of CRISPR RNA structures. We further note that the two non-Canonical types appear more often in archaeal or thermophilic organisms, suggesting an impact of environment on the Cas6-mediated CRISPR RNA processing. Elevated temperature can “melt” repeats for Cas6 to reshape them into a catalytically active conformation. It is especially interesting to understand how Cas6 can recognize and cleave the two classes of repeat RNA that lack the Canonical stem-loop.
Figure 2.
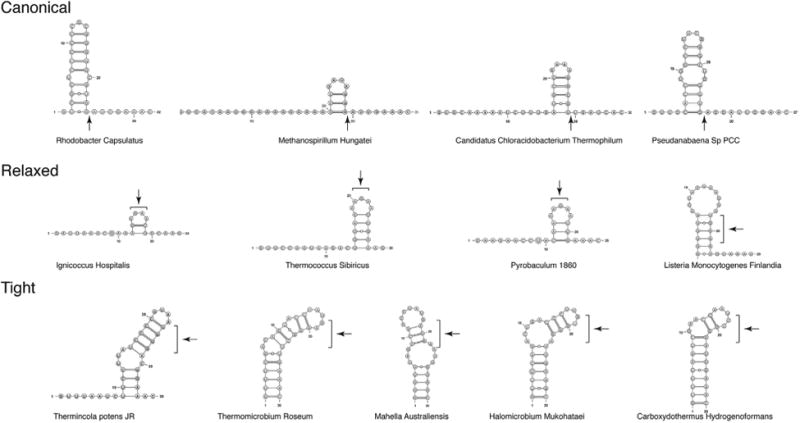
Representative CRISPR repeat RNA structures predicted by mFold. The structures are categorized into three classes. The Canonical class comprises repeat RNAs that form a stable stem-loop (−19.0kcal/mol < ΔG°<4.8kcal/mol) immediately upstream of the putative cleavage site (indicated by arrows). The Relaxed class comprises repeat RNAs that have unstable structures (ΔG° > 0 kcal/mol) with no defined locations. The putative cleavage sites (8–11 nts from 3′ end) of these RNA are indicated by brackets. The Tight class comprises repeat RNA that also form stable and long stem loops (−14.5 kcal/mol < ΔG° < 3.8 kcal/mol) but unlike the Canonical class, reveal no cleavage sites at the end of the stem.
Cas6 contains a conserved fold
Sequence analysis of Cas6 proteins indicates shared features of Cas6 with the well-known ferredoxin-like fold [16]. The ferredoxin-like fold comprises two sets of β-α-β supersecondary structure motifs that are arranged into a single, four-stranded anti-parallel β-sheet packed against two α-helices (β1–β4 and α1–α2). The ferreodoxin-like fold is the most abundant protein fold known and comprises a superfamily of RNA Recognition Motif (RMM) proteins including several mRNA splicing factors [17], Fragile-X syndrome protein [18, 19] and Amyotrophic Lateral Sclerosis (ALS)-associated protein FUS [20]. Unlike RRM, however, Cas6 comprise a tandem ferredoxin-like fold arranged nearly in orthogonal and connected by a short loop from the last β-strand, β4 of the N-terminal to the first β-strand, β5, of the C-terminal ferredoxin-like fold. The N-terminal ferredoxin-like fold of Cas6 invariably contains an inserted α-helix, αA, following its first α-helix, α1. The C-terminal ferredoxin-like fold also contains variable insertions following β5 (β5_loop) and β6 (β6– β7 loop), respectively. Furthermore, a glycine-rich loop (G-loop) between α4 and the last β-strand, β8, is situated at the interface between the two ferredoxin-like folds (Fig. 1). These elements enable Cas6 to bind and cleave a wide range of CRISPR repeat RNA as described below.
Cas6 stabilizes the canonical stem-loop at the cleavage site
All currently known Cas6-RNA complex structures show that Cas6 locates the cleavage site by binding a stem-loop immediately upstream of the cleavage site regardless of the structure when RNA is in isolation. The repeat RNA of both Pseudomonas aeruginosa and Thermus thermophilus are predicted by mFold to form a stem-loop containing 4–5 base pairs that are indeed observed in the presence of Cas6 [21, 22]. Whereas those of Sofulobus solfataricus and Methanococcus maripaludis either lack the conserved stem-loop or have a short and unstable stem-loop [11, 23]. However, upon association with Cas6, both S. solfataricus and M. maripaludis RNA did conform to the same rule of stem-loop-mediated cleavage by Cas6. Consistently, biochemical analysis of the four Cas6-RNA systems shows that the stem-loop preceding the cleavage site is important to formation of the catalytically productive structure of the RNA. These results suggest that Cas6 has an intrinsic ability to fold RNA, which likely provides a significant source of rate enhancement.
