Abstract
Objective
Lysine acetylation is an important post-translational modification that regulates metabolic function in skeletal muscle. The acetyltransferase, general control of amino acid synthesis 5 (GCN5), has been proposed as a regulator of mitochondrial biogenesis via its inhibitory action on peroxisome proliferator activated receptor-γ coactivator-1α (PGC-1α). However, the specific contribution of GCN5 to skeletal muscle metabolism and mitochondrial adaptations to endurance exercise in vivo remain to be defined. We aimed to determine whether loss of GCN5 in skeletal muscle enhances mitochondrial density and function, and the adaptive response to endurance exercise training.
Methods
We used Cre-LoxP methodology to generate mice with muscle-specific knockout of GCN5 (mKO) and floxed, wildtype (WT) littermates. We measured whole-body energy expenditure, as well as markers of mitochondrial density, biogenesis, and function in skeletal muscle from sedentary mice, and mice that performed 20 days of voluntary endurance exercise training.
Results
Despite successful knockdown of GCN5 activity in skeletal muscle of mKO mice, whole-body energy expenditure as well as skeletal muscle mitochondrial abundance and maximal respiratory capacity were comparable between mKO and WT mice. Further, there were no genotype differences in endurance exercise-mediated mitochondrial biogenesis or increases in PGC-1α protein content.
Conclusion
These results demonstrate that loss of GCN5 in vivo does not promote metabolic remodeling in mouse skeletal muscle.
Keywords: Acetylation, GCN5, Mitochondria, SIRT1, Deacetylase, PGC-1α
Abbreviations: AMP, adenosine monophosphate; Ca2+, calcium; CBP, CREB-binding protein; Cre-MCK, creatine kinase promoter; CHO, carbohydrate; DAC, deacetylase; ETC, electron transport chain; ExT, endurance exercise training; FA, fatty acid; GA, gastrocnemius; GCN5, general control of amino acid synthesis 5; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HKII, hexokinase 2; KAT, acetyltransferase; mKO, muscle knockout; LCAD, long chain acyl CoA dehydrogenase; MCAD, medium-chain acyl-CoA dehydrogenase; Mef2, myocyte enhance factor 2; Myf6, myogenic factor 6; MyoG, myogenin; NAD+, nicotinamide-adenine dinucleotide; PARPs, poly (ADP-ribose) polymerases; PCAF, p300/CBP-associated factor; PDH, pyruvate dehydrogenase; PGC-1α, peroxisome proliferator activated receptor-γ coactivator-1α; Pln, plantaris; Q, quadriceps; RQ, respiratory quotient; SDH, succinate dehyrogenase; SIRT, sirtuin; TA, tibialis anterior; TCA, tricarboxylic acid; Tfam, mitochondrial-specific transcription factor A; TRI, triceps; WT, wildtype; mHZ, muscle heterozygous
Highlights
-
•
Development of a novel muscle-specific GCN5 knockout (mKO) mouse model.
-
•
GCN5 mKO does not affect body composition or 24 h whole-body metabolism.
-
•
GCN5 mKO mice do not exhibit changes in basal mitochondrial abundance or respiratory capacity.
-
•
Exercise-induced mitochondrial biogenesis in skeletal muscle is not enhanced in GCN5 mKO mice.
1. Introduction
For over 50 years endurance exercise has been known to induce mitochondrial adaptations in skeletal muscle [1], [2], [3], [4]. While the mechanisms by which contraction initiates mitochondrial biogenesis remain to be fully elucidated, it is clear that perturbation in allosteric factors such as calcium (Ca2+)1, adenosine monophosphate (AMP), nicotinamide-adenine dinucleotide (NAD+), and acetyl-CoA are important initiators of the adaptive response [5], [6], [7], [8]. Accordingly, considerable research has focused on the transduction pathways that are modulated by these metabolic intermediates and their contribution to exercise-induced mitochondrial adaptations in skeletal muscle. Reversible lysine acetylation has emerged as an important posttranslational modification that links cellular flux to the adaptive responses [9], [10]. An underlying reason for this is that NAD+ is the primary substrate for the sirtuin class of deacetylases (DACs), which remove acetyl groups from lysine residues [11], whilst acetyl-CoA is the substrate for acetyltransferases (KATs), which add acetyl groups to lysine residues [12]. However, despite the high prevalence of lysine acetylation in skeletal muscle [13], [14] much remains unknown regarding its contribution to the remodeling of skeletal muscle, both at rest and also in response to endurance exercise [15].
Peroxisome proliferator activated receptor-γ coactivator-1α (PGC-1α) is an important contributor to mitochondrial biogenesis and function in skeletal muscle [2], [5], [6], with its transcriptional activity being regulated, at least in part, by reversible acetylation [16], [17], [18]. Deacetylation by sirtuin 1 (SIRT1) is a potent activator of PGC-1α [16], [17], [19], [20], while general control of amino acid synthesis 5 (GCN5) acetylates and inhibits its transcriptional activity [8], [17], [18], [21]. GCN5 and PGC-1α form a complex in PGC-1α immunoprecipitates from Fao hepatocytes, while GCN5 overexpression in HEK293 cells represses PGC-1α intrinsic transcriptional activity [18]. In relation to skeletal muscle, overexpression of GCN5 in C2C12 myotubes represses PGC-1α-mediated induction of mitochondrial and fatty acid metabolism genes [17]. Moreover, we previously reported that deacetylation of PGC-1α following acute endurance exercise occurs in conjunction with a reduction in both the nuclear presence of GCN5 and its association with PGC-1α [8]. This relationship was maintained in both the presence and absence of SIRT1 deacetylase activity, suggesting that exercise-induced deacetylation of PGC-1α occurs as a result of reduced GCN5-PGC-1α interaction, rather than solely through SIRT1-dependent deacetylation of PGC-1α [8].
Taken together, these data implicate GCN5 as an important negative regulator of PGC-1α transcriptional activity in skeletal muscle and, by extension, mitochondrial biogenesis [8], [17], [18], [21]. However, no studies to date have directly investigated the contribution of GCN5 to skeletal muscle metabolism and mitochondrial function in vivo. Accordingly, we used CreLoxP methodology to generate mice with muscle-specific knockout of GCN5 (mKO). We hypothesized that mKO mice would display increased mitochondrial biogenesis in skeletal muscle as compared to their floxed/wildtype (WT) littermates and that mitochondrial adaptations to endurance exercise training (ExT) would be enhanced in mKO mice.
