Abstract
Alzheimer’s disease (AD) is a genetically heterogeneous neurodegenerative disorder caused by fully penetrant single gene mutations in a minority of cases, while the majority of cases are sporadic or show modest familial clustering. These cases are late-onset and likely result from the interaction of many genes and the environment. More than thirty loci have been implicated in AD by a combination of linkage, genome-wide association and whole genome/exome sequencing. We have learned from these studies that perturbations in endolysosomal, lipid metabolism and immune response pathways substantially contribute to sporadic AD pathogenesis. We review here current knowledge about functions of AD susceptibility genes, highlighting cells of the myeloid lineage as drivers of at least part of the genetic component in late-onset AD. Although targeted resequencing utilized for the identification of causal variants has discovered coding mutations in some AD-associated genes, a lot of risk variants lie in non-coding regions. Here we discuss the use of functional genomics approaches that integrate transcriptomic, epigenetic and endophenotype traits with systems biology in order to annotate genetic variants, and to facilitate discovery of AD risk genes. Further validation in cell culture and mouse models will be necessary to establish causality for these genes. This knowledge will allow mechanism-based design of novel therapeutic interventions in AD and promises coherent implementation of treatment in a personalized manner.
Keywords: Alzheimer’s disease, functional genomics, genome-wide association studies, lipid metabolism, endolysosomal pathway, immune response
Introduction
Alzheimer’s disease (AD) is the most common form of neurodegeneration characterized clinically by the presence of short-term memory loss, impaired judgement and problem solving as well as changes in mood and behavior, together resulting in significant familial and social burden. Pathologically AD is characterized by the accumulation of extracellular amyloid-β (Aβ) plaques and hyperphosphorylation of tau protein aggregated in intraneuronal neurofibrillary tangles (1). There is also substantial neuronal loss in hippocampus and entorhinal cortex (2), and marked gliosis (3). AD affects approximately 5% of people over 65 years old and prevalence doubles with every 5 years of increasing age (4). Initial insight into the pathogenesis of AD came from genetic studies of early-onset familial forms that are caused by mutations in amyloid-β precursor protein (APP) (5), presenilin 1 (PSEN1) (6) and presenilin 2 (PSEN2) (7, 8) inherited as an autosomal dominant trait (9). These findings led to the proposal of “amyloid cascade hypothesis”, which postulates that dysregulation of Aβ peptide production and degradation underlies the pathological and behavioral changes observed in AD patients (10). However, the majority of cases are late-onset AD (LOAD) and sporadic with an unknown cause. According to twin studies the heritability of AD is ~58% (11), suggesting that both genetic and non-genetic variation influence disease, e.g. environmental and epigenetic factors, somatic mutations. Indeed, a study of identical twins discordant for AD has shown reduced DNA methylation in temporal neocortex neuronal nuclei of the AD-affected twin (12).
Early-onset Alzheimer’s Disease
The main factors influencing early-onset AD are coding mutations or copy number changes in genes that regulate Aβ production and degradation. Aβ is generated by sequential cleavage of APP by β- and γ-secretases. Overproduction of Aβ is a recognized AD risk factor observed in Down syndrome cases that possess chromosome 21 trisomy encompassing APP locus (9), and APP duplication cases because of copy number changes (13, 14). Most APP pathogenic mutations occur around the Aβ cleavage sites affecting APP processing by secretases, e.g. APP-KM670/671NL (15) or APP-E682K (16) at the β-secretase cleavage site, which increase Aβ production. Mutations in the Aβ sequence have the potential to affect its biophysical properties, such as hydrophobicity and aggregation rate, while C-terminal Aβ mutations at the γ-secretase site influence the Aβ42 to Aβ40 ratio (17). Mutations in PSEN1 and PSEN2 that form the active core of γ-secretase complex, affect endopeptidase- or carboxypeptidase-like activity, shifting production of Aβ40 and Aβ42 to longer and more neurotoxic species, e.g. Aβ43 in the case of PSEN1-R278I (18, 19) or PSEN1-L435F (20), which also shows a dramatic reduction in total Aβ production. Thus, the toxic dysfunction mechanism is used to describe AD-related genetic changes in γ-secretase (19). Indeed, evaluation of heterozygous null PSEN1 mutation in genome-edited induced pluripotent stem cells (hiPSc)-derived human neurons shows reduced level of γ-secretase, but no effect on Aβ levels, supporting the toxic gain of function model (21). Interestingly, a study of 138 mutations in PSEN1 concluded there is no correlation between Aβ42/Aβ40 ratio and AD age-at-onset, based on the in vitro assay used in this study, suggesting that Aβ levels may not be the sole driving factor and that other genetic and environmental factors contribute to disease progression (22). To date sequencing of β-secretase (BACE1) has not identified mutations that influence AD risk (23). However, two rare coding variants in α-secretase ADAM10 (Q170H and R181G) have been reported in familial LOAD (24), and both mutations show impaired activity due to incorrect ADAM10 folding and elevated plaque load in APP transgenic mice (25).
