Abstract
Purpose
To determine global protein expression changes in the lens of the GSH-deficient LEGSKO mouse model of age-related cataract for comparison with recently published gene expression data obtained by RNA-Seq transcriptome analysis.
Methods
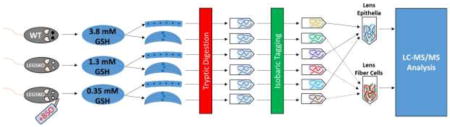
Lenses were separated into epithelial and cortical fiber sections, digested with trypsin, and labeled with isobaric tags (10-plex TMT). Peptides were analyzed by LC-MS/MS (Orbitrap Fusion) and mapped to the mouse proteome for relative protein quantification.
Results
1871 proteins in lens epithelia and 870 proteins in lens fiber cells were quantified. 40 proteins in LEGSKO epithelia, 14 proteins in LEGSKO fiber cells, 22 proteins in buthionine sulfoximine (BSO)-treated LEGSKO epithelia, and 55 proteins in BSO-treated LEGSKO fiber cells had significantly (p<0.05, FDR<0.1) altered protein expression compared to WT controls. HSF4 and MAF transcription factors were the most common upstream regulators of the response to GSH-deficiency. Many detoxification proteins, including aldehyde dehydrogenases, peroxiredoxins, and quinone oxidoreductase, were upregulated but several glutathione S-transferases were downregulated. Several cellular stress response proteins showed regulation changes, including an upregulation of HERPUD1, downregulation of heme oxygenase, and mixed changes in heat shock proteins. NRF2-regulated proteins showed broad upregulation in BSO-treated LEGSKO fiber cells, but not in other groups. Strong trends were seen in downregulation of lens specific proteins, including β- and γ-crystallins, lengsin, and phakinin, and in epithelial-mesenchymal transition (EMT)-related changes. Western blot analysis of LEGSKO lens epithelia confirmed expression changes in several proteins.
Conclusions
This dataset confirms at the proteomic level many findings from the recently determined GSH-deficient lens transcriptome and provides new insight into the roles of GSH in the lens, how the lens adapts to oxidative stress, and how GSH affects EMT in the lens.
Keywords: LEGSKO mouse, cataract, glutathione, oxidative stress, NRF2, epithelial-mesenchymal transition, isobaric tagging, TMT, proteomics
Graphical Abstract
INTRODUCTION
Glutathione (GSH) is an essential antioxidant for defending against oxidative stress in the lens and, therefore, preventing cataract formation [1]. With age, the lens becomes depleted in GSH due to a loss of activity in GSH synthesis enzymes [2] and a growing barrier to GSH diffusion to the lens nucleus [3]. The resulting low GSH levels accelerate cataract formation and play an important role in the pathogenesis of age-related cataract [4–5].
In order to better study the role of GSH in the lens and cataract formation, the Lens Glutathione Synthesis KnockOut (LEGSKO) mouse was developed, in which the first enzyme in the GSH synthetic pathway, the gamma-glutamylcysteine synthetase catalytic subunit (GCLC), was conditionally knocked out in the lens using a lox/Cre system [4]. These knockout mouse lenses lack all GSH synthesis, which results in partially suppressed levels of GSH (1–1.5 mM) in the lens. This residual lens GSH content stems from an adjacent GSH reservoir in the vitreous humor [6]. This GSH reservoir can be depleted by treating mice systemically with the GCLC inhibitor buthionine sulfoximine (BSO), further reducing the knockout lens GSH content [6].
In order to determine the genetic response to GSH deficiency in the lens, an in-depth RNA-Seq analysis was carried out on the lens epithelia and cortical fiber cells of wild-type (WT) (GSH ~3.8 mM), LEGSKO (GSH ~1.3 mM), and BSO-treated LEGSKO mice (GSH ~0.35 mM) [7]. GSH levels were determined by LC-MS/MS analysis of whole lenses [6]. Transcriptomic analysis revealed numerous gene expression changes, including upregulation of detoxification genes such as aldehyde dehydrogenases and metallothioneins, activation of epithelial-mesenchymal transition signaling, a loss of vision gene expression (including crystallins), and changes in lipid homeostasis and transport processes.
In the present study, relative protein expression of WT, LEGSKO, and BSO-treated LEGSKO mouse lens epithelia and cortical fiber cells was performed using a tandem mass tag (TMT) quantitative proteomic approach. This quantitative shotgun approach was used to determine the degree to which the transcriptomic changes are translated to protein expression changes and to determine any additional changes not revealed by RNA-Seq analysis. Here we present data showing that the lens GSH-deficient proteome confirms many results from its transcriptome counterpart, reveals differing oxidative stress responses in epithelia and fiber cells, and further confirms the role of low GSH levels in lens epithelia-mesenchymal transition (EMT) signaling.
METHODS
Animal Work
Lenses from LEGSKO and wild-type (WT) mice, all of C57Bl/6 genetic background and age-matched at 3 months, were used in this study. Groups were as follows: three male WT mice, three male LEGSKO mice, and three male LEGSKO mice exclusively receiving drinking water containing 10 mM buthionine sulfoximine (BSO) for 1 month prior to sample collection. Mice were housed under diurnal lighting conditions and allowed free access to food and water. All animals were used in accordance with the guidelines of the International Guiding Principles for Biomedical Research Involving Animals and experimental protocols for this study were approved by the Institutional Animal Care and Use Committee (IACUC) of Case Western Reserve University.
Tissue Dissection
Mice were sacrificed by CO2 asphyxiation. Eyes were removed and placed in ice-cold 50 mM HEPES, 150 mM NaCl, pH 7.4 buffer. Lenses were removed by cutting away sclera and then separated into epithelial/capsule and cortical fiber sections in ice-cold HEPES/NaCl buffer using forceps. Any visible fiber cells attached to the capsule were carefully removed using forceps. Matching tissue samples from both eyes of each mouse were pooled together in CHELEX-treated, nitrogen-flushed 50 mM triethyl ammonium, pH 7.4 buffer (TEAB) and homogenized using a pellet pestle motor (Kimble-Chase, Rockwood, TN). Samples were centrifuged at 37,000xg for 10 minutes to precipitate insoluble proteins and the supernatant was saved. Pellets were resuspended in TEAB buffer and sonicated for 30 rounds of 5 sec bursts at 40% power. Samples were again centrifuged, supernatant was removed, and the pellet was resuspended in TEAB and sonicated. This preparation method was effective at solubilizing >99.6% of lens protein (Supp. Fig. 1). All three supernatants were pooled together for each sample and a bicinchoninic acid (BCA) protein assay was performed using bovine serum albumin as a standard (ThermoFisher Scientific, Waltham, MA) to normalize sample input. Samples were nitrogen-flushed, sealed tightly with parafilm, and sent overnight on ice to Oregon Health & Science University for further processing.
Protein digestion
20 and 50 μg portions of epithelial or cortical fiber samples, respectively, were trypsin digested. Diluted samples were brought to a volume of 48 μl in 50 mM TEAB buffer, 2 μl of 1% ProteaseMAX™ detergent (PM) (Promega Corporation, Madison, WI) was added, and samples were shaken for 30 min at 37°C. One μl of 0.5M DTT was then added, samples were incubated for 20 min at 56°C with shaking, 2.7 μl of 0.55 M iodoacetamide was added, and samples were incubated for an additional 20 min at room temperature in the dark. 1 ul of additional 1% PM was then added, followed by addition of 1 or 2.5 μg of sequencing grade modified trypsin (Promega) to epithelial and cortical fibers samples respectively, and the final volume of each sample was adjusted to 100 μl by addition of 50 mM TEAB buffer. After a 3-hour incubation at 37°C, 5 μl of 10% trifluoroacetic acid (TFA) was added, incubation was performed for 5 min at 37°C, samples were centrifuged at 16,000 × g for 15 min, and supernatants were removed.
Peptides were then solid phase extracted using MicroSpin C18 columns (The Nest Group, Southborough, MA). Columns were first conditioned by passing 100 μl of 80% acetonitrile (ACN), 0.1% TFA through twice by centrifugation at 2,000 × g, equilibrated using two 100 μl portions of 5% ACN, 0.1% TFA, and peptide digests were then loaded. Columns were then washed using two 100 μl portions of 5% ACN, 0.1% TFA, and peptides eluted in 100 μl of 80% ACN, 0.1% TFA. A peptide assay was then performed using a Pierce™ colorimetric peptide assay (ThermoFisher); 7.3 and 20 μg of peptide from each epithelial and cortical fiber cell digest, respectively, were then dried by vacuum centrifugation in preparation for tandem mass tagging (TMT™) (ThermoFisher).