2. Materials and methods
2.1. Generation of mKO mouse
To generate mKO mice, mice harboring LoxP sites flanking exons 3–19 of the GCN5 gene (Figure 1A) [22] (referred to as GCN5flox/flox, and kindly provided by Dr. Sharon Dent, The University of Texas MD Anderson Cancer Center, Smithville TX, USA) were crossed with mice expressing Cre recombinase under the control of the muscle creatine kinase promoter (Cre-MCK); after Cre-mediated recombination, exons 3–19 are removed [22]. Our breeding strategy was to breed GCN5flox/flox mice with GCN5flox/+ (‘+’ refers to a WT allele) mice, with one breeder being Cre-MCK positive, and the other Cre-MCK negative. This provided littermates that were Cre-MCK positive or negative and GCN5flox/flox or GCN5flox/+; mice that were GCN5flox/+ and Cre-MCK positive are heterozygous for loss of GCN5, and are referred to as mHZ. Mice negative for Cre-MCK are referred to as wildtype (WT) and were used as controls for all experiments. Mice were housed on a 12:12 h light–dark cycle, and all experiments were conducted in 13-wk-old littermates. All experiments were approved by and conducted in accordance with the Animal Care Program at the University of California, San Diego.
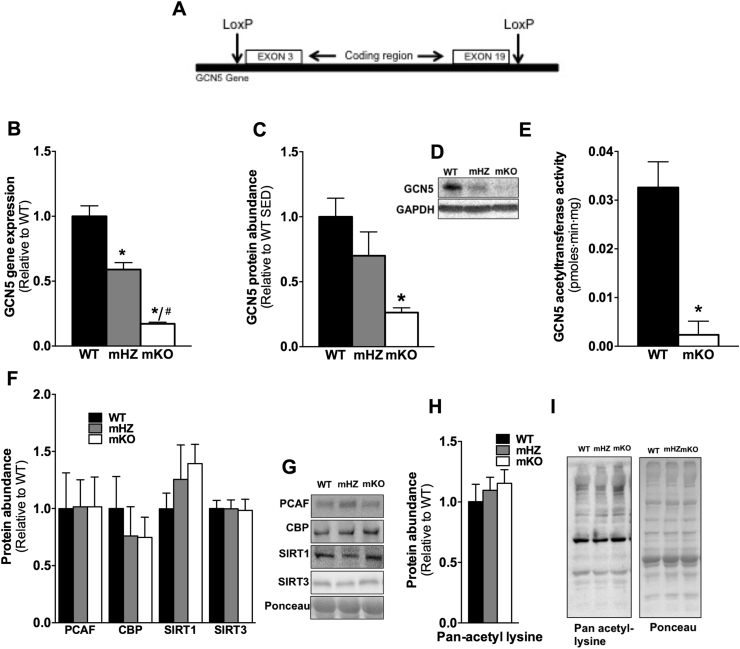
Figure 1.
Efficient gene deletion in skeletal muscle of GCN5 (mKO). A) Schematic of LoxP sites flanking exons 3–19 of the GCN5 gene. B) GCN5 gene expression is diminished in mHZ and mKO compared to WT mice. C) Quantitation of GCN5 protein abundance in skeletal muscle of WT, mHZ, and mKO mice. D) Representative image of GCN5 protein abundance from whole-cell muscle lysates in WT, mHZ and mKO mice. D) GCN5 acetyltransferase activity is reduced in mKO compared to WT mice. E) Quantitation and F) representative images of protein abundance of PCAF, CBP, SIRT1, and SIRT3 in skeletal muscle from WT, mHZ and mKO mice. G) Quantitation and H) representative images of whole-cell lysine acetylation in skeletal muscle of WT, mHZ, and mKO mice. All values were normalized to the actin band on the ponceau except for total lysine-acetylation, which was normalized to whole ponceau, and all values are presented relative to WT. Data represent n = 3–6/genotype. Data presented as mean ± SEM. *Significantly different to WT; p < 0.05, #Significantly different to mHZ; p < 0.05.
2.2. Tissue collection
Tissues were excised from fasted (4 h) and anesthetized mice. Skeletal muscles (gastrocnemius [GA], quadriceps [Q], triceps [TRI], tibialis anterior [TA], plantaris [Pln]), heart, liver, and epididymal adipose tissue (AT) were rinsed in sterile saline, blotted dry, weighed, and frozen in liquid nitrogen. The TA that was used for sectioning was pinned on cork and frozen in liquid nitrogen-cooled isopentane. All tissues were stored at −80 °C for subsequent analysis.
2.3. RNA extraction and cDNA synthesis for the quantitation of GCN5 gene expression
RNA was extracted from the Q muscle of WT, mHZ and mKO mice using the standard TRIzol method, and cDNA synthesized, as previously described [8]. The expression of GCN5 in each sample was normalized to values of the reference control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the comparative (2−ΔΔCT) method and expressed relative to WT values. Primer sequences were: GCN5 forward, 5′-CAG GTC AAG GGC TAT GGC AC-3′ and reverse, 5′-GAT AGC GGC TCT TGG GCA C-3′, GAPDH forward 5′-TGGAAAGCTGTGGCGTGAT-3′ and reverse, 5′-TGCTTCACCACCTTCTTGAT-3'.
2.4. Skeletal muscle nuclear isolation
Isolated nuclear fractions were prepared from GA muscle using a commercially available kit (78835: NE-PER; Thermo Fisher Scientific, Waltham, MA, USA) with the addition of 4.8% cOmplete Mini protease inhibitor mixture (Sigma–Aldrich, St. Louis, MO, USA), 1 μM trichostatin A, 10 mM nicotinamide, 1 mM 1,4-Dithiothreitol, 1% Phosphatase Inhibitor Cocktail 2 (Sigma–Aldrich), 1% Phosphatase Inhibitor Cocktail 3 (Sigma–Aldrich).
2.5. GCN5 acetylatransferase activity
GCN5 specific acetyltransferase activity was determined in nuclear fractions from the GA using an immunoprecipitation HAT assay kit (17–284, Merck Millipore, Billerica, MA, USA). For this, the primary antibody in the kit was replaced with a GCN5 primary antibody (607201; Biolegend, San Diego, CA, USA) and the peptide substrate was replaced with Histone HS Peptide (12–403; Merck Millipore).
2.6. Targeted array
Total RNA was isolated from the TRI muscle using a combination of TRIzol (Thermo Fisher Scientific) and Promega ReliaPrep RNA Tissue Miniprep System (Promega, Madison, WI, USA) according to manufacturer instructions. Isolated RNA was reverse transcribed using the RT2 First Strand Kit (SABiosciences-Qiagen, Frederick, MD, USA). A 384-well custom-designed PCR array containing pre-optimized and pre-validated primers was developed in collaboration with SABiosciences. Genes representing the functional groups: electron transport chain (ETC), metabolism, mitochondrial remodeling, mitochondrial protein transport, transcription, mitochondrial transcription, DACs, KATs, angiogenesis, poly ADP ribose polymerases (PARPs), and myogenesis. The arrays also contained a replicate positive PCR control, reaction lacking reverse transcriptase, a mouse DNA positive control and a panel of housekeeping genes including GAPDH, Actb, Hsp90ab1, B2m, Tbp. PCR was performed on the Bio Rad CFX384 thermo cycler following the manufacturer's protocol. Expression of each gene was normalized to the respective average value of the panel of housekeeping genes using the comparative (2−ΔΔCT) method and expressed relative to WT.