Late-Onset Alzheimer’s Disease
Genetic risk factors play a critical role in AD susceptibility. The “common disease – common variant” hypothesis proposes that a combination of multiple common variants and environmental factors underlie disease risk (26). Technological advances in high-throughput genotyping and sequencing allow testing of tens of thousands of control and patient samples that can be used to conduct genome-wide association studies (GWAS). GWAS report genetic variants and loci that are enriched in populations with a disease trait compared to unaffected individuals. Several GWAS of AD were performed (27–31) and later combined in a meta-analysis (32) reporting more than twenty AD susceptibility loci in European populations (33). APOE is the most significant risk factor confirmed in all studies across populations. While the largest GWAS have been performed in European populations, a GWAS in African Americans identified variants in APOE and ABCA7 as genome-wide significant (34). A GWAS in Asian populations identified AD-associated genome-wide significant variants in or near APOE and SORL1 (35). Given the vastly different sizes of the datasets in these GWAS, European cohorts remain the most powerful for gene discovery. Nevertheless, it is important to establish the contribution of these loci to risk in other populations. Half of identified susceptibility loci have minor alleles that are protective, thereby increasing AD age-at-onset. Furthermore, the known risk loci do not fully explain the genetic component of AD estimated at 58% based on twin studies (11).
Functional Genomics
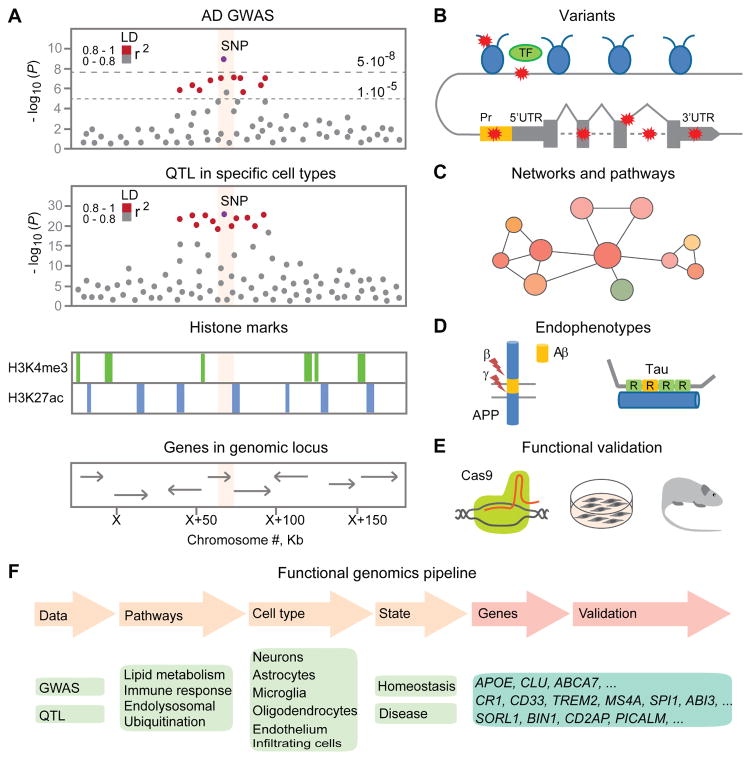
GWAS are extremely useful as a way to identify association for multifactorial traits such as AD that have genetic and environmental components and don’t rely on family history like linkage studies. However, large sample sizes are required and most signals have small effect. Furthermore, associations are reported for an index single nucleotide polymorphism (SNP) with the lowest P value, but in reality can be driven by any variant in the linkage disequilibrium (LD) block (33). Loci identified by GWAS often contain multiple genes that could all contribute to disease association, or only one of the genes in the locus could be causative. As a result labeling a gene within a locus in Manhattan association plots could be misleading (Figure 1), since it suggests assignment of causation, which is not possible based on GWAS alone.