TMT™ Analysis
Peptides were reconstituted in 25 μl of 100 mM TEAB buffer by shaking for 15 min. TMT™ 10-plex reagents were freshly prepared by dissolving in anhydrous ACN at a concentration of 16 μg/μl, 12 μl of each reagent added, and samples were incubated for one hour at room temperature. Since there were 9 epithelial and cortical fiber samples, the 130C tag was not used. Two μl of each reaction mixture from epithelial and cortical fiber cell samples were then mixed, 2 μl of 5% hydroxylamine added, and the combined sample incubated for a further 15 min. The mixture was then dried down, dissolved in 5% formic acid, and 2 μg of peptide was analyzed by a single 2 hour LC-MS/MS method using an Orbitrap Fusion as described below. This run was performed to normalize the total reporter ion intensity of each multiplexed sample and check labeling efficiency. The remaining samples were quenched by addition of 2 μl of 5% hydroxylamine as above, then combined in 1:1:1:1:1:1:1:1:1 ratios based on total reporter ion intensities determined during the normalization run, and dried down in preparation for two-dimensional reverse-phase liquid chromatography mass spectrometry analysis (2D-LC-MS/MS).
Final multiplexed samples were reconstituted in 5% formic acid and 60 μg portions of epithelial and cortical fiber samples were separated using a Dionex NCS-3500RS UltiMate RSLCnano UPLC system (ThermoFisher). Digests were injected onto a NanoEase 5 μm XBridge BEH130 C18 300 μm × 50 mm column (Waters Corporation, Milford, MA) at 3 μl/min in a mobile phase containing 10 mM ammonium formate (pH 9). Peptides were eluted by sequential injection of 20 μl volumes of 14, 17, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 50 and 90% ACN in 10 mM ammonium formate (pH 9) at 3 μl/min flow rate. Eluted peptides were diluted with mobile phase containing 0.1% formic acid at a 24 μl/min flow rate and delivered to an Acclaim PepMap 100 μm × 2 cm NanoViper C18, 5 μm trap (ThermoFisher) on a switching valve. After 10 min of loading, the trap column was switched on-line to a PepMap RSLC C18, 2 μm, 75 μm × 25 cm EasySpray column (ThermoFisher). Peptides were then separated at low pH in the second dimension using a 7.5–30% ACN gradient over 90 min in mobile phase containing 0.1% formic acid at 300 nl/min flow rate. Each second dimension LC run required 2 hours for separation and re-equilibration, so each 2D LC-MS/MS method required 34 hours for completion.
Tandem mass spectrometry data was collected using an Orbitrap Fusion Tribrid instrument configured with an EasySpray NanoSource (ThermoFisher). Survey scans were performed in the Orbitrap mass analyzer (resolution = 120,000), and data-dependent MS2 scans performed in the linear ion trap using collision-induced dissociation (normalized collision energy = 35) following isolation with the instrument’s quadrupole. Reporter ion detection was performed in the Orbitrap mass analyzer (resolution = 60,000) using MS3 scans following synchronous precursor isolation (SPI) of the top 10 ions in the linear ion trap, and higher-energy collisional dissociation in the ion-routing multipole (normalized collision energy = 65). The MSn scans used default automatic gain control ion targets provided in the TMT™ SPI template and variable numbers of MS2 scans between survey scans every 3 sec to automatically optimize data collection. Singly charged precursor ions were excluded and dynamic exclusion settings were 10 PPM m/z tolerances for 30 sec durations.
Bioinformatics
RAW instrument files were processed using Proteome Discoverer (PD) version 1.4.1.14 (ThermoFisher). For each experiment, raw files from the 17 fractions were merged and searched with the SEQUEST HT search engine with a Mus musculus Swiss-Prot protein database downloaded from www.UniProt.org in October 2016 (16,831 entries with an additional 179 common contaminant sequences). Searches were configured with static modifications for the TMT™ reagents (+229.163 Da) on lysines and N-termini, carbamidomethyl (+57.021 Da) on cysteines, dynamic modifications for oxidation of methionine residues (+15.9949 Da), parent ion tolerance of 1.25 Da, fragment mass tolerance of 1.0005 Da, monoisotopic masses, and trypsin cleavage (max 2 missed cleavages). Searches used a reversed sequence decoy strategy to control peptide false discovery and identifications were validated by Percolator software [9]. Search results and TMT™ reporter ion intensities were exported from Proteome Discoverer as text files and processed with in-house Python scripts.
Only peptide spectral matches (PSMs) uniquely matching a single protein entry with q scores <0.05, accurate masses within 10 PPM, and trimmed average reporter ion intensity peak heights greater than 500 were used for quantification. The individual reporter ion intensities from all PSMs were summed to create total protein intensities. Differential protein abundances between groups were determined by comparing the total reporter ion intensities using the Bioconductor package edgeR [10]. EdgeR was developed for serial analysis of gene expression data but its modeling is flexible enough to handle a variety of other data types such as TMT reporter ion intensities [11–13].
Additional data normalizations [14] multiple testing corrections, and calculation of false discovery rates were performed within edgeR. Only results with false discovery rate (FDR) < 0.1 were considered significant. Protein annotations from UniProt Swiss-Prot database records for the quantified proteins were added using in-house scripts.
Immunoblot assay
Lens capsules were collected and lysed on ice for 10 min in lysis buffer (20 mM Tris, pH 7.5, 1 mM EDTA, 1mM EGTA, 150 mM NaCl, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM α-glycerolphosphate, 1 mM Na3VO4, 1 mg/ml leupeptin, and 1 mM PMSF). Protein concentration from the supernatant was determined by protein BCA assay (ThermoFisher). The protein extract was further processed for immunoblot analysis and probed for γD-crystallin (Sigma-Aldrich), BFSP2, EEF1D, TOMM34 and DPYSL3 (Proteintech, Rosemont, IL) and LGSN (gift generously provided by Dr. Graeme Wistow at NIH). We found no GAPDH expression changes relevant to total protein in our working models and therefore all data were normalized to the level of GAPDH [8].
RESULTS
Overview of Protein Expression Changes Resulting from GSH Depletion in the Lens
EdgeR clustering reports showed that lens epithelia samples had much more intersample variance than fiber cell samples (Supp. Fig. 2). This is likely due to the significantly smaller amount of lens epithelia/capsule tissue that could be collected compared to fiber cells, which make up the bulk of the lens. One BSO-treated LEGSKO epithelia sample showed particularly high deviation from the other two samples in its group, clustering more closely to WT samples than other BSO-treated LEGSKO samples, and was eliminated from the analysis (Supp. Fig. 2B).
Complete results, including analyses with and without the BSO-treated outlier sample, can be found in Appendix 1 for epithelia samples and Appendix 2 for fiber cell samples.
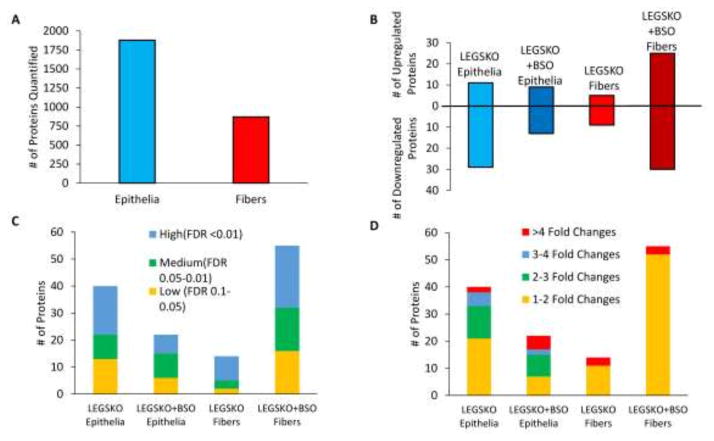
Altogether, 1871 proteins in lens epithelia and 870 proteins in lens fiber cells were quantified (Fig. 1A). The smaller number of quantifiable proteins in lens fiber cells is expected due to their very high abundance of crystallins, which makes quantification of less abundant proteins more difficult. Of the quantified proteins, 111 in total had significantly (FDR < 0.1) altered expression compared to WT controls, which included 40 proteins in LEGSKO epithelia, 22 proteins in BSO-treated LEGSKO epithelia, 14 proteins in LEGSKO fiber cells, and 55 proteins in BSO-treated LEGSKO fiber cells (Fig. 1B). All sample groups had a fairly even distribution of high (FDR < 0.01), medium (FDR 0.05-0.01), and low (FDR 0.1–0.05) candidate protein expression changes (Fig. 1C). Epithelium sample groups showed a wider range of expression fold changes, whereas fiber cells had many minor changes (fold change 1–2) and only a few major changes (fold change >4), with no medium changes (Fig. 1D).