2.7. Energy expenditure and body composition
Body composition was measured by magnetic resonance imaging (MRI, EchoMRI, Houston, TX, USA), and energy expenditure and voluntary cage activity were assessed using the Comprehensive Lab Animals Monitoring System (CLAMS, Columbus Instruments, Columbus, OH, USA), as previously described [23]. For CLAMS, measurements were made for 3 consecutive days, and the values presented are the average diurnal values from the 12 h light and dark phases recorded on day 2 and 3.
2.8. Myosin heavy chain composition
Myosin heavy chain (MHC) composition was measured in GA, Pln, and Q muscle as previously described [24], [25].
2.9. High-resolution respirometry
High-resolution respirometry was performed using an Oroboros O2K (Oroboros Instruments, Innsbruck, Austria) as previously described [23], with minor adjustments. Specifically, the following substrate-uncoupler-inhibitor titration (SUIT) protocol was used: 0.4 mM malate, 0.2 mM octanoylcarnitine, 5 mM ADP, 10 μM cytochrome c, 5 mM pyruvate, 10 mM glutamate, 10 mM succinate, followed by 0.5 μM titrations carbonyl cyanide m-chloro phenyl hydrazone, 0.5 μM rotenone, and 2.5 μM antimycin A. Any samples showing a greater than 10% increase in respiratory flux after addition of cytochrome c were excluded from the study. Respiratory flux was normalized to muscle fiber weight. All reagents were obtained from Sigma–Aldrich.
2.10. Succinate dehydrogenase activity
Succinate dehydrogenase (SDH) activity was measured by incubation of muscle sections in medium consisting of: 1.5 mM nitro blue tetrazolium, 5 mM ethylenediaminetetraacetic acid, 48 mM succinic acid, 0.75 mM sodium azide, 30 mM methyl-phenylmethlyl sulfate, phosphate buffered to pH 7.6, for 10 min. SDH intensity was quantified in whole muscle sections of the TA with an average of 604 fibers per cross-section. Images were acquired on a Brightfield microscope (Leica DM6000, Leica Microsystems, Buffalo Grove, IL, USA) at a magnification of 10× using a (Leica DFC295 camera, Leica Microsystems) camera. Muscle sections were analyzed using image J software (ImageJ, National Institutes of Health, Bethesda, MD, USA) and mean grey values were subtracted from 255.
2.11. Endurance exercise training (ExT)
10-week old mice had free access to a running wheel for 20 days, as previously described [8], [23]. After 20 d, the running wheel was removed, and, 24 h following its removal, tissues were excised from fasted mice (4 h), as described above. Time spent running, distance run, and average speed were noted daily throughout ExT at 1300 h.
2.12. Immunoblotting
Equivalent amounts of protein (30 ug) were separated on 7.5–12.5% gels by SDS-page as previously described [26], proteins transferred to BioTrace NT nitro-cellulose membranes (Pall Life Sciences, Pensacola, FL, USA), blocked in 3% milk/TBST and incubated overnight in primary antibodies and subsequently incubated for 1 h at room temperature in relevant secondary antibodies. Chemiluminescence horseradish peroxidase reagent kit (Merck-Millipore) was used to quantify protein content following IgG binding. Images were captured with a G: Box Chemi-XR5 (Syngene (A Division of Synoptics Ltd.), Cambridge, UK) imaging system while blot bands were quantified using GeneTools software (Syngene).
2.13. Antibodies
All primary antibodies were used at a concentration of 1:1000 dilution unless otherwise stated. Antibodies for SIRT1 (3931), CREB-binding protein (CBP, 7389; 1:500), GAPDH (2118) hexokinase 2 (HKII, 2867), pyruvate dehydrogenase (PDH, 2784), SIRT3 (5490) p300/CBP-associated factor (PCAF, 3378), and GCN5 (3305; 1:500) were from Cell Signaling Technologies; mitochondrial-specific transcription factor A (Tfam, SAB1401383), complex I–V (OXPHOS, Ab MS664/62830) and acetyl-lysine (ab193) were from Abcam (Cambridge, UK); cytochrome-c (cyt-c, BD556433) was from BD Biosciences (Oxford, UK), long chain acyl CoA dehydrogenase (LCAD, 1:5000), and medium-chain acyl-CoA dehydrogenase (MCAD, 1:5000) were kind gifts from Prof Jerry Vockley, University of Pittsburgh, USA. PGC-1α (AB3242) was from Merck-Millipore; myogenic factor 6 (Myf6, sc-301, 1:750) and myocyte enhance factor 2 (pan-Mef2, sc-313, 1:10000) were from Santa Cruz Biotechnology, Dallas, TX, USA. Myogenic factor 4 (myogenin) F5D was deposited to the Developmental Studies Hybridoma Bank by Woodring E. Wright, University of Southwestern Medical Center, Dallas, TX, USA. Secondary antibodies were used at a concentration of 1:10000 in TBST. Anti-rabbit (7074) and anti-mouse (7076) antibodies were from Cell Signaling Technologies.
2.14. Statistical analysis
Statistical analyses were performed using Prism 6 (GraphPad Software Incorporated, La Jolla, CA, USA). Unpaired t-test, one-way analysis of variance (ANOVA) or two-way ANOVA, where appropriate, with Holm-Bonferroni correction for multiple comparisons was used to determine differences between WT, heterozygous (mHZ) and mKO mice. Values are presented as mean ± SEM and are expressed relative to the WT group unless otherwise stated. Statistical significance was set at p < 0.05.
3. Results
3.1. Generation and validation of the GCN5 mKO mouse model
Skeletal muscle gene expression of GCN5 was ∼40 and ∼85% lower in mHZ and mKO mice, respectively, as compared to WT mice (Figure 1B). Following this, GCN5 protein abundance was ∼30% and 75% lower in mHZ and mKO, respectively, vs. WT mice (Figure 1C). In line with the gene and protein expression data, GCN5 acetyltransferase activity was absent in mKO vs. WT mice (Figure 1E). Notably, there was no compensatory increase in PCAF (also known as KAT2B), which is highly homologous to GCN5 [27], [28], [29], [30], at the protein (Figure 1F, G) or gene level (Figure 2G). Further, there was no up regulation of other KAT family members CBP or E1a-binding protein (p300), down-regulation of sirtuin 1 gene expression (Figure 2H), or alterations in CBP, SIRT1 or SIRT3 (Figure 1F, G) protein abundance, or alterations in whole cell lysine acetylation in mHZ or mKO compared with WT controls (Figure 1H, I).