Figure 1. Schematic representation of a multidimensional approach for fine-mapping risk variants in Alzheimer’s disease-associated loci.
(A) An illustration of a locus-specific association signal from a genome-wide association study (GWAS) of Alzheimer’s disease (AD), e.g. Manhattan association plot (top panel). Each dot represents a single nucleotide polymorphism (SNP), with the X-axis showing the chromosomal position and Y-axis showing the association P values on the -log10 scale. SNPs are colored (in red) by pairwise linkage disequilibrium (LD) pattern with the most strongly associated SNP. The regional association signal from a quantitative trait loci (QTL) study in specific cell populations (e.g., peripheral monocytes or macrophages) (middle panel), that may show expression, splicing and methylation QTL. The functional annotation of the genome with histone marks and the genomic position with the genes (bottom panel), e.g. H3K4me3 marks promoter regions, H3K27ac marks enhancer regions.
(B) Classification of SNPs in coding and non-coding regions by mechanism of action that may affect expression, splicing or protein function due to mutations, insertions and deletions. The coding variants change protein sequence. The non-coding variants may influence protein levels by modulating transcription factor binding at the intronic/distal enhancer or promoter regions, changing histone methylation and acetylation, splicing, miRNA, long noncoding RNA binding and stability, or structural variation. TF: transcription factor. UTR: untranslated region.
(C) Gene co-expression network and pathway analysis using large-scale transcriptomic or proteomic datasets.
(D) Endophenotypes relevant to AD: amyloid-β (Aβ) plaque load measured with positron emission tomography (PET) tracer, Aβ42 and tau/p-tau181 levels in cerebrospinal fluid (CSF), neuropathological changes.
(E) Functional validation of genetic hits using genome editing tools (e.g. CRISPR/Cas9) in cell culture and mouse models.
(F) Integrating AD GWAS with functional genomics approaches can help prioritize candidate genes, biological pathways and cell types, which in turn can help generate novel hypothesis for experimental validation. Genes listed are discussed in the review.
It is the goal of functional genomics to make sense of genomic and transcriptomic data to uncover the mechanisms underlying SNP associations with disease. Foremost, association of genetic variation with AD endophenotypes that characterize disease progression and correlate with pathology can help prioritize SNPs that modify AD risk. As such, cerebrospinal fluid (CSF) levels of Aβ42 and tau/p-tau181, and pathological traits in brain tissue such as plaque and tangle density, brain atrophy and cognitive impairment have been used as quantitative traits in genetic association analyses (36). For example, APOE4 genotype is the strongest marker associated with accelerated grey matter atrophy as well as lower Aβ42 and higher tau/p-tau181 levels in CSF (37–39).
Genomic regions where differences in gene expression are associated with SNP genetic variation are named expression quantitative trait loci (QTL) (40). Gene expression has been analyzed in normal human tissue (GTEx Consortium (41)), regional brain tissue (BRAINEAC database (42)) and prefrontal cortex of aging and demented people (ROS/MAP project (43, 44)). The disadvantage of these datasets is that tissues are not homogenous and thus underrepresented cell populations may be beyond the resolution of current datasets. Indeed, attempts to identify brain tissue eQTLs corresponding to AD GWAS loci have not produced compelling associations, with marginal results that do not replicate across datasets. Analyses of primary cell-type specific expression from the Immune Variation (ImmVar) project have shown that AD susceptibility alleles are enriched among eQTLs in monocytes, but not T cells (45). Based on this observation, evaluation of eQTLs in primary cells at baseline and under stimulated conditions in patient samples may help decipher the causal relationship between genetic and phenotypic variation.