Figure 1. Overview of the GSH-Deficient Lens Proteome.
(A) Total number of mapped proteins in lens epithelia and fiber cells. (B). Number of significantly (FDR < 0.1) up- and down-regulated proteins for each comparison. (C) FDR ranking of significantly altered proteins. (D) Fold change ranking of significantly altered proteins. All comparisons are relative to WT control.
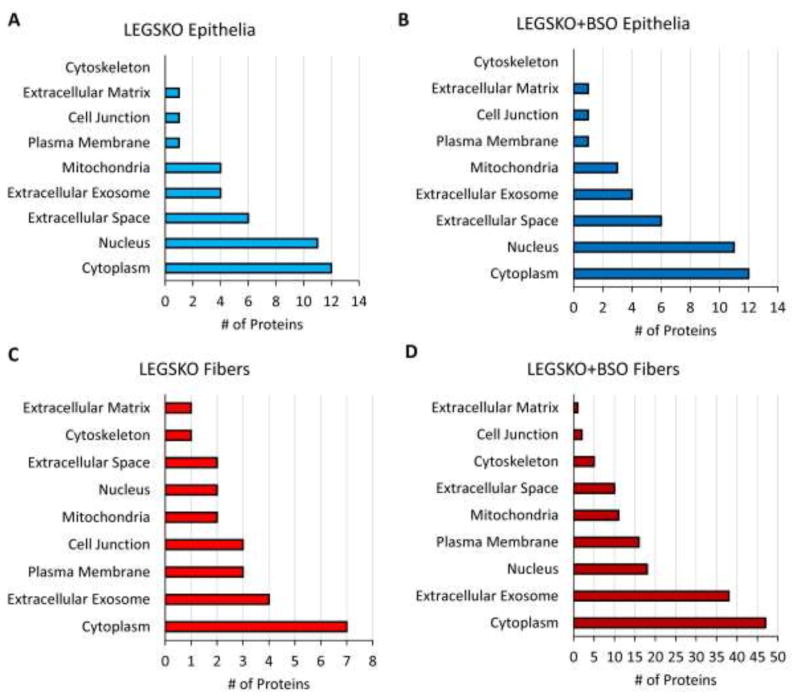
Proteins with significantly altered expression were localized to a variety of organelles and cellular regions, with the cytoplasm being the most common (Fig. 2). The nucleus was the next most common source of proteins in epithelium samples, whereas extracellular exosomes were the second major source in lens fiber cells.
Figure 2. Cellular Localization of Significantly Modulated Proteins (FDR< 0.1) in the GSH-Deficient Lens Proteome.
(A) LEGSKO epithelia. (B) BSO-treated LEGSKO epithelia. (C) LEGSKO fiber cells. (D) BSO-treated LEGSKO fiber cells. All comparisons are relative to WT control.
Major Trends in the GSH-Deficient Lens Proteome
Ingenuity Pathway Analysis (IPA) software (Qiagen, Hilden, Germany) was used to determine the major upstream regulators and molecular and cellular functions of the proteomic response to GSH deficiency in the lens (Table 1). It should be noted that upstream regulator analysis does not guarantee the activation or involvement of specific protein but rather lists the proteins that are the most likely regulators of the responses seen in the proteome. Due to the sensitivity of the proteomic analysis, these proteins may be missing from the lists of quantified proteins.
Table 1.
Top Upstream Regulators and Molecular and Cellular Functions of Gene Expression Changes in GSH-Deficient Lenses.
| LEGSKO Epithelia | BSO-Treated LEGSKO Epithelia | LEGSKO Fiber Cells | BSO-Treated LEGSKO Fiber Cells |
|---|---|---|---|
| Upstream Regulators (p-value) | |||
|
| |||
| HSF4(5.32E-09) | HSF4(2.84E-10) | STIM1(4.23E-05) | HSF4(1.98E-08) |
| MAF(2.76E-06) | MAF(1.52E-07) | FGF1(5.56E-05) | MAF(2.31E-07) |
| GLUL(3.47E-06) | ANGPT2(7.22E-04) | NRF2(1.14E-04) | MAP2K1/2(1.67E-06) |
| MKNK1(3.89E-05) | ORC4(9.44E-04) | SOD1(2.79E-04) | GNA12(1.67E-06) |
| BDNF(2.84E-04) | ORC3(9.44E-04) | SQSTM1(3.60E-04) | APP(1.78E-06) |
|
| |||
| Molecular and Cellular Functions (p-value range) | |||
|
| |||
| Amino Acid Metabolism (1.82E-02 – 9.84E-06) | Cell Morphology (4.35E-02 – 7.47E-04) | Carbohydrate Metabolism (1.60E-02 – 2.42E-06) | Post-Translational Modification (4.93E-09 - 4.93E-09) |
| Post-Translational Modification (5.50E-03 – 9.84E-06) | Cellular Assembly and Organization (4.35E-02 – 8.47E-04) | Small Molecule Biochemistry (4.71E-02 – 2.42E-06) | Protein Degradation (4.93E-09 - 4.93E-09) |
| Small Molecule Biochemistry (4.67E-02 – 9.84E-06) | Cellular Compromise (3.99E-02 – 8.47E-04) | Vitamin and Mineral Metabolism (4.60E-03 – 2.42E-06) | Free Radical Scavenging (7.75E-03 – 9.15E-07) |
| Cellular Assembly and Organization (3.96E-02 – 5.50E-04) | Cellular Development (4.49E-02 – 8.47E-04) | Drug Metabolism (9.19E-03 – 4.28E-06) | DNA Replication, Recombination, and Repair (7.75E-03 – 3.79E-06) |
| Cellular Function and Maintenance (4.14E-02 – 5.50E-04) | Cellular Function and Maintenance (4.49E-02 – 8.47E-04) | Protein Synthesis (9.19E-03 – 4.28E-06) | Cell Morphology (1.03E-02 – 5.15E-06) |
Common top upstream regulators for both GSH-deficient lens epithelium groups, as well as for BSO-treated LEGSKO fiber cells, were heat shock transcription factor 4 (HSF4) and transcription factor MAF, two essential transcription factors that govern lens development and crystallin expression [15].
The other major upstream regulators implicated in the LEGSKO lens epithelia GSH deficiency response were glutamate-ammonia ligase (GLUL), MAP kinase interacting serine/threonine kinase 1 (MKNK1), and brain-derived neurotrophic factor (BDNF). GLUL, which was 2-fold downregulated in LEGSKO epithelia (Appendix 1), synthesizes glutamine from glutamate and ammonia and also regulates glutaminase (GLS), which acts oppositely by breaking glutamine down into glutamate and ammonia. GLS was 1.9-fold upregulated in LEGSKO lens epithelia. Taken together, these changes are strongly indicative of decreasing cytosolic glutamine and increasing cytosolic glutamate and ammonia, possibly as an attempt to increase GSH synthesis. MKNK1 plays roles in both cellular stress response and transformation. BDNF is a transcription factor that plays a role in neural development and functioning, so its role in the lens is unclear.
The other upstream regulators implicated in the BSO-treated LEGSKO epithelia response are angiopoietin-2 (ANGPT2) and origin recognition complex (ORC) 3 and 4. ANGPT2 is a regulator of cellular morphology and organization while ORC3 and 4 are involved in DNA replication and cellular proliferation. These results imply potentially enhanced proliferation and transformation in these cells.
LEGSKO lens fiber cells, which did not indicate HSF4 or MAF as primary upstream regulators, had primary upstream regulators of stromal interaction protein 1 (STIM1), fibroblast growth factor 1 (FGF1), nuclear factor (erythroid-derived 2)-like 2 (NRF2), superoxide dismutase 1 (SOD1), and sequestosome 1 (SQSTM1). The transcription factor NRF2 is considered to be the master regulator of the antioxidant response and GSH synthesis/utilization [16]. When combined with SOD1, which detoxifies superoxide anion, these regulators imply major changes to the oxidative stress response. Fibroblast growth factor 1 (FGF1) is a transcription factor that regulates many pathways, including lens development and EMT [17]. STIM1 and SQSTM1 regulate calcium homeostasis and selective autophagy, respectively. SQSTM1 has been shown to be active in the mammalian lens, where it plays a role in the oxidative stress response [18].
In addition to HSF4 and MAF, BSO-treated LEGSKO lens fiber cells also showed upstream regulators of dual specificity mitogen-activated protein kinase kinase 1 (MAP2K1/2), guanine nucleotide-binding protein subunit alpha-12 (GNA12), and amyloid precursor protein (APP). MAP2K1/2 and GNA12 regulate many processes including cellular differentiation, proliferation, and migration and thus have a clear relation to EMT signaling. APP is best known for its role in Alzheimer Disease and has an unclear role here.