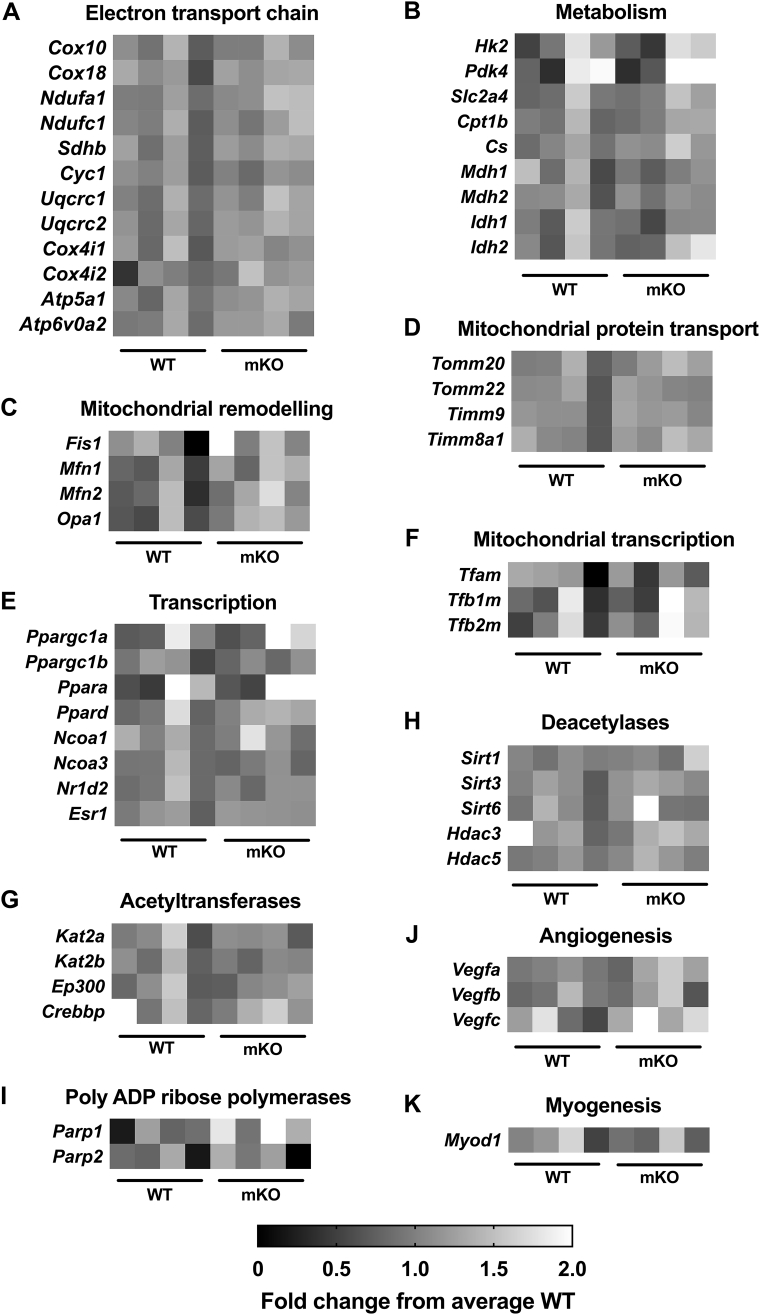
Figure 2.
Targeted array reveals minimal differences in skeletal muscle gene expression between WT and GCN5 mKO. A–K) Basal gene expression in WT and mKO TRI muscle. Heat maps represent individual fold-change in gene expression from average WT in WT and mKO mice. Data are organized into functional groups: A) electron transport chain, B) metabolism, C) mitochondrial remodeling, D) mitochondrial protein transport, E) transcription, F) mitochondrial transcription, G) acetyltransferases, H) deacetylases, I) poly ADP ribose polymerases, J) angiogenesis, K) myogenesis. Data represent individual fold change from average WT, n = 4/genotype.
3.2. Skeletal muscle gene expression of metabolic, angiogenic, and mitochondrial genes is not affected by loss of GCN5
Targeted gene arrays for a wide variety of targets from the functional groups: ETC, metabolism, mitochondrial remodeling, mitochondrial protein transport, transcription, mitochondrial transcription, DACs, KATs, angiogenesis, PARPs, and myogenesis revealed no differences between mKO and WT mice (p > 0.05; Figure 2A–K).
3.3. Body mass, composition, tissue weights, and energy expenditure are comparable between WT, mHZ, and mKO mice
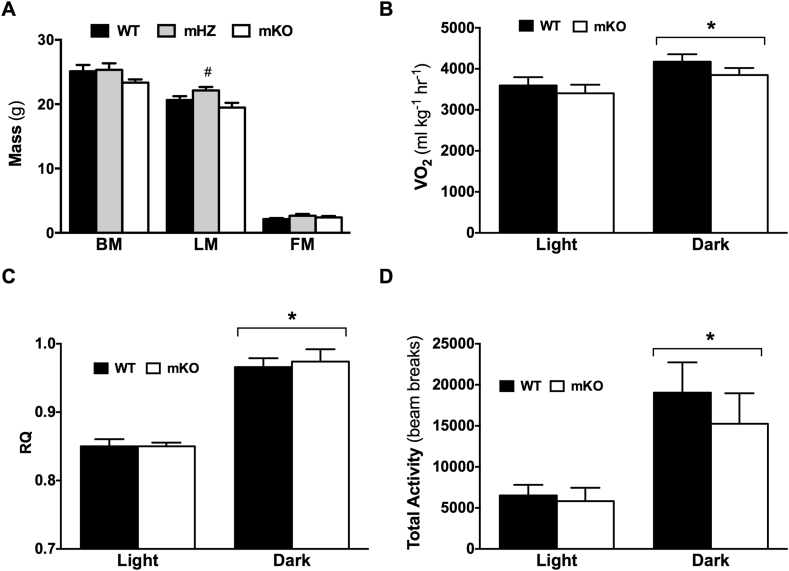
There were no differences in body mass or body fat between WT, mHZ, and mKO mice (Figure 3A). Lean mass was higher in mHZ compared to WT and mKO mice (p < 0.05). Heart, liver, and epididymal fat pad weights did not differ between genotypes in the basal state (Table 1), nor did whole-body diurnal rhythms in VO2, respiratory quotient (RQ), or total activity (i.e., all x axis beam breaks) differ between WT and mKO (Figure 3B–D).
Figure 3.
Loss of GCN5 does not alter body composition, in vivo metabolism or energy expenditure. A) Body mass (BM), lean mass (LM), and fat mass (FM) determined by MRI for WT, mHZ, and mKO mice. B-D) In vivo measurements were made using the Comprehensive Lab Animals Monitoring System over 3 consecutive days. Data represent averages for the light and dark phases of day 2 and 3 for WT and mKO mice. B) VO2 and C) respiratory quotient (RQ) were measured by indirect calorimetry, while D) total activity was measured as all x-axis beam breaks. Data represent n = 5–12/genotype. Data presented as mean ± SEM. *Significantly different to light phase; p < 0.05, #Significantly different to WT and mKO; p < 0.05.
Table 1.