Analyses of genomic sequence can provide information to categorize functional SNPs if found in regulatory regions (Figure 1), which include any of the elements involved in transcription and translation, such as enhancers, promoters, untranslated regions, introns, histone marks, etc. and lead to changes in chromatin state causing changes in expression or mRNA splicing captured by expression, splicing and methylation QTL (46, 47). AD-related methylation changes have been detected near known GWAS genes ABCA7 and BIN1 and novel genes ANK1, RHBDF2, CDH23 and RPL13 (48, 49). A study of chromatin state alterations in human samples found an upregulation of immune response genes and regulatory regions that are targeted by SPI1, a myeloid specific transcription factor (50). Furthermore, protein QTL can be used to map loci that affect protein abundance, which when coupled with GWAS can reveal networks of protein-protein interactions (51). Other epigenomic datasets are being generated by consortia such as PsychENCODE (52), the NIH ROADMAP Epigenomics (53), BLUEPRINT Epigenome (54), Accelerating Medicines Partnership for AD (AMP-AD) (55) and CommonMind (56), and will facilitate large-scale integrative functional genomics analyses.
A complementary approach to functional genomics uses systems biology to infer multi-scale networks, which are effective in identifying co-expressed gene modules enriched in functional categories. These gene modules can be used to generate hypotheses for further experimental testing. However, prior knowledge is rarely verified experimentally and annotations lack context- and cell-specific functions of each gene, thus prohibiting modeling of dynamic processes, such as disease progression. Analyses of networks in samples from AD patients versus control individuals revealed differentially regulated nodes of immune-related genes, governed by TYROBP (57), an adaptor protein DAP12 expressed in microglia that is required for TREM2 signaling. Whole genome sequencing in patients with sporadic early-onset AD has identified rare coding variants in TYROBP that perturb expression levels of TREM2 and TYROBP in vitro (58), confirming the significance of this module in AD risk. A proteomic study of cortical tissue from AD patients reported enrichment of AD GWAS candidates in microglial protein networks, supporting a causal role for myeloid cells in AD (59).
While GWAS enable the identification of common variants, usually with small effect size, other approaches are needed to identify rare variants. The most commonly used approaches are whole exome (WES) and whole genome sequencing (WGS). WGS provides the most comprehensive survey of the genome including regulatory regions not covered by WES. Like GWAS these studies may be performed in large unrelated cohorts, in isolated populations or in families. When studying rare variants, one advantage of families is that a rare variant discovered in one family member will be enriched in the remaining family members allowing analysis of segregation with disease. Most of the published WES/WGS studies have been relatively small and have focused on families/isolated populations. deCODE Genetics has used WGS in the Icelandic population to identify rare variants in APP (60), TREM2 (61, 62) and ABCA7 (63) that influence risk of AD. The TREM2 (64, 65) and ABCA7 (66, 67) findings have been widely replicated. Other studies using late onset AD families have identified PLD3 (68), UNC5C (69) and AKAP9 (70), but these await replication in larger cohorts.
Analysis of GWAS and gene expression data has highlighted four pathways enriched for AD association: cholesterol metabolism, immune response, regulation of endocytosis and protein ubiquitination (71). Below we discuss a selection of genes that fall into these categories reviewing experimental evidence for their contribution to AD (Table 1).
Table 1. Overview of genomic risk loci and genes with common and rare variants identified through Alzheimer's disease (AD) linkage studies, genome-wide association studies (GWAS) and whole genome /whole exome sequencing (WGS/WES).
Identification of a functional variant in a specific causal gene has only been established for a few loci. In those cases the putative gene and variant are named in the table with additional information about the cell type-specific gene expression and related biological processes. The odds ratio from a case-control study tells about the association of disease outcome with the presence of certain genotype: increased (risk) or decreased (protection) chances of getting the disease for an individual with one risk allele versus having no risk alleles. Adjacent genes were selected with dbSNP database by searching for the tagging SNP. Definitive conclusion about SNP association requires replication in independent datasets. QTL: quantitative trait loci.