The most common molecular and cellular functions of significantly altered proteins in LEGSKO lens epithelia included amino acid metabolism, post-translational modification, small molecule biochemistry, cellular assembly and organization, and cellular function and maintenance. The major functions of altered proteins in the BSO-treated LEGSKO lens epithelium proteome were highly indicative of transformation and included cell morphology, cellular assembly and organization, cellular compromise, cellular development, and cellular function and maintenance. The major functions of the LEGSKO lens fiber cell proteome were the most metabolically-focused and included carbohydrate metabolism, small molecule biochemistry, vitamin and mineral metabolism, drug metabolism, and protein synthesis. The major functions of proteins in the BSO-treated LEGSKO lens fiber cell proteome were the most indicative of a typical cellular stress response and include post-translational modification, protein degradation, free radical scavenging, cell morphology, and DNA replication, recombination, and repair.
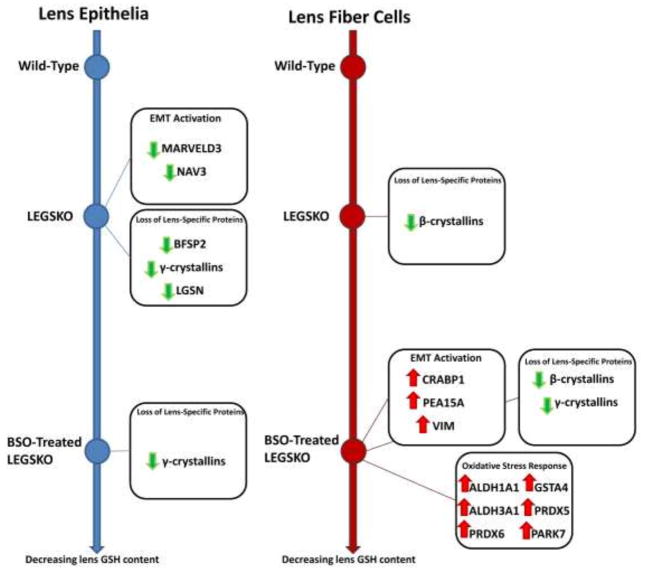
The major trends found in this proteomic analysis are also outlined as a schematic in Fig. 3, with the individual protein expression changes addressed in depth in the following sections.
Figure 3. Schematic Representation of Major Trends in the GSH-Deficient Lens Proteome.
Color and direction of arrows indicate direction of regulation change. Green arrows = downregulation, red arrows = upregulation. Line graphs represent lenticular GSH content which decreases from top to bottom of the figure.
Comparison to RNA-Seq Analysis
The trends discussed above fit well with those found in the GSH-deficient lens transcriptome [7], with the exception of major shifts in lipid homeostasis and transport systems, which were seen in the transcriptome but not the proteome of the GSH-deficient lens. Specific overlap in significant gene and protein expression changes was also investigated (Table 2). The GSH-deficient proteome recapitulates many of the same findings as the corresponding transcriptome, including upregulation of aldehyde dehydrogenases, downregulation of crystallins and other vision-related proteins, and an increase in EMT markers, although the specific changes were not always in the same sample groups when comparing the proteome to the transcriptome.
Table 2.
Overlapping mRNA and Protein Expression Changes in GSH-Deficient Mouse Lenses.
| Gene Symbol |
Description | mRNA Fold Change | Protein Fold Change | ||||||
|---|---|---|---|---|---|---|---|---|---|
| LEGSKO Epithelia |
LEGSKO + BSO Epithelia |
LEGSKO Fiber Cells |
LEGSKO + BSO Fiber Cells |
LEGSKO Epithelia |
LEGSKO + BSO Epithelia |
LEGSKO Fiber Cells |
LEGSKO + BSO Fiber Cells |
||
| Ace | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | 1.90 | 2.20 | 1.70 | |||||
| Aldh1a1 | aldehyde dehydrogenase family 1, subfamily A1 | 1.23 | 2.18 | 2.66 | 1.17 | ||||
| Aldh3a1 | aldehyde dehydrogenase family 3, subfamily A1 | 1.63 | 2.15 | 1.38 | |||||
| Anxa3 | annexin A3 | 1.46 | 1.49 | 1.32 | |||||
| Car2 | carbonic anhydrase 2 | 1.64 | 2.16 | 1.96 | 1.66 | ||||
| Crip2 | cysteine rich protein 2 | −2.50 | −2.97 | ||||||
| Crybb1 | crystallin, beta B1 | −1.56 | −1.22 | ||||||
| Cryga | crystallin, gamma A | −2.63 | −3.38 | −1.73 | |||||
| Crygb | crystallin, gamma B | −1.85 | −2.90 | ||||||
| Cryge | crystallin, gamma E | −2.08 | −2.32 | −2.94 | −1.88 | −2.59 | −1.78 | ||
| Crygn | crystallin, gamma N | −1.93 | −1.47 | ||||||
| Dpysl3 | dihydropyrimidinase-like 3 | 2.26 | 1.76 | 1.66 | 1.74 | ||||
| Fabp5 | fatty acid binding protein 5, epidermal | −1.69 | −1.95 | ||||||
| G6pdx | glucose-6-phosphate dehydrogenase X-linked | 1.20 | 1.42 | 1.25 | |||||
| Gls | glutaminase | −1.30 | 1.91 | ||||||
| Gsta4 | glutathione S-transferase, alpha 4 | 1.24 | 1.41 | ||||||
| Gsto1 | glutathione S-transferase omega 1 | 1.44 | −1.41 | ||||||
| Gstp1 | glutathione S-transferase, pi 1 | −1.24 | −1.34 | ||||||
| Hist1h1d | histone cluster 1, H1d | 1.76 | −1.83 | ||||||
| Hist2h3c2 | histone cluster 2, H3c2 | 2.17 | −1.64 | ||||||
| Hmox1 | heme oxygenase (decycling) 1 | −1.69 | −1.70 | ||||||
| Nadk | NAD kinase | 1.37 | 1.26 | ||||||
| Nap1l1 | nucleosome assembly protein 1-like 1 | −1.20 | 1.49 | ||||||
| Plec | plectin | −1.25 | −1.16 | ||||||
| Prdx5 | peroxiredoxin 5 | 1.32 | 1.23 | ||||||
| Pygl | liver glycogen phosphorylase | 1.65 | 2.10 | 1.34 | |||||
| Sri | sorcin | 1.34 | 1.33 | 1.25 | |||||
| Synm | synemin, intermediate filament protein | −1.40 | −3.93 | ||||||
| Tst | thiosulfate sulfurtransferase, mitochondrial | 1.27 | 1.68 | ||||||
| Tubb2a | tubulin, beta 2A class IIA | −1.79 | −1.24 | −1.24 | |||||
| Vim | vimentin | 1.49 | 1.49 | 1.25 | |||||
In most cases, expression changes in counterpart transcripts and proteins were very similar in both fold change and direction, with only a few exceptions. One discrepancy was GLS, which showed 1.3-fold downregulation in BSO-treated LEGSKO lens fiber cells at the mRNA level but 1.9-fold upregulation in LEGSKO lens epithelia at the protein level. Glutathione S-transferase omega 1 (GSTO1) was 1.4-fold upregulated in BSO-treated LEGSKO epithelia at the mRNA level but 1.4-fold downregulated in LEGSKO fiber cells at the protein level. Histone subunits H1d and H3c2 were ~2-fold upregulated in BSO-treated LEGSKO epithelia at the mRNA level but similarly downregulated in LEGSKO fiber cells at the protein level. Nucleosome assembly protein 1 like 1 (NAP1L1) was 1.2-fold downregulated in BSO-treated LEGSKO fiber cells at the mRNA level but 1.5-fold upregulated in LEGSKO epithelium at the protein level. All of these discrepancies were between different lens tissue types and could just represent differences between the responses found in GSH-deficient lens epithelia and fiber cells.