Body and tissue weights in sedentary and ExT mice.
| SED |
ExT |
|||||
|---|---|---|---|---|---|---|
| WT | mHZ | mKO | WT | mHZ | mKO | |
| BW (g) | 25.1 ± 1.0 | 25.3 ± 1.0 | 23.3 ± 0.5 | 21.3 ± 1.7a | 24.0 ± 0.6a | 22.4 ± 0.6a |
| TA (mg) | 42.4 ± 1.9 | 40.6 ± 1.5 | 39.1 ± 1.0 | 39.1 ± 1.0 | 39.6 ± 1.6 | 40.8 ± 1.0 |
| % BW | 1.69 ± 0.04 | 1.60 ± 0.05 | 1.68 ± 0.04 | 2.77 ± 1.05 | 1.68 ± 0.08 | 1.82 ± 0.03 |
| GA (mg) | 110.6 ± 5.8 | 108.4 ± 5.1 | 94.8 ± 3.1 | 99.0 ± 2.7 | 106.1 ± 4.3 | 95.7 ± 3.7 |
| % BW | 4.39 ± 0.13 | 4.27 ± 0.11 | 4.07 ± 0.13 | 6.98 ± 2.65 | 4.45 ± 0.22 | 4.26 ± .10 |
| Q (mg) | 168.2 ± 9.5 | 164.8 ± 8.4 | 156.0 ± 3.6 | 148.6 ± 2.7 | 157.1 ± 8.8 | 159.4 ± 4.7 |
| % BW | 6.67 ± 0.23 | 6.48 ± 0.17 | 6.70 ± 0.16 | 11.30 ± 4.90 | 6.55 ± 0.34 | 7.1 ± 0.19 |
| Heart (mg) | 117.7 ± 5.8 | 112.1 ± 5.6 | 100.6 ± 3.1 | 125.3 ± 2.6 | 123.8 ± 3.2 | 118.5 ± 4.1 |
| % BW | 4.70 ± 0.19 | 4.39 ± 0.14 | 4.32 ± 0.13 | 9.23 ± 3.78 | 5.17 ± 0.14 | 5.29 ± 0.14 |
| Liver (mg) | 1166.6 ± 39.4 | 1112.4 ± 46.5 | 1036.6 ± 29.2 | 1212.4 ± 37.7 | 1320.8 ± 57.6 | 1228.3 ± 62.9 |
| % BW | 46.55 ± 0.70 | 43.84 ± 0.88 | 44.26 ± 1.09 | 48.54 ± 4.63a | 55.02 ± 1.81a | 52.98 ± 1.50a |
| Epi fat (mg) | 293.3 ± 22.1 | 348.9 ± 35.7 | 334.1 ± 35.8 | 158.2 ± 10.6 | 184.8 ± 17.7 | 164.8 ± 24.2 |
| % BW | 11.59 ± 0.63 | 13.54 ± 1.09 | 14.25 ± 1.34 | 6.82 ± 0.40a | 7.68 ± 0.64a | 7.03 ± 0.87a |
Main effect for effect for exercise training (ExT); p < 0.05. SED, sedentary; ExT, endurance exercise training; BW, body weight; TA, tibialis anterior; GA, gastrocnemius; Q, quadriceps, Epi fat; epididymal fat.
3.4. Loss of GCN5 does not affect myosin heavy chain (MHC) composition
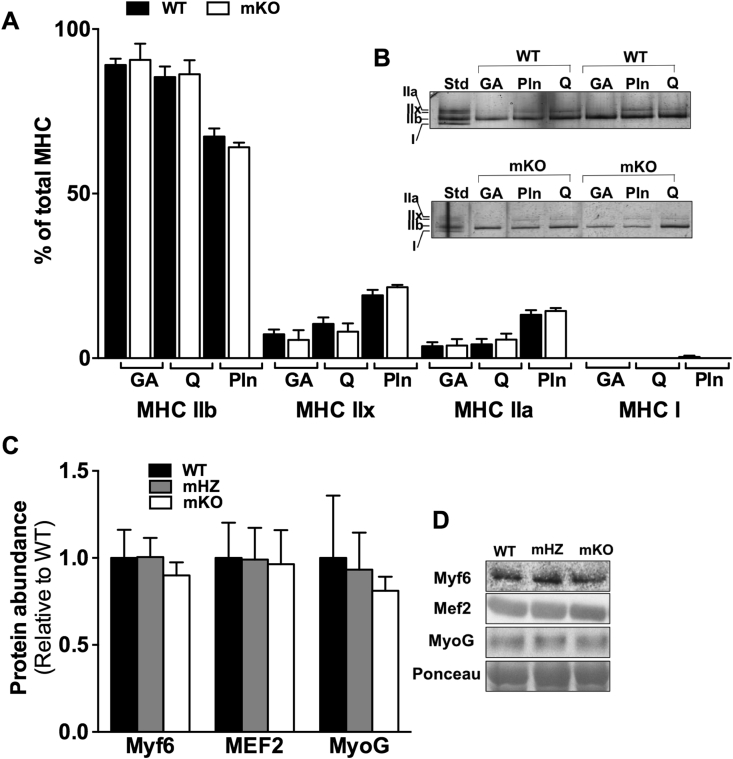
Type I, IIa, IIx, and IIb MHC composition in the GA, Pln, and Q was comparable between WT and mKO (Figure 4A, B).
Figure 4.
Loss of GCN5 does not affect skeletal muscle MHC isoform composition. A, B) Skeletal muscle Type IIb, IIx, IIa, and I MHC isoform composition were not different between mKO versus WT mice. A) Quantitation of MHC isoform in the gastrocnemius (GA), quadriceps (Q), and plantaris (Pln) of WT and mKO mice. B) Representative silver stain image for MHC expression in skeletal muscle of WT and mKO mice. C) Quantitation of muscle regulatory factors, Myf6, Mef2, and myogenin, protein abundance in GA whole-cell muscle lysates. D) Representative images of protein abundance of muscle regulatory factors. Values are normalized to the actin band on the ponceau and presented relative to WT. Data represent n = 5–8/genotype. Data presented as mean ± SEM.
3.5. Markers of skeletal muscle development are unaffected by loss of GCN5
Myf6, Mef2, and myogenin protein abundance did not differ between mKO, mHZ, and WT mice (Figure 4C, D). Further, MyoD gene expression was not different between mKO and WT mice (Figure 2K).
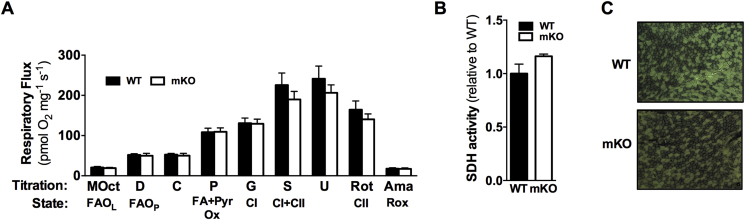
3.6. Skeletal muscle maximal respiratory capacity and succinate dehydrogenase (SDH) enzyme activity are not affected by loss of GCN5
As determined in permeabilized TA fiber bundles using high-resolution respirometry, there were no differences in maximal respiratory capacity, ETC capacity, leak respiration (in the absence of adenylates), residual oxygen consumption, and oxidative phosphorylation-coupling efficiency between WT and mKO mice (Figure 5A). Further, there were no differences in SDH activity in the TA of WT vs. mKO mice (Figure 5B, C).
Figure 5.