Unlike canonical RRM that bind single-stranded RNA with their β-sheet surface, Cas6 primarily use their α-helices and insertion elements to stabilize the stem-loop. The first α-helices of both ferredoxin-like motifs, α1 and α3, as well as the G-loop located between α1 and α3 form an exclusively helical surface for the descending strand and the minor groove whereas β5_loop is positioned to interact with the major groove (Fig. 1). The β6_β7 loop is wedged between the ascending and the descending strand and is thus important to formation of the RNA fork at the base of the stem (Fig. 1). Furthermore, residues from β6_β7 loop and α1 form the active site around the scissile phosphate group at the base of the RNA stem. Therefore, the juxtaposition of α1 and α3 and the varying structures of both β5 and β6 insertions enable Cas6 to recognize individual stem-loop structures of different length and sequences. This observed mode of RNA binding explains the requirement for the tandem ferredoxin-like fold.
Hypothesis: Cas6 stabilizes relaxed repeats through bivalent interactions
Though Cas6 readily recognizes stably folded stem-loop RNA, the question arises how it recognizes and cleaves the repeat lacking a stable stem-loop at the cleavage site. Recent crystal structures of two Cas6 bound with their respective substrate RNA reveal surprising interactions between Cas6 and the RNA beyond the cleavage site described above. In the 1:1 Cas6-RNA complex of Pyrococcus furiosus (Pf), the first 12 nucleotides of the repeat binds tightly and specifically to Cas6 but the rest of the 25 nucleotides are disordered (Fig. 3) [24]. This structure explains the critical importance of the first eight nucleotides in PfCas6 processing activity [25]. It, however, does not address directly how PfCas6 is able to cleave the RNA 21 nucleotides away from the 5′ end. Because the first 12 nucleotides are bound to the β-sheet side of PfCas6, a wrap-around model was first proposed in which the β-sheet face of the protein anchors the 5′ end whereas its α-helical face binds the 3′ end including the cleavage site [24]. This model suggests that PfCas6 is required to interact with two motifs of the repeat RNA, likely to prevent non-productive folding of the RNA (Fig. 3).
Figure 3.
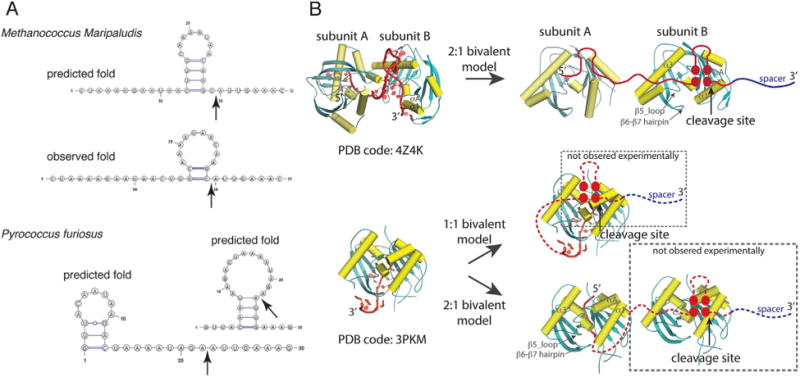
Examples of the observed bivalent interactions for the Relaxed repeat RNA. A: The predicted and the observed (for M. maripaludis only) repeat RNA folds. Top, a single fold is predicted for the M. maripaludis repeat in comparison with its observed fold when bound with Cas6 in a crystal structure (PDB code: 4Z4K). Bottom, two folds of equal stability are predicted for the P. furiosus repeat. B: Crystal structures of M. maripaludis (top) and P. furiosus (bottom) Cas6 bound with their respective repeat RNA and illustration of the bivalent binding models. Two M. maripaludis Cas6 subunits interact with the two RNA motifs of a single repeat RNA (2:1 bivalent model). P. furiosus Cas6 interacts with the first 12 nucleotides of the repeat with the rest of the RNA not observed. This leads to two possible models: The 1:1 bivalent binding model (wrap-around model) and the 2:1 bivalent binding model. Note that dashed frames indicate portions of the models not observed experimentally.