Lens GSH Deficiency Induces Differential Expression of Protective Proteins
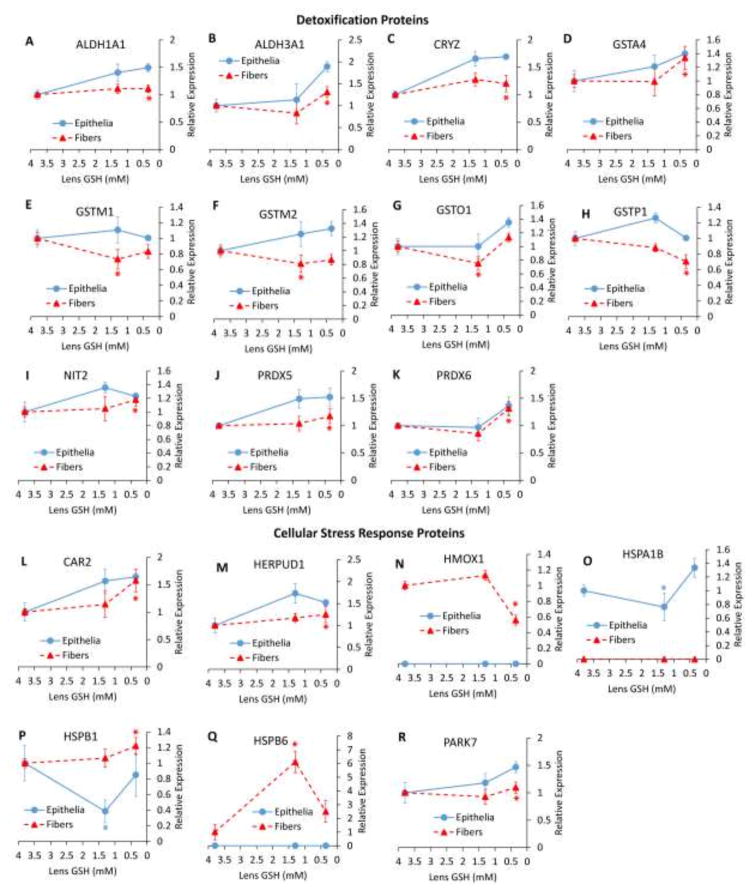
Expression of potentially protective proteins in GSH-deficient lenses was analyzed and proteins were grouped generally into categories of detoxification (including antioxidants and drug-metabolizing enzymes) and cellular stress response proteins. The expression of these proteins is presented as a function of decreasing lenticular GSH (Fig. 4).
Figure 4. Relative Abundance Changes in Detoxification and Cellular Stress Response Proteins.
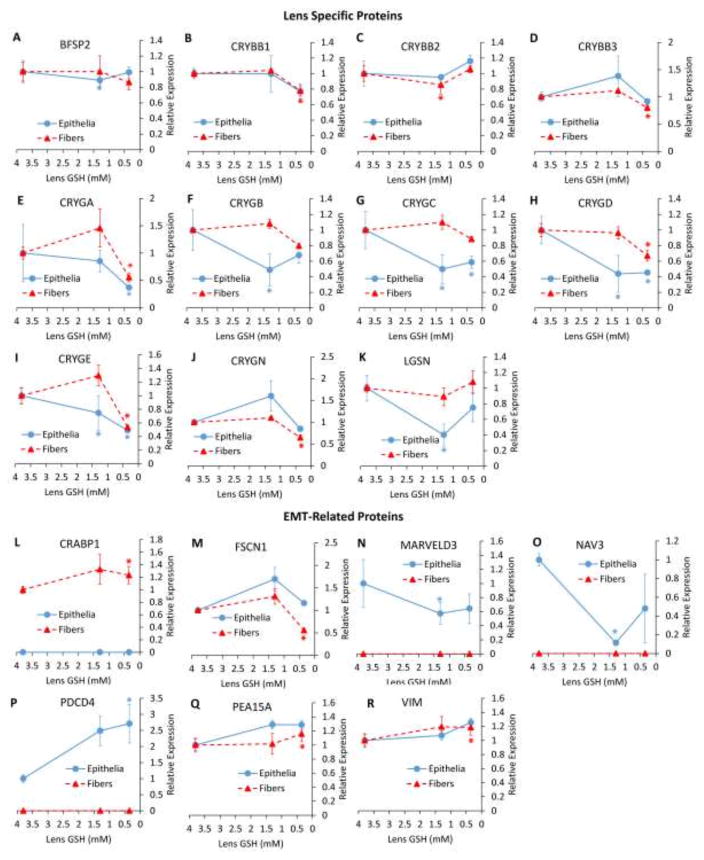
(A–K) Protein expression changes relating to detoxification. (L–R) Protein expression changes relating to the cellular stress response. (A) aldehyde dehydrogenase 1 family member A1 (ALDH1A1), (B) aldehyde dehydrogenase family 3 member A1 (ALDH3A1), (C) quinone oxidoreductase (CRYZ), (D) glutathione S-transferase alpha 4 (GSTA4), (E) glutathione S-transferase mu 1 (GSTM1), (F) glutathione S-transferase mu 2 (GSTM2), (G) glutathione S-transferase omega 1 (GSTO1), (H) glutathione S-transferase pi 1 (GSTP1), (I) omega-amidase (NIT2), (J) peroxiredoxin 5 (PRDX5), (K) peroxiredoxin 6 (PRDX6), (L) carbonic anhydrase 2 (CAR2), (M) homocysteine inducible ER protein with ubiquitin like domain 1 (HERPUD1), (N) heme oxygenase 1 (HMOX1), (O) heat shock protein family A (HSP70) member 1B) HSPA1B, (P) heat shock protein family B member 1 (HSPB1), (Q) heat shock protein family B member 6 (HSPB6), (R) Parkinson disease protein (PARK7). Values are means +/− SD. 3.8 mM GSH = WT, 1.3 mM GSH = LEGSKO, 0.35 mM GSH = BSO-treated LEGSKO. * = significant (FDR < 0.1) change. Fold change and significance is relative to WT.
Significant detoxification protein expression changes occurred only in fiber cells and were primarily found in the BSO-treated LEGSKO lens. These changes include modest upregulation of aldehyde dehydrogenases 1a1 and 3a1 (Fig. 4A and B), as well as peroxide-reducing peroxiredoxins (PRDX) 5 and 6 (Fig. 4J and K), omega-amidase (NIT2)(Fig. 4I), and quinone oxidoreductase (CRYZ)(Fig. 5C). Glutathione S-transferases showed mixed changes with GSTM1 (Fig. 4E), GSTM2 (Fig. 4F), and GSTO1 (Fig. 4G) all being slightly downregulated in LEGSKO lens fiber cells, and GSTA4 (Fig. 4D) and GSTP1 (Fig. 4H) being up- and down-regulated, respectively, in BSO-treated LEGSKO lens fiber cells.
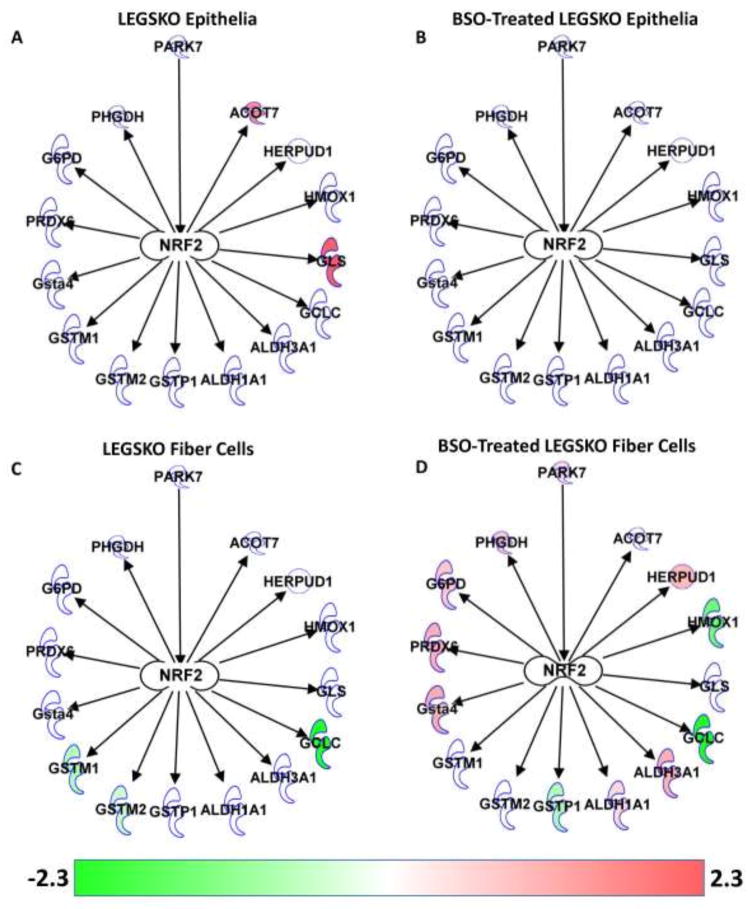
Figure 5. NRF2-Regulated Protein Expression Changes.