High-resolution respirometry and SDH activity in WT and mKO TA muscle. A) Respiratory flux normalized to muscle fiber weight in the presence of MOct (leak respiration in the absence of adenylates), ADP (D), cytochrome c (C; mitochondrial integrity), pyruvate (P), glutamate (G; complex I (CI) capacity), succinate (S; complex I + complex II (CII) capacity), carbonyl cyanide m-chloro phenyl hydrazone (U; maximal respiration), rotenone (Rot; complex II capacity), and Ama (residual oxygen consumption (Rox)). B) Quantitation (data presented relative to WT) and C) representative images of SDH activity in WT and mKO TA muscle. Data represent n = 5–7/genotype. Data presented as mean ± SEM.
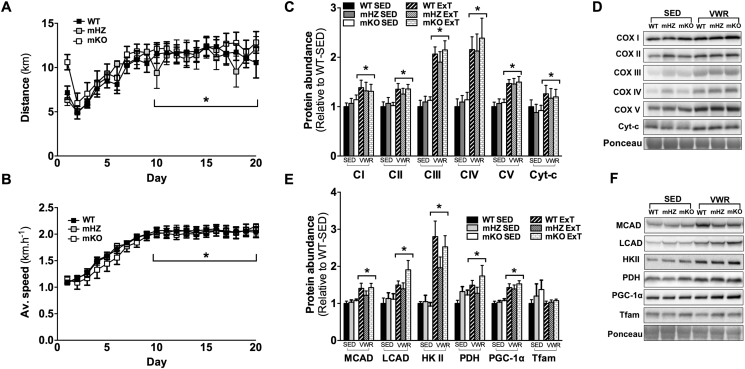
3.7. Loss of GCN5 does not affect mitochondrial content or adaptations to ExT
Average 24 h running distance and running speed were comparable between genotypes (Figure 6A, B); as expected, daily distance and average speed increased over the first 10 d of training (p < 0.05). Although skeletal muscle weights were similar between sedentary (SED) and ExT groups, epididymal fat weight was lower and liver weight higher (p < 0.05) following ExT, with no genotype differences observed (Table 1). At the molecular level, ExT increased the skeletal muscle abundance of mitochondrial markers (∼1.3–2.5 fold), including complexes I-V, cyt-c, MCAD, LCAD, HKII, PDH, PGC-1α and SIRT3, as compared to SED (p < 0.05; Figure 6C–F); there were no genotype differences within SED or ExT groups (p > 0.05).
Figure 6.
Loss of GCN5 in skeletal muscle does not inhibit the ability for exercise training or exercise-induced mitochondrial adaptation A) Average 24 h running distance and B) average 24 h running speed during 20 days ExT in WT, mHZ, and mKO. C, D) (C) Quantitation (D) and representative images of protein abundance of electron transport chain complexes (complex I (CI), complex II (CII), complex III (CIII), complex IV (CIV), and complex V (CV)) and cytochrome-c (cyt-c) in TRI muscle from SED and ExT WT, mHZ, and mKO mice. E, F) (E) Quantitation (F) and representative images of protein abundance of MCAD, LCAD, HKII, PDH, PGC-1α, and TFAM in TRI muscle from SED and ExT WT and mKO mice. All values were normalized to the actin band on the ponceau and all values are presented relative to WT-SED. (Data represent n = 5–9/genotype). Data presented as mean ± SEM. *Significant effect for exercise; p < 0.05.
4. Discussion
The role of reversible lysine acetylation in skeletal muscle metabolism has received considerable interest in recent years [7], [8], [13], [17], [31], [32]. To date, the contribution of DACs, particularly SIRT1 and SIRT3, to the positive metabolic adaptations in skeletal muscle to exercise training have been heavily studied [6], [7], [8], [31]. In contrast, very little is known about the contribution of KATs to skeletal muscle metabolism and remodeling in vivo. To address this, we developed a conditional mouse model in which GCN5 activity is deleted in muscle. The primary reason for focusing on GCN5 is that previous in vitro studies have demonstrated its key role in acetylating and inhibiting PGC-1α, thereby opposing the actions of SIRT1 [17], [18], [21]. Our results reveal that whole-body energy expenditure, skeletal muscle morphology, mitochondrial protein abundance, and maximal respiratory capacity are comparable between sedentary mKO, mHZ, and WT mice, as is the induction of skeletal muscle mitochondrial biogenesis in response to endurance exercise training.
Reversible acetylation is a major mechanism by which the transcriptional activity of PGC-1α is regulated [16], [20], [21], [33], [34]. In elegant cell-based studies, a role for SIRT1 in modulating the transcriptional capacity of PGC-1α via its deacetylation has been well documented [6], [17], [20], [34], while its role in vivo remains controversial [6], [8], [16], [31], [35], [36]. In fact, studies in bona fide skeletal muscle provide little support for a direct role of SIRT1 in modulating skeletal muscle mitochondrial biogenesis [8], [31]. In contrast, GCN5 acetylates PGC-1α in vitro and inhibits its transcriptional activity [8], [17], [18], [21], with overexpression of GCN5 in C2C12 myotubes leading to repression of PGC-1α-mediated induction of mitochondrial and fatty acid metabolism genes [17]. Moreover, in Fao hepatocytes [17] and skeletal muscle [8] GCN5 and PGC-1α coimmunoprecipitate, while GCN5 overexpression in HEK293 cells represses PGC-1α intrinsic transcriptional activity [18]. The repressive effects of GCN5-mediated acetylation on PGC-1α appear to be due to altered cellular distribution of PGC-1α, with hyperacetylation segregating PGC-1α from target promoters and towards nuclear foci [18]. With these findings in mind, we expected that mitochondrial function and abundance, whole body energy expenditure, and exercise training-induced mitochondrial adaptations would be enhanced in our mKO mouse. To our surprise, however, neither partial (i.e. mHZ) nor complete (i.e. mKO) loss of GCN5 altered any of these parameters.
A potential explanation for the lack of effect of the loss of GCN5 on skeletal muscle metabolic or mitochondrial remodeling may be due to compensation from other KATs. PCAF shares ∼73% homology with the GCN5 gene [27], [28], [29], [30], whilst GCN5 and p300 have numerous substrates in common [37]. Indeed, GCN5/PCAF whole-body double KO mice (GCN5−/− PCAF−/−) die earlier than GCN5 whole-body KO mice, inferring shared functions between these two proteins [38]. Furthermore, an often overlooked observation in the study of Lerin et al. [18], which identified GCN5 as the specific acetyltransferase of PGC-1α, is the fact that PCAF induced acetylation of PGC-1α, albeit to a lesser extent than GCN5. Further, in vitro, PCAF-mediated acetylation of two lysine residues (K328 and K450) on PGC-1α accelerates its proteasomal degradation and suppresses the transcriptional activity of PGC-1α [39]. In liver of fasted and diabetic mice, PCAF acetylated PGC-1α, while liver-specific knockdown of PCAF increased PGC-1α abundance and activity and led to an increase in hepatic glucose output [39]. Notably, however, whether GCN5 and PCAF acetylate and/or recognize the same lysine residues or whether PCAF interacts with PGC-1α in skeletal muscle is unknown. Embryonic redundancy between GCN5 and p300 also exists, with mice heterozygous for null alleles of both GCN5 and p300 demonstrating reduced viability compared to mice heterozygous for either single mutation [40]. Additionally, while p300 has been shown to induce acetylation of PGC-1α [34], it can also prompt a conformational change that increases (not decreases) PGC-1α transcriptional activity, in vitro [41]. Importantly, however, we recently demonstrated that loss of p300 in skeletal muscle does not impact mitochondrial function or metabolic adaptations to endurance exercise [23]. In the present study, although there was no up-regulation of PCAF protein content or gene expression, or p300 gene expression in mHZ and mKO mice, redundancy or overlapping functions of GCN5 with PCAF and/or p300 could explain the lack of differences we see between mKO/mHZ and WT mice. To this end, clearly more work is required to understand the redundancy between these KAT family members in skeletal muscle in vivo.