| Chr | SNPs | Closest gene name | Genes adjacent to the SNP | Putative gene with functional significance | Cell type-specific express ion of putative gene | Biological processes | Odds ratio meta-analysis for minor allele with 95% confidence interval (CI) (common/ rare variant) | References | |
|---|---|---|---|---|---|---|---|---|---|
| Loci | |||||||||
| Chr1 | rs6656401 | CR1 | CR2, CR1, CR1L | CR1 intragenic copy number variations leading to longer isoform overproduction | Microglia | Immune response and phagocytosis | 1.18 (1.14–1.22) | (32) | |
| Chr2 | rs6733839 | BIN1 | BIN1, CYP27C1 | BIN13 base pair insertion | Ubiquitous | Endocytosis | 1.22 (1.18– 1.25) | (32) | |
| Chr2 | rs35349669 | INPP5D | NEU2, INPP5D, ATG16L1 | / | / | / | 1.08 (1.05– 1.11) | (32) | |
| Chr5 | rs2074612 | HBEG F | CYSTM1 , PFDN1, HBEGF | / | / | / | 1.08 (1.05– 1.11) | (138, 139) | |
| Chr5 | rs190982 | MEF2 C | TMEM161B, MIR9-2, LINC00461, MEF2C, MEF2C-AS1 | / | / | / | 0.93 (0.90– 0.95) | (32) | |
| Chr6 | rs10948363 | CD2AP | TNFRSF21, CD2AP, ADGRF2, ADGRF4, OPN5 | / | / | / | 1.10 (1.07– 1.13) | (32) | |
| Chr6 | rs9271192 | HLA-DRB1 / HLA-DRB5 | C6orf10, BTNL2, HLA-DRA, HLA-DRB5, HLA-DRB6, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DQA2, HLA-DQB2, HLA-DOB | / | / | / | 1.11 (1.08– 1.15) | (32) | |
| Chr7 | rs11771145 | EPHA1 | ZYX, EPHA1, EPHA-AS1, TAS2R62P, TAS2R60 | / | / | / | 0.90 (0.88– 0.93) | (32) | |
| Chr7 | rs2718058 | NME8 | GPR141 , NME8, SFRP4, EPDR1 | / | / | / | 0.93 (0.90– 0.95) | (32) | |
| Chr7 | rs1476679 | ZCWP W1 | GPC2, STAG3, PVRIG, GATS, SPDYE3 , PILRB, PILRA, ZCWPW1, MEPCE, C7orf61 | PILRB expression QTL | Microglia | Inflammatory signal transduction | 0.91 (0.89– 0.94) | (32, 141) | |
| Chr8 | rs9331896 | CLU | EPHX2, CLU, SCARA3 | CLU rare coding variants and insertions / deletions | Astrocytes | Lipid metabolis m | 0.86 (0.84– 0.89) | (32) | |
| Chr8 | rs28834970 | PTK2 B | TRIM35, PTK2B, CHRNA2 | / | / | / | 1.10 (1.08– 1.13) | (32) | |
| Chr10 | rs7920721 | ECHDC3 | USP6NL , ECHDC3, PROSE R2 | / | / | / | 1.07 (1.04– 1.10) | (138, 139) | |
| Chr11 | rs10838725 | CELF1 | DDB2, ACP2, NR1H3, MADD, MYBPC3, SPI1, SLC39A13, PSMC3, RAPSN, CELF1, PTPMT1, KBTBD4, NDUFS3, FAM180B, C1QTN F4, MTCH2, AGBL2, FNBP4, NUP160 | SPI1 expression QTL | Myeloid cells | Myeloid lineage determination | 1.08 (1.05– 1.11) | (32, 136) | |
| Chr11 | rs983392 | MS4A6A | OOSP2, MS4A3, MS4A2, MS4A6A , MS4A4E , MS4A4A , MS4A6E , MS4A7, MS4A14 , MS4A5, MS4A1 | MS4A4A, MS4A6A expression QTL | Microglia | Chemosensory receptors | 0.90 (0.87– 0.92) | (32, 136) | |
| Chr11 | rs10792832 | PICAL M | CCDC83, PICALM , EED | / | / | / | 0.87 (0.85– 0.89) | (32) | |
| Chr11 | rs11218343 | SORL1 | SORL1 | SORL1 rare and common variants | Ubiquitous | Endocytosis and sorting | 0.77 (0.72– 0.82) / 5.03 (2.02– 14.99) | (32, 142) | |
| Chr14 | rs17125944 | FERM T2 | ERO1A, PSMC6, STYX, GNPNAT1, FERMT2 | / | / | / | 1.14 (1.09– 1.19) | (32) | |
| Chr14 | rs10498633 | SLC24 A4 / RIN3 | SLC24A4, RIN3, LGMN | / | / | / | 0.91 (0.88– 0.94) | (32) | |
| Chr15 | rs59685680 | SPPL2 A | USP8, USP50, TRPM7, SPPL2A | / | / | / | 0.92 (0.89– 0.95) | (138) | |
| Chr15 | rs74615166 | TRIP4 | CSNK1G1, KIAA0101, TRIP4, ZNF609 | / | / | / | 1.31 (1.19– 1.44) | (143) | |
| Chr17 | rs2632516 | BZRA P1-AS1 | TSPOAP1, BZRAP1 -AS1, SUPT4H1, RNF43 | / | / | / | 0.92 (0.91– 0.94) | (139) | |