(A) LEGSKO lens epithelia. (B) BSO-treated LEGSKO lens epithelia. (C) LEGSKO lens fiber cells. (D) BSO-treated LEGSKO lens fiber cells. Arrows indicate activation. Color indicates fold change of gene relative to WT, as indicated by the color bar. Diagram created using Ingenuity Pathway Analysis software (Qiagen, Hilgen, Germany).
In cellular stress response proteins, mixed changes were found in heat shock proteins. HSPA1B was modestly downregulated in LEGSKO lens epithelia and undetectable in fiber cells (Fig. 4O). Conversely, HSPB6 was >6-fold upregulated in LEGSKO lens fiber cells but was undetectable in lens epithelia (Fig. 4Q). Most strikingly, HSPB1 was 1.2-fold upregulated in BSO-treated LEGSKO lens fiber cells but >2-fold downregulated in LEGSKO lens epithelia (Fig. 4P). These data suggest major differences in the profile of heat shock proteins utilized in the epithelial and fiber cell compartments of the lens.
Other cellular stress response changes included a nearly 2-fold downregulation of heme oxygenase 1 (HMOX1), a well-established stress response protein [19], in BSO-treated LEGSKO lens fiber cells, but this protein was undetectable in lens epithelia (Fig. 4N). Carbonic anhydrase 2 (CAR2) (Fig. 4L), homocysteine inducible ER protein with ubiquitin like domain 1 (HERPUD1)(Fig. 4M), and Parkinson disease protein 7 (PARK7)(Fig. 4R) were all modestly upregulated in BSO-treated LEGSKO lens fiber cells. CAR2, which was also upregulated in the GSH-deficient lens transcriptome (Table 2) [7], regulates acid-base balance through the bicarbonate buffer system and may be upregulated in order to accommodate for the loss of GSH, which is acidic and normally present at millimolar levels within the lens. HERPUD1 is a chaperone involved in the unfolded protein response. PARK7 has been suggested to have multiple protective functions, including free radical scavenging, redox-sensitive chaperone activity, and stabilization of NRF2 [20]. However, PARK7 is also considered an oncogene and enhances cellular transformation.
Analysis of NRF2-Regulated Changes Reveals Activation in BSO-Treated LEGSKO Fiber Cells
Given the relation of many of the above changes to NRF2 activity and NRF2 being indicated as a major upstream regulator of the proteomic response in LEGSKO lens fiber cells (Table 1), the expression of NRF2-related proteins was analyzed and overlaid on a diagram showing the relation between molecules (Fig. 5).
The only NRF2 regulated proteins detected as upregulated in LEGSKO lens epithelia were GLS (also known as GLSK) and cytosolic acyl coenzyme A thioester hydrolase (ACOT7) (Fig. 5A), neither of which have a direct role in mediating oxidative stress and are metabolically important enzymes regulated by a variety of factors. Intriguingly, the upregulation of these proteins was not found in BSO-treated LEGSKO lens epithelia, nor were regulation changes in any other NRF2-regulated genes (Fig. 5B). As noted previously, there was a modest downregulation of GSTM1 and 2 in LEGSKO lens fiber cells (Fig. 5C). GCLC also showed strong downregulation but this is simply due to the LEGSKO mouse being an engineered GCLC knockout. The most robust NRF2-related response was in BSO-treated LEGSKO lens fiber cells, which showed upregulation of ALDH1A1, ALDH3A1, GSTA4, HERPUD1, and PRDX6 and downregulation of HMOX1 and GSTP1 (Fig. 5D). Additionally, glucose-6-phosphate dehydrogenase X-linked (G6PDX) and phosphoglycerate dehydrogenase (PHGDH), which produce NAD(P)H and NADH, respectively, and have been demonstrated to be regulated by NRF2 [21–22], were modestly upregulated. Nicotinamide nucleotides produced by these enzymes are used to carry out the activities of various antioxidants, including glutathione reductase and aldehyde dehydrogenases. Also upregulated exclusively in BSO-treated LEGSKO lens fiber cells was PARK7, which is not regulated by NRF2 but, rather, stabilizes NRF2 and helps to activate its transcriptional activity. This could mean that PARK7 is an essential factor for mediating an NRF2-regulated oxidative stress response in the lens.
Lens GSH Deficiency Induces EMT-Related Protein Expression Changes
As was found in the GSH-deficient lens transcriptome [7], the GSH-deficient lens proteome showed a broad downregulation of β- and γ-crystallins in both lens epithelia and fiber cells (Fig. 6B–J). Additionally, the essential lens structural proteins lengsin (LGSN) and beaded filament structural protein 2 (BFSP2), also known as phakinin, were downregulated in LEGSKO lens epithelia (Fig. 6A and K). Downregulation of such essential and highly specific lens proteins is strongly indicative of a loss of proper differentiation in lens cells, likely resulting from EMT.
Figure 6. Relative Abundance Changes in Lens Specific and EMT-Related Proteins.
(A–K) Protein expression changes in lens specific proteins. (L–S) Protein expression changes relating to EMT. (A) beaded filament structural protein/phakinin (BFSP2), (B) β-crystallin B1 (CRYBB1), (C) β-crystallin B2 (CRYBB2), (D) β-crystallin B3 (CRYBB3), (E) γ-crystallin A (CRYGA), (F) γ-crystallin B (CRYGB), (G) γ-crystallin C (CRYGC), (H) γ-crystallin D (CRYGD), (I) γ-crystallin E (CRYGE), (J) γ-crystallin N (CRYGN), (K) lengsin (LGSN), (L) cellular retinoic acid binding protein 1 (CRABP1), (M) fascin actin-bundling protein (FSCN1), (N) MARVEL domain containing 3 (MARVELD3), (O) neuron navigator 3 (NAV3), (P) programmed cell death protein 4 (PCDC4), (W) astrocytic phosphoprotein PEA-15 (PEA15A), (R) vimentin (VIM). Values are means +/− SD. 3.8 mM GSH = WT, 1.3 mM GSH = LEGSKO, 0.35 mM GSH = BSO-treated LEGSKO. * = significant (FDR < 0.1) change. Fold change and significance is relative to WT.
Many changes were also found in proteins with a known association to EMT. Most notably, the classical EMT marker vimentin (VIM) was slightly upregulated in BSO-treated LEGSKO fiber cells (Fig. 6R). Other traditional EMT markers, such as α-smooth muscle actin (α-SMA), type I collagen, and fibronectin could not be quantitated due to a lack of unique peptides in these proteins, which share close homology with many other family members, and/or low abundance.
However, a number of other EMT-related changes were noted. Cellular retinoic acid-binding protein 1 (CRABP1)(Fig. 5L), and phosphoprotein enriched in astrocytes 15A (PEA15A)(Fig. 6Q), both of which enhance proliferation and transformation, were slightly upregulated in BSO-treated LEGSKO fiber cells. Marvel domain containing 3 (MARVELD3)(Fig. 6N), a tight junction protein that is downregulated during EMT, and neuron navigator 3 (NAV3)(Fig. 6O), a tumor suppressor downregulated in various malignancies, were downregulated in LEGSKO epithelia and undetectable in fiber cells.
There were several changes that appeared to work counter to activation of EMT. Programmed cell death protein 4 (PDCD4)(Fig. 6P), a tumor and transformation suppressor, was 2.7-fold upregulated and in BSO-treated LEGSKO epithelia. Fascin actin bundling protein 1 (FSCN1)(Fig. 6M), which plays a major role in cell migration, was slightly downregulated in BSO-treated LEGSKO epithelia. However, the overall trend of protein expression changes implies activation of EMT signaling.
Western Blot Analysis of Lens Epithelia Supports Proteomic Findings
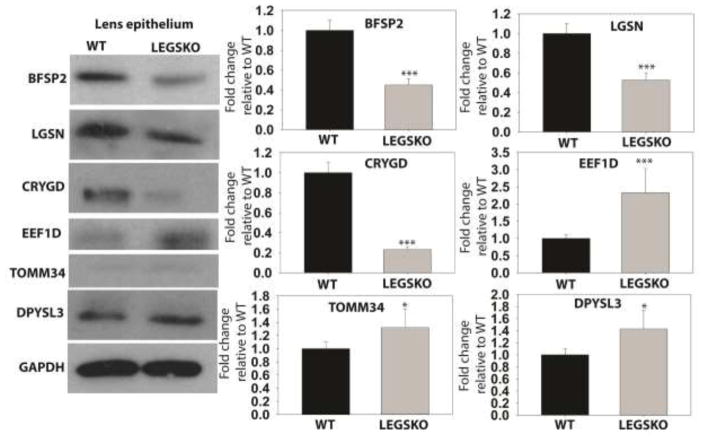
To support the veracity of the GSH-deficient lens proteome, expression of several proteins was measured by Western blot in LEGSKO and WT lens epithelia (Fig. 7). The results of this analysis aligned closely with the proteomic analysis, with BFSP2, LGSN, and CRYGD being significantly (p < 0.001) downregulated and elongation factor 1-delta (EEF1D), translocase of outer mitochondrial membrane (TOMM34), and dihydropyrimidinase-related protein 3 (DPYSL3) being significantly (p < 0.05) upregulated in the LEGSKO lens compared to WT control. These results demonstrate the reproducibility of the proteomic database presented here.