GCN5 is fundamental to development, as evidenced by the embryonic lethality seen in whole body KO and whole body GCN5hat/hat (embryos with mutant GCN5 KAT activity) mice [38], [42]. In these mice, severe retardation is evident by 8.5 days post coitum (dpc) and death at 10.5 and 16.5 dpc in the KO and KOhat/hat, respectively [38], [42]. This retardation is characterized, in part, by the failure to form the paraxial mesoderm and somites [38], [42], which go on to form skeletal muscle [43]. As GCN5 whole body KO embryos are not viable beyond ∼10 dpc, a point in development in which early myogenic regulatory genes are beginning to be expressed [44], [45], [46], the contribution of GCN5 to skeletal muscle development is currently unknown. Nonetheless, as embryonic lethality coincides with failure to form somites, there is reason to consider that GCN5 may be important for development of myogenic precursor cells. Also, acetylation of MyoD, one of the earliest markers of myogenic commitment, by PCAF and/or p300 is considered critical for execution of the skeletal muscle program [46], [47], [48], and suggests that lysine acetylation has a role in skeletal muscle development. In our mKO model, cre-mediated KO of GCN5 would occur after induction of MCK at ∼13 dpc [44]. Considering mKO and mHZ mice demonstrate no overt differences in muscle weights, fiber type, or myogenic genes as compared to WT littermates, our data suggest that, in and of itself, GCN5 is not required for muscle development in vivo. Finally, GCN5 was originally identified as a transcriptional regulator due to its ability to acetylate histones and modify chromatin structure [49] Indeed, it is now considered to be the catalytically active KAT of multiple histone acetyltansferase complexes [10]. With this in mind, we expected to see differences in skeletal muscle gene expression between our WT and mKO mice. Somewhat surprisingly, however, we found no influence of the loss of GCN5 on the expression of ∼50 different genes indicative of a wide variety of functions in skeletal muscle. These findings could simply be due to the fact that GCN5 is not important for skeletal muscle development or transcriptional control, or as discussed above, could be due to redundancy and compensation from other KATs.
5. Conclusion
While in vitro data provide a mechanistic link between GCN5 acetyltransferase activity and metabolism, our results suggest that loss of GCN5 in muscle does not enhance in vivo basal or ExT-induced metabolic adaptation. Further, we show that GCN5 is not required for adult skeletal muscle development nor does it alter myosin heavy chain composition, whole cell lysine acetylation or gene expression in skeletal muscle. Given the homology between PCAF and GCN5 [27], [28], [29], [30] and their demonstrated overlapping functions during embryogenesis [38], as well as commonality in substrates between p300 and GCN5 [37], it will be of high interest in future studies to probe the separate and combined effects of GCN5, p300 and/or PCAF on skeletal muscle biology.
Acknowledgements
This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) New Investigator Award (BB/L023547/1) to A.P., National Institutes of Health (NIH) Grants R01 AG043120 and P30 DK063491 (Pilot and Feasibility Award from the UCSD/University of California, Los Angeles Diabetes Research Center) to S.S., a postdoctoral fellowship from the UC San Diego Frontiers of Innovation Scholars Program to S.S., S.A.L., and K.S., an NIH T32 (AR060712) Pre-Doctoral Fellowship and Graduate Student Researcher Support from the UC San Diego Institute of Engineering in Medicine and the Office of Graduate Studies to V.F.M., and a postdoctoral fellowship from the Swiss National Science Foundation to K.S.
Contributor Information
Simon Schenk, Email: sschenk@ucsd.edu.
Andrew Philp, Email: a.philp@bham.ac.uk.
Conflicts of interest
None.
References
- 1.Baar K., Wende A.R., Jones T.E., Marison M., Nolte L.A., Chen M. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. The FASEB Journal. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 2.Geng T., Li P., Okutsu M., Yin X., Kwek J., Zhang M. PGC-1α plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. American Journal of Physiology – Cell Physiology. 2010;298:C572. doi: 10.1152/ajpcell.00481.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibala M.J., Little J.P., van Essen M., Wilkin G.P., Burgomaster K.A., Safdar A. Short-term sprint interval versus traditional endurance training: similar initial adaptations in human skeletal muscle and exercise performance. The Journal of Physiology. 2006;575:901–911. doi: 10.1113/jphysiol.2006.112094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holloszy J.O. Biochemical adaptations in muscle: effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. Journal of Biological Chemistry. 1967;242:2278–2282. [PubMed] [Google Scholar]
- 5.Akimoto T., Pohnert S.C., Li P., Zhang M., Gumbs C., Rosenberg P.B. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. The Journal of Biological Chemistry. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 6.Cantó C., Jiang L.Q., Deshmukh A.S., Mataki C., Coste A., Lagouge M. Interdependence of AMPK and SIRT1 for Metabolic adaptation to fasting and exercise in skeletal Muscle. Cell Metabolism. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurd B.J., Perry C.G.R., Heigenhauser G.J.F., Spriet L.L., Bonen A. High-intensity interval training increases SIRT1 activity in human skeletal muscle. Applied Physiology, Nutrition, and Metabolism. 2010;35:350–357. doi: 10.1139/H10-030. [DOI] [PubMed] [Google Scholar]
- 8.Philp A., Chen A., Lan D., Meyer G.A., Murphy A.N., Knapp A.E. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-γ Coactivator-1α (PGC-1α) deacetylation following endurance exercise. Journal of Biological Chemistry. 2011;286:30561–30570. doi: 10.1074/jbc.M111.261685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (New York, NY) 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 10.Lee K.K., Workman J.L. Histone acetyltransferase complexes: one size doesn't fit all. Nature Reviews Molecular Cell Biology. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 11.Finnin M.S., Donigian J.R., Pavletich N.P. Structure of the histone deacetylase SIRT2. Nature Structural Biology. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 12.Hodawadekar S.C., Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–5540. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]