| Chr17 | rs77493189 | SCIM P | ZFP3, ZNF232, USP6, ZNF594, SCIMP, RABEP1 , NUP88, RPAIN, C1QBP, DHX33 | / | / | / | 1.11 (1.07– 1.15) | (138) | |
| Chr18 | rs8093731 | DSG2 | DSG3, DSG2, DSG2-AS1 | / | / | / | 0.73 (0.62– 0.86) | (32) | |
| Chr19 | rs4147929 | ABCA7 | CNN2, ABCA7, HMHA1, POLR2E | ABCA7 rare and common loss-of-function mutations and deletions | Ubiquitous | Lipid metabolism and phagocytosis | 1.15 (1.11– 1.19) / 2.81 (1.89– 4.20) | (32, 86) | |
| Chr19 | rs429358 , rs7412 | APOE | APOE | ε4 genotype | Ubiquitous, major in astrocytes and microglia | Lipid metabolism and phagocytosis | ~3–12 | (72, 73) | |
| Chr19 | rs3865444 | CD33 | SIGLEC9, SIGLEC7, CD33, SIGLECL1 | CD33 alternative splicing of IgV domain | Microglia | Immune response and phagocytosis | 0.94 (0.91– 0.96) | (32) | |
| Chr20 | rs7274581 | CASS4 | AURKA, CSTF1, CASS4, RTFDC1, GCNT7 | / | / | / | 0.88 (0.84– 0.92) | (32) | |
| Genes | |||||||||
| Chr4 | rs137875858 | UNC5 C | UNC5C | UNC5C T835M |
Neuron s | Response to neurotoxic stimuli, cell death | / | (69) | |
| Chr6 | rs75932628 | TREM2 | TREM2 | TREM2 R47H |
Microglia | Immune response and phagocytosis | ~3–5 | (61, 62) | |
| Chr6 | rs3747742 | TREML2 | TREML2 | TREML2 S144G |
Microglia | Immune response | 0.91 (0.86– 0.97) | (144) | |
| Chr7 | rs144662445, rs149979685 | AKAP9 | AKAP9 | AKAP9 I2546M, S3767L |
Ubiquitous | Kinase signaling | 3.61 (1.51– 9.00) | (70) | |
| Chr15 | rs61751103, rs145518263 | ADAM10 | ADAM10 | ADAM10 Q170H, R181G |
Ubiquit ous | APP processing to Aβ | / | (24) | |
| Chr16 | rs72824905 | PLCG2 | PLCG2 | PLCG2 P522R |
Microglia | Phospholipase signaling | 0.68 (CI not reported) | (137) | |
| Chr17 | rs616338 | ABI3 | ABI3 | ABI3 S209F |
Microglia | Cell growth | 1.43 (CI not reported) | (137) | |
| Chr19 | rs145999145 | PLD3 | PLD3 | PLD3 V232M |
Ubiquit ous | Unknown | 2.75 (2.05– 3.68) | (68) | |
| Chr21 | rs63750847 | APP | APP | APP A673T |
Ubiquit ous | APP processing to Aβ | ~0.2 | (60) | |
Lipid Metabolism
Apolipoprotein E (APOE) is the most important genetic AD risk factor influencing prevalence and age-at-onset. APOE association was originally identified from linkage studies and explains 15–20% of AD heredity. Two coding SNPs define six APOE genotypes – ε2/ε2, ε2/ε3, ε3/ε3, ε2/ε4, ε3/ε4, ε4/ε4 listed from lowest to highest risk for AD (72, 73). APOE is the major apolipoprotein expressed in human brain primarily by astrocytes, is involved in cholesterol homeostasis and has been extensively studied in AD (74). APOE influences Aβ plaque load in an isoform-specific manner in APP transgenic mice, with highest amyloid-β deposition in human knock-in APOE4 genotype lines compared to APOE3 and APOE2 (75). This effect can be explained by decreased Aβ clearance and/or facilitation of Aβ fibrillogenesis that is due to isoform-dependent differences, because APOE4 shows lower binding of Aβ and is degraded more rapidly through lipoprotein receptors (74, 76, 77). APOE contributes to synapse pruning by astrocytes (78) and together with Clusterin (CLU) is induced after injury in astrocytes and microglia promoting neuronal survival (74). CLU is primarily expressed in astrocytes and is involved in lipid transport, apoptosis and immune response. The minor allele of rs1113600, located in the intron of CLU is associated with reduced AD risk (28, 29), however, no eQTL was found in the locus (79).