The Cas6-RNA complex from Meth-anococcus maripaludis (Mm) [11] reveals another bivalent binding mechanism. The crystal structure of MmCas6-RNA complex shows a 2:1 stoichiometry (Fig. 3). One protein stabilizes the cleavage site stem-loop that only contains two base pairs and the other binds to a 5′-motif analogous to the loop portion of the cleavage site stem-loop (Fig. 3). Computational analysis of the isolated RNA reveals a stable stem-loop adjacent to the cleavage that, however, would be inhibitory to correct processing (Fig. 3). Thus, we can rationalize that the bivalent binding mode of MmCas6 is a mechanism for eliminating non-productive RNA structures. PfCas6 may also adapt this mechanism, rather than the previously proposed wrap-around model, by requiring two enzymes, one binding the 5′ sequence and one binding the 3′ cleavage site (Fig. 3). As a further extension, similar mechanisms employed by PfCas6 and MmCas6 may apply to all those CRISPR RNA in the Relaxed category (Fig. 2, Relaxed).
This hypothesis may be tested by studying the structure and function of additional Cas6-RNA pairs. The fact that the Relaxed type of repeats has no predictable secondary structures suggests that they adopt multiple unstable structures when in isolation including one that is capable of binding Cas6. Whereas structures of Cas6-RNA complexes are straightforward to characterize, the ensemble of the RNA structures is more difficult to resolve. One possible strategy is to combine biophysical and biochemical measurement with computational methods in analyzing the structure ensemble. Such information is needed to determine if these RNA either form inhibitory or have no productive structures and to compare the RNA structures to that when Cas6 is bound. Though bivalent binding is a compelling model based on observed examples, other binding models may be revealed from studying additional Cas6-RNA systems in the Relaxed category.
Hypothesis: Cas6 unfolds tight repeats
The tight repeats appear to be thermo-dynamically stable when in isolation (Fig. 2, Tight), which makes it difficult to apply previously learned principles of Cas6-repeat RNA interactions. If we assume that the sites of cleavage are similarly 7–11 nts from the 3′ end of the repeat, they would most likely fall within stems or internal loops (Fig. 2, Tight). How can then Cas6 facilitate phosphodiester bond cleavage within these regions? Since there has not been any study of Cas6 associated with the Tight repeats, we can only speculate their mechanism of processing. One possibility is for Cas6 to unwind the lower half of the tight stem, leading to the same binding mode as observed for the Canonical stem-loops. The bivalent binding mechanism proposed for processing Relaxed repeats (Fig. 3B) may also be used to aid formation of the alternative fold. The frequent appearance of internal loops at 7–11 nts from the 3′ end within the Tight repeats and the AU-rich lower stem seem to support this hypothesis (Fig. 2, Tight). The predicted structures by homology modeling of Cas6 associated with the Tight repeats also support this model. These Cas6 seem to contain long insertions in both β5_loop and β6_β7 loop regions that may be used for disrupting base pairing.
It would be interesting to test this hypothesis by analyzing structures of Cas6 proteins and the Tight repeat RNA. Unlike the Relaxed type, the Tight repeat RNA likely form single stable stem-loops in the absence of Cas6, which should be easily verified experimentally. The structure of the Cas6-bound Tight repeats can be determined by structural biology methods such as x-ray crystallography and Nuclear Magnetic Resonance. Comparing the structures of a Tight repeat before and after Cas6 binding will reveal structural changes that occur in both RNA and Cas6. Due to possible alternative structures of RNA or Cas6-RNA complexes, complementary biochemical and biophysical experiments should be performed to confirm the observed structures in solution. Having a firm understanding of Tight repeat RNA structure in the presence and absence of Cas6 provides the basis for understanding the mechanism of their processing.
Conclusions and outlook
The analysis presented here reveals a previously unexpected diversity in RNA substrates of Cas6, and raises tantalizing questions about Cas6-RNA interactions. The common thread of Cas6-RNA interaction is the Cas6-faciliated formation of the catalytically active conformation of the phosphodiester bond at the cleavage site, regardless of the RNA starting structure. The different degree of structural variations between Cas6 and the repeat RNA reflects the difference in physicochemical properties of the two types of macromolecules, and suggests an evolutionary advantage of proteins over RNA in meeting the challenges of both binding and catalysis.