- 13.Lundby A., Lage K., Weinert B.T., Bekker-Jensen D.B., Secher A., Skovgaard T. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Reports. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryder D.J., Judge S.M., Beharry A.W., Farnsworth C.L., Silva J.C., Judge A.R. Identification of the acetylation and ubiquitin-modified proteome during the progression of skeletal muscle atrophy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philp A., Rowland T., Perez-Schindler J., Schenk S. Understanding the acetylome: translating targeted proteomics into meaningful physiology. American Journal of Physiology – Cell Physiology. 2014;307:C763. doi: 10.1152/ajpcell.00399.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1[alpha] and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 17.Gerhart-Hines Z., Rodgers J.T., Bare O., Lerin C., Kim S.-H., Mostoslavsky R. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. The EMBO Journal. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerin C., Rodgers J.T., Kalume D.E., Kim S.-H., Pandey A., Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1a. Cell Metabolism. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Amat R., Planavila A., Chen S.L., Iglesias R., Giralt M., Villarroya F. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. The Journal of Biological Chemistry. 2009;284:21872–21880. doi: 10.1074/jbc.M109.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canto C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coste A., Louet J.-F., Lagouge M., Lerin C., Antal M.C., Meziane H. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1α. Proceedings of the National Academy of Sciences. 2008;105:17187–17192. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin W., Srajer G., Evrard Y.A., Phan H.M., Furuta Y., Dent S.Y. Developmental potential of Gcn5(-/-) embryonic stem cells in vivo and in vitro. Developmental Dynamics : An Official Publication of the American Association of Anatomists. 2007;236:1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- 23.LaBarge S.A., Migdal C.W., Buckner E.H., Okuno H., Gertsman I., Stocks B. p300 is not required for metabolic adaptation to endurance exercise training. The FASEB Journal. 2016;30:1623–1633. doi: 10.1096/fj.15-281741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talmadge R.J., Roy R.R. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. Journal of Applied Physiology (Bethesda, Md : 1985) 1993;75:2337–2340. doi: 10.1152/jappl.1993.75.5.2337. [DOI] [PubMed] [Google Scholar]
- 25.White A.T., Philp A., Fridolfsson H.N., Schilling J.M., Murphy A.N., Hamilton D.L. High-fat diet-induced impairment of skeletal muscle insulin sensitivity is not prevented by SIRT1 overexpression. American Journal of Physiology Endocrinology and Metabolism. 2014;307:E764–E772. doi: 10.1152/ajpendo.00001.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philp A., Perez-Schindler J., Green C., Hamilton D.L., Baar K. Pyruvate suppresses PGC1α expression and substrate utilization despite increased respiratory chain content in C2C12 myotubes. American Journal of Physiology – Cell Physiology. 2010;299:C240–C250. doi: 10.1152/ajpcell.00438.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrera J.E., Bergel M., Yang X.-J., Nakatani Y., Bustin M. The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. Journal of Biological Chemistry. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- 28.Jin Q., Zhuang L., Lai B., Wang C., Li W., Dolan B. Gcn5 and PCAF negatively regulate interferon-β production through HAT-independent inhibition of TBK1. EMBO Reports. 2014;15:1192. doi: 10.15252/embr.201438990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy Z., Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 30.Xu W., Edmondson D.G., Roth S.Y. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Molecular and Cellular Biology. 1998;18:5659–5669. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurd B.J., Yoshida Y., Lally J., Holloway G.P., Bonen A. The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its overexpression reduces mitochondrial biogenesis. The Journal of Physiology. 2009;587:1817–1828. doi: 10.1113/jphysiol.2008.168096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White A.T., Schenk S. NAD(+)/NADH and skeletal muscle mitochondrial adaptations to exercise. American Journal of Physiology Endocrinology and Metabolism. 2012;303:E308–E321. doi: 10.1152/ajpendo.00054.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominy J.E., Jr., Lee Y., Gerhart-Hines Z., Puigserver P. Nutrient-dependent regulation of PGC-1alpha's acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochimica et Biophysica Acta. 2010;1804:1676–1683. doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} The Journal of Biological Chemistry. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 35.Chabi B., Adhihetty P.J., O'Leary M.F., Menzies K.J., Hood D.A. Relationship between Sirt1 expression and mitochondrial proteins during conditions of chronic muscle use and disuse. Journal of Applied Physiology (Bethesda, Md : 1985) 2009;107:1730–1735. doi: 10.1152/japplphysiol.91451.2008. [DOI] [PubMed] [Google Scholar]
- 36.Philp A., Schenk S. Unraveling the complexities of SIRT1-mediated mitochondrial regulation in skeletal muscle. Exercise and Sport Sciences Reviews. 2013;41:174–181. doi: 10.1097/JES.0b013e3182956803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han Z., Chou C.W., Yang X., Bartlett M.G., Zheng Y.G. Profiling cellular substrates of lysine acetyltransferases GCN5 and p300 with orthogonal labeling and click chemistry. ACS Chemical Biology. 2017;12:1547–1555. doi: 10.1021/acschembio.7b00114. [DOI] [PubMed] [Google Scholar]
- 38.Xu W., Edmondson D.G., Evrard Y.A., Wakamiya M., Behringer R.R., Roth S.Y. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nature Genetics. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 39.Sun C., Wang M., Liu X., Luo L., Li K., Zhang S. PCAF improves glucose homeostasis by suppressing the gluconeogenic activity of PGC-1alpha. Cell Reports. 2014;9:2250–2262. doi: 10.1016/j.celrep.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 40.Phan H.M., Xu A.W., Coco C., Srajer G., Wyszomierski S., Evrard Y.A. GCN5 and p300 share essential functions during early embryogenesis. Developmental Dynamics : An Official Publication of the American Association of Anatomists. 2005;233:1337–1347. doi: 10.1002/dvdy.20445. [DOI] [PubMed] [Google Scholar]
- 41.Puigserver P., Adelmant G., Wu Z., Fan M., Xu J., Malley B. Activation of PPARγ coactivator-1 through transcription factor docking. Science (New York, NY) 1999;286:1368. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 42.Bu P., Evrard Y.A., Lozano G., Dent S.Y.R. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Molecular and Cellular Biology. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S. The formation of skeletal muscle: from somite to limb. Journal of Anatomy. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyons G.E., Muhlebach S., Moser A., Masood R., Paterson B.M., Buckingham M.E. Developmental regulation of creatine kinase gene expression by myogenic factors in embryonic mouse and chick skeletal muscle. Development (Cambridge, England) 1991;113:1017–1029. doi: 10.1242/dev.113.3.1017. [DOI] [PubMed] [Google Scholar]
- 45.Ott M.O., Bober E., Lyons G., Arnold H., Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development (Cambridge, England) 1991;111:1097. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- 46.Roth J.F., Shikama N., Henzen C., Desbaillets I., Lutz W., Marino S. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. The EMBO Journal. 2003;22:5186–5196. doi: 10.1093/emboj/cdg473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puri P.L., Avantaggiati M.L., Balsano C., Sang N., Graessmann A., Giordano A. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. The EMBO Journal. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sartorelli V., Puri P.L., Hamamori Y., Ogryzko V., Chung G., Nakatani Y. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Molecular Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 49.Roth S.Y., Denu J.M., Allis C.D. Histone acetyltransferases. Annual Review of Biochemistry. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]