CLU can bind Aβ and influence fibril formation in vitro. Deficiency of either APOE or CLU in APP transgenic mice does not affect Aβ deposition, but significantly reduces fibrillar Aβ in brain. Interestingly, double APOE and CLU knock-out increases plaque load, possibly through reduced clearance of Aβ in brain parenchyma corroborated by higher Aβ levels in CSF (80). The risk allele of rs1113600 has been associated with lower white matter integrity (81) and reduced connectivity between hippocampus and frontal cortex (82) in healthy individuals. Targeted resequencing of CLU in one study identified rare coding variations in the β-chain that were enriched in AD patients independent from rs1113600 association (83). These non-synonymous mutations and small insertion-deletions were subsequently shown to be associated with altered cellular localization and diminished extracellular secretion of CLU (84).
ATP-binding cassette transporter A7 (ABCA7) is universally expressed in brain and involved in lipid transport modulating lipid efflux. ABCA7 is a highly replicated genetic risk factor for AD in individuals of European and African American ancestry (30, 32, 34). Deep sequencing in African Americans carrying risk and protective alleles at ABCA7 led to the identification of a 44 base pair deletion in LD with the high risk allele, which results in a frameshift (85). Other analyses in European descent cohorts have identified several rare variants in ABCA7 resulting in frameshift mutations and deletions, missense or splicing site variants that are enriched in AD cases, presumably leading to early stop codons and loss-of-function alleles (63, 86). Overexpression of ABCA7 potentiates phagocytosis in macrophages (87) and decreases neuronal APP processing in vitro (88). ABCA7 deficiency accelerates amyloid-β deposition in APP-J20 (89) or APP/PS1 (90) mouse models of amyloidosis without effect on cognition, and in humans loss-of-function alleles are associated with cortical and hippocampal atrophy (91).
Regulation of Endocytosis
Cellular trafficking has long been implicated in AD pathogenesis corroborated by the association of the sortilin-related receptor L (SORL1) with AD in case-control studies (92). LOAD GWAS identified rs11218343, a common variant in SORL1 in European (32) and Asian populations (35). Rare variants in SORL1 were also found in several families with autosomal dominant early-onset AD (93). Overexpression of SORL1 in cell lines reduces Aβ production through increased retention of APP in the Golgi (94), while overexpression of the AD associated SORL1-G511R variant results in decreased binding and turnover of Aβ (95). Ablation of SORL1 in APP/PS1 mice leads to increased plaque deposition, similar to the effect of SORL1 KO on endogenous murine Aβ production (96).
Bridging integrator 1 (BIN1) participates in the endocytic trafficking of synaptic vesicles through membrane remodeling in neurons (97). The index SNP rs6733839 in the BIN1 locus has been associated with AD risk in different populations (32, 98, 99). Fine-mapping of the BIN1 locus identified rs59335482, a 3 base pair insertion ~28 kb upstream of BIN1, that is associated with higher AD risk, increased transcriptional activity in vitro using a luciferase assay, and higher BIN1 levels (