A more difficult question to address is whether variations in repeat RNA structure are correlated with the functional fitness of the host cells. The known enzyme kinetics data for Cas6 associated with both the Canonical and the Relaxed repeats show some evidence for such correlation. The cleavage rate constants for the Canonical repeats are in general a few fold greater than those for the Relaxed repeats [21–23, 26, 27] and are similar to the observed cleavage rate constants of ribozymecatalyzed reactions [28]. The ribozymecatalyzed rate constants are believed to correspond to that of RNA conformational changes necessary to facilitate catalysis [29], suggesting that, even for the pre-formed RNA substrates, the activity of Cas6 is limited by conformational changes in RNA. The slower cleavage rates for the Relaxed repeat RNA – and presumably even slower for the Tight repeat RNA – suggest additional transitional processes required for these RNA to reach their catalytic conformation. It is not yet known which of the three functional pathway (spacer acquisition, RNA processing, and target degradation) limits the efficiency of CRISPR-Cas immunity. However, to the extend that CRISPR RNA processing is the rate-limiting step, cells harboring Canonical repeat RNA would have more efficient CRISPR-Cas immunity responses than those with non-Canonical repeat RNA. Though less efficient at the step of repeat RNA processing, cells harboring the non-Canonical repeats could compensate the loss by optimizing other functions such as target degradation or promoting non-CRISPR-Cas-mediated immune responses. Understanding the implications of different types of Cas6-RNA interactions requires both in vitro as well as cellular observations.
Acknowledgments
This work was supported by NIH grant R01 GM099604 to H.L.
We thank the Undergraduate Research Opportunity Program at Florida State University (M.R.) and all Li Lab members for helpful discussions and training provided for M.R.
Footnotes
Authors' contributions: H.L. designed the experiments. J.S and M.R. performed RNA structure analysis.J.S., M.R., G.Y., and H.L. analyzed the results. H.L. wrote the manuscript. All authors edited the manuscript.
The authors have declared no conflict of interest.
References
- 1.Shen LX, Cai Z, Tinoco I., Jr RNA structure at high resolution. Faseb J. 1995;9:1023–33. doi: 10.1096/fasebj.9.11.7544309. [DOI] [PubMed] [Google Scholar]
- 2.Ivica NA, Obermayer B, Campbell GW, Rajamani S, et al. The paradox of dual roles in the RNA world: resolving the conflict between stable folding and templating ability. J Mol Evol. 2013;77:55–63. doi: 10.1007/s00239-013-9584-x. [DOI] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–86. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 6.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–7. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H. Structural principles of CRISPR RNA processing. Structure. 2015;23:13–20. doi: 10.1016/j.str.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Makarova KS, AravindL, WolfYI, KooninEV Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R, Li H. The mysterious RAMP proteins and their roles in small RNA-based immunity. Protein Sci. 2012;21:463–70. doi: 10.1002/pro.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao Y, Richter H, Sun S, Sharma K, et al. A non-stem-loop CRISPR RNA is processed by dual binding Cas6. Structure. 2016;24:547–54. doi: 10.1016/j.str.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange SJ, Alkhnbashi OS, Rose D, Will S, et al. CRISPRmap: an automated classification of repeat conservation in pro-karyotic adaptive immune systems. Nucleic Acids Res. 2013;41:8034–44. doi: 10.1093/nar/gkt606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornton JM, Orengo CA, Todd AE, Pearl FM. Protein folds, functions and evolution. J Mol Biol. 1999;293:333–42. doi: 10.1006/jmbi.1999.3054. [DOI] [PubMed] [Google Scholar]
- 17.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–26. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen E, Joseph S. Fragile X mental retardation protein: a paradigm for translational control by RNA-binding proteins. Biochimie. 2015;114:147–54. doi: 10.1016/j.biochi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musco G, Kharrat A, Stier G, Fraternali F, et al. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat Struct Biol. 1997;4:712–6. doi: 10.1038/nsb0997-712. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Schwartz JC, Cech TR. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015;43:7535–43. doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haurwitz RE, Sternberg SH, Doudna JA. Csy4 reliesonan unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–32. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–53. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ShaoY, Li H. Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6. Structure. 2013;21:385–93. doi: 10.1016/j.str.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R, Preamplume G, Terns MP, Terns RM, et al. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–64. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carte J, Pfister NT, Compton MM, Terns RM, et al. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–8. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokolowski RD, Graham S, White MF. Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system. Nucleic Acids Res. 2014;42:6532–41. doi: 10.1093/nar/gku308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeks J, Sokolowski RD, Graham S, Liu H, et al. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem J. 2013;452:223–30. doi: 10.1042/BJ20130269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9:907–18. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc Chem Res. 2008;41:1027–35. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]