Figure 7. Confirmation of proteomic changes by Western blot analysis.
Left = Representative images of Western blot analysis. Right = Quantification of relative protein expression. All values are normalized to GAPDH loading control and WT values are set to 1.0. Values are means ± SD. n = 3. * = p < 0.05, *** = p < 0.001.
DISCUSSION
This study provides, to our knowledge, the first comparative proteomic analysis based on GSH depletion in any tissue. These data present key insight into the lenticular response to GSH deficiency and oxidative stress, as discussed below. However, when evaluating the meaning of these changes, it is important to consider that the LEGSKO mouse lens represents a model of chronic GSH depletion, which exerts its effect soon after birth. Thus any change in this model represents chronic genetic adaptation to sustained oxidative stress. In contrast, BSO was administered for only one month and, therefore, represents acute oxidative stress added onto the chronic stress stemming from low GSH levels.
Comparison to Other Studies of Stressed Lens Proteomes
While there have been a number of excellent proteomic studies of the lens [23–25], most have focused on changes to post-translational modifications (PTMs) of proteins or description of the normal lens proteome, rather than relative abundances of proteins under conditions of stress, making a direct comparison to this study difficult. These studies detected many of the same established lens proteins found in the Appendices, such as AQP0/MIP and AQP5, BFSP1 and BFSP2, and Cx46 and Cx50, indicating proper isolation of lens tissues in this study.
One recent study examined the proteomic changes that occur in Type I and II diabetic cataract (DC) rat model lenses [26]. Primarily, this study reported that DC lenses show a decrease in the abundance of crystallins and other lens structural proteins, including LGSN and BFSPs, closely matching what was found in the present study. Interestingly, VIM abundance was actually decreased, indicating that the loss of traditional lens proteins may be unrelated to EMT pathology in DC lenses. Another discrepancy was the downregulation of annexins ANXA3 and ANXA5 in DC lenses, whereas ANXA2 and ANXA5 were upregulated in GSH-deficient lenses (Appendix 2). Intriguingly, DC lenses were found to have decreased GSH levels, which may explain some of the similarity between the proteomes. Unfortunately, DC lenses were analyzed in total rather than in local fractions, limiting the conclusions that can be drawn from this comparison.
A study of proteomic changes in newborn lenses of K6W mutant ubiquitin (i.e. ER stressed) mice revealed several strikingly similar changes to the GSH-deficient lens proteome, including downregulation of γ-crystallins and HSPB1 [27]. Another proteomic study of the Cx46 knockout mouse, which develops cataract shortly after birth, revealed downregulation of crystallins, BFSPs, and heat shock protein HSPB1 and upregulation of ANXA1 [28]. Again, both studies extracted protein from whole lenses, limiting the interpretation of the data.
An in vitro study analyzed the proteome of cultured human lens epithelial cells after exposure to 500 μM H2O2 [29]. Changes found included an upregulation of HSPA8 and VIM and oxidation of peroxiredoxin I (PRDXI), showing clear similarity to the results found in our study. Human lens epithelial cells exposed to microwave radiation showed upregulation of two ribosomal proteins and heat shock protein HSP70 [30].
Taken together, these findings imply a general blueprint for the lens response to stress, which includes a loss of crystallins and other important lens proteins, such as BFSPs and LGSN. These changes are accompanied by mixed changes in heat shock proteins and annexins, with downregulation of HSPB1 being a common event. VIM was upregulated by all cases of oxidative stress but unchanged or downregulated by other stressors, implying that activation of EMT signaling in the lens may be specific to oxidative stress and not part of a general stress response.
NRF2-Mediated Oxidative Stress Response
We were previously surprised to find a very muted oxidative stress response in the GSH-deficient lens transcriptome [7]. Aldh1a1, Aldh3a1, Mt1, and Hmox1 are all upregulated by NRF2 activation [31] but, while Aldh1a1, Aldh3a1, and Mt1 were upregulated in the GSH-deficient lens transcriptome, Hmox1 was downregulated. Furthermore, Txnip, which is downregulated by NRF2 activation [32], was upregulated in GSH-deficient lenses. Based on these results, and a lack of regulation changes in other NRF2-regulated genes (including GSH-related genes), we speculated that NRF2 may not be activated by GSH deficiency in the lenses, as would have been expected.
The GSH-deficient lens proteome clarifies these results. IPA analysis indicates NRF2 as a major upstream regulator of the response in LEGSKO lens fiber cells (Table 1), though it should be noted that this is just based on the number of NRF2-regulated proteins that showed significant expression changes, regardless of the direction of the change. IPA analysis also indicated NRF2 as an upstream regulator of the response in the BSO-treated LEGSKO lens fiber cell proteome, although it was not one of the top 5 regulators based on corrected p-value (data not shown). However, NRF2 was not determined to be an upstream regulator at all in the epithelial samples.
These trends can be seen in analyzing the expression of NRF2-regulated proteins (Fig. 5). Only BSO-treated LEGSKO lens fibers appeared to show a robust NRF2-mediated response. This group was also the only one to show upregulation of the NRF2 activator PARK7. This could indicate that PARK7 is critical for NRF2 activation in the lens and that PARK7 is only upregulated under conditions of severe GSH-deficiency in the lens. These hypotheses will need to be tested in model systems to confirm this paradigm.
It remains unclear why HMOX1 and GSTP1 were downregulated in BSO-treated LEGSKO fiber cells, which otherwise seemed to show a clear NRF2-mediated response. Additionally, while BSO treatment appeared to induce NRF2 activation in LEGSKO lens fiber cells, it appeared to have an opposite effect on LEGSKO lens epithelia, which showed modest upregulation of two NRF2-regulated genes in the untreated group that was lost in the BSO-treated group (Fig. 5). However, this may simply be due to having to eliminate one of the BSO-treated samples as an outlier, leading to a decrease in statistical model performance.
Overall, it is clear that lens fiber cells are capable of mounting a traditional NRF2-mediated oxidative stress response to GSH deficiency when sufficiently stressed while lens epithelia appear to lack this ability.
Activation of EMT in the GSH-Deficient Lens
The upregulation of VIM found in this proteomic study is the clearest sign that GSH-deficient lenses are truly undergoing EMT. α-smooth muscle actin, which we previously demonstrated is expressed in GSH-deficient lenses [7], could not be detected in this analysis because of its high similarity to other forms of actin, preventing it from producing any unique tryptic peptides that could be used for quantification. This proteomic data also confirms a loss of crystallins and other lens marker genes, as seen in the GSH-deficient lens transcriptome, indicating transformation. While it could be argued that this decrease in protein abundance was due to a loss of solubility for these proteins, we found that our cell lysis technique could solubilize >99.6% of all lens proteins (Supp. Fig. 1). Thus, this loss of lens marker proteins is likely a regulated process and indicative of improper differentiation.
Limitations of this Study
While the data presented in this study provide broad insight into proteomic changes in the GSH-deficient lens, there are several limitations that must be considered when interpreting this information. Firstly, the coverage of the proteomic results is limited to sufficiently abundant proteins. This excludes many proteins that may be playing an important role in lens homeostasis, particularly in the lens epithelia where only a small amount of tissue could be collected. The small sample sizes may have also limited the detection of small but significant changes in the lens proteome.
Additionally, quantified proteins must feature unique tryptic peptides, excluding many proteins of interest with highly similar family members, such as α-SMA and metallothioneins. This analysis also does not give information on post-translational modification (PTM) in the lens. PTMs of crystallins and other lens proteins are tightly linked to cataract formation and the activities of many proteins are governed by PTMs. Thus, data on PTMs of lens protein may be equally important to expression data in some cases and should also be considered.
Conclusions
The data presented here give strong insight into the mechanisms of adaptation in the GSH-deficient lens. These proteomic data confirm the relevance of the previously reported GSH-deficient lens transcriptome and align with other proteomic studies of stressed lenses. These results suggest that severely GSH-deficient lenses show activation of EMT signaling and rely on NRF2 activation, possibly mediated by PARK7, to mount an oxidative stress response.
Supplementary Material
Highlights.
Lens proteomes were compared across three different levels of GSH.
Lens epithelia and fiber cells differ in their oxidative stress response.
Proteomic data confirms changes found in the GSH-deficient lens transcriptome.
Lens GSH-deficiency results in EMT-related proteomic remodeling.
Acknowledgments
Research funding provided by NIH grants EY07099 to VMM, EY024553 to XF, T32 EY007157 to the Visual Sciences Research Center at Case Western Reserve University, T32 EY024236 to the Cole Eye Institute at the Cleveland Clinic, P30 EY10572 to the Casey Eye Institute at the Oregon Health & Science University, and S10 OD012246 to LLD. We are very grateful to Dr. Graeme Wistow for his generosity in providing us with the lengsin (LGSN) antibody.
Footnotes
Conflicts of Interest: The authors claim no conflicts of interest related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
WORKS CITED
- 1.Giblin FJ. Glutathione: a vital lens antioxidant. J Ocul Pharmacol Ther. 2000;16:121–35. doi: 10.1089/jop.2000.16.121. [DOI] [PubMed] [Google Scholar]
- 2.Rathbun WB, Schmidt AJ, Holleschau AM. Activity loss of glutathione synthesis enzymes associated with human subcapsular cataract. Invest Ophthalmol Vis Sci. 1993 May;34(6):2049–54. [PubMed] [Google Scholar]
- 3.Sweeney MH, Truscott RJ. An impediment to glutathione diffusion in older normal human lenses: a possible precondition for nuclear cataract. Exp Eye Res. 1998;67:587–595. doi: 10.1006/exer.1998.0549. [DOI] [PubMed] [Google Scholar]
- 4.Fan X, Liu X, Hao S, Wang B, Robinson ML, Monnier VM. The LEGSKO mouse: a mouse model of age-related nuclear cataract based on genetic suppression of lens glutathione synthesis. PLoS One. 2012;7:e50832. doi: 10.1371/journal.pone.0050832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey JW, Pinarci EY, Penugonda S, Karacal H, Ercal N. In vivo inhibition of l-buthionine-(S, R)-sulfoximine-induced cataracts by a novel antioxidant, N-acetylcysteine amide. Free Radic Biol Med. 2011;50(6):722–9. doi: 10.1016/j.freeradbiomed.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Whitson JA, Sell DR, Goodman MC, Monnier VM, Fan X. Evidence of Dual Mechanisms of Glutathione Uptake in the Rodent Lens: A Novel Role for Vitreous Humor in Lens Glutathione Homeostasis. Invest Ophthalmol Vis Sci. 2016;57(8):3914–25. doi: 10.1167/iovs.16-19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitson JA, Zhang X, Medvedovic M, Chen J, Wei Z, Monnier VM, Fan X. Transcriptome of the GSH-Depleted Lens Reveals Changes in Detoxification and EMT Signaling Genes, Transport Systems, and Lipid Homeostasis. Invest Ophthalmol Vis Sci. 2017;58(5):2666–84. doi: 10.1167/iovs.16-21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Z, Whitson J, Zhang AD, Srinivasagan R, Kavanagh TJ, Yan H, Fan X. Reduced glutathione level promotes epithelial-mesenchymal transition in lens epithelial cells via a Wnt/β-catenin-mediated pathway: relevance for cataract therapy. Am J Pathol. 2017 doi: 10.1016/j.ajpath.2017.07.018. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–5. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 10.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huan J, Hornick NI, Goloviznina NA, Kamimae-Lanning AN, David LL, Wilmarth PA, Mori T, Chevillet JR, Narla A, Roberts CT, Loriaux MM. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia. 2015;29(12):2285–95. doi: 10.1038/leu.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Midgett M, López CS, David L, Maloyan A, Rugonyi S. Increased Hemodynamic Load in Early Embryonic Stages Alters Endocardial to Mesenchymal Transition. Front Physiol. 2017;8:56. doi: 10.3389/fphys.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plubell DL, Wilmarth PA, Zhao Y, Fenton AM, Minnier J, Reddy AP, Klimek J, Yang X, David LL, Pamir N. Extended Multiplexing of Tandem Mass Tags (TMT) Labeling Reveals Age and High Fat Diet Specific Proteome Changes in Mouse Epididymal Adipose Tissue. Mol Cell Proteomics. 2017;16(5):873–90. doi: 10.1074/mcp.M116.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cvekl A, McGreal R, Liu W. Lens Development and Crystallin Gene Expression. Prog Mol Biol Transl Sci. 2015;134:129–67. doi: 10.1016/bs.pmbts.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Ma Q. Role of Nrf2 in Oxidative Stress and Toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–26. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAvoy JW, Chamberlain CG, de Iongh RU, Richardson NA, Lovicu FJ. The role of fibroblast growth factor in eye lens development. Ann N Y Acad Sci. 1991;638:256–74. doi: 10.1111/j.1749-6632.1991.tb49036.x. [DOI] [PubMed] [Google Scholar]
- 18.Brennan L, Khoury J, Kantorow M. Parkin elimination of mitochondria is important for maintenance of lens epithelial cell ROS levels and survival upon oxidative stress exposure. Biochim Biophys Acta. 2017;1863(1):21–32. doi: 10.1016/j.bbadis.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryter SW, Choi AM. Heme oxygenase-1: redox regulation of a stress protein in lung and cell culture models. Antioxid Redox Signal. 2005;7(1–2):80–91. doi: 10.1089/ars.2005.7.80. [DOI] [PubMed] [Google Scholar]
- 20.Cao J, Lou S, Ying M, Yang B. DJ-1 as a human oncogene and potential therapeutic target. Biochem Pharmacol. 2015;93(3):241–50. doi: 10.1016/j.bcp.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 21.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, Cantley LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47(12):1475–81. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller CJ, Gounder SS, Kannan S, Goutam K, Muthusamy VR, Firpo MA, Symons JD, Paine R, 3rd, Hoidal JR, Rajasekaran NS. Disruption of Nrf2/ARE signaling impairs antioxidant mechanisms and promotes cell degradation pathways in aged skeletal muscle. Biochim Biophys Acta. 2012;1822(6):1038–50. doi: 10.1016/j.bbadis.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Han J, David LL, Schey KL. Proteomics and phosphoproteomics analysis of human lens fiber cell membranes. Invest Ophthalmol Vis Sci. 2013;54(2):1135–43. doi: 10.1167/iovs.12-11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Schey KL. Proteomic Analysis of Lipid Raft-Like Detergent-Resistant Membranes of Lens Fiber Cells. Invest Ophthalmol Vis Sci. 2015;56(13):8349–60. doi: 10.1167/iovs.15-18273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bassnett S, Wilmarth PA, David LL. The membrane proteome of the mouse lens fiber cell. Mol Vis. 2009;15:2448–63. [PMC free article] [PubMed] [Google Scholar]
- 26.Su S, Leng F, Guan L, Zhang L, Ge J, Wang C, Chen S, Liu P. Differential proteomic analyses of cataracts from rat models of type 1 and 2 diabetes. Invest Ophthalmol Vis Sci. 2014;55(12):7848–61. doi: 10.1167/iovs.14-15175. [DOI] [PubMed] [Google Scholar]
- 27.Shang F, Wilmarth PA, Chang M, Liu K, David LL, Caceres MA, Wawrousek E, Taylor A. Newborn Mouse Lens Proteome and Its Alteration by Lysine 6 Mutant Ubiquitin. J Proteome Res. 2014;13(3):1177–89. doi: 10.1021/pr400801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoehenwarter W, Tang Y, Ackermann R, Pleissner K, Schmid M, Stein R, Zimny-Arndt U, Kumar NM, Jungblut PR. Identification of Proteins that Modify Cataract of the Eye Lens. Proteomics. 2008;8(23–24):5011–24. doi: 10.1002/pmic.200800380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paron I, D’Elia A, D’Ambrosio C, Scaloni A, D’Aurizio F, Prescott A, Damante G, Tell G. A proteomic approach to identify early molecular targets of oxidative stress in human epithelial lens cells. Biochem J. 2004;378(3):929–37. doi: 10.1042/BJ20031190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li HW, Yao K, Jin HY, Sun LX, Lu DQ, Yu YB. Proteomic analysis of human lens epithelial cells exposed to microwaves. Jpn J Ophthalmol. 2007;51(6):412–6. doi: 10.1007/s10384-007-0483-9. [DOI] [PubMed] [Google Scholar]
- 31.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 32.He X, Ma Q. Redox regulation by nuclear factor erythroid 2-related factor 2: gatekeeping for the basal and diabetes-induced expression of thioredoxin-interacting protein. Mol Pharmacol. 2012;82(5):887–97. doi: 10.1124/mol.112.081133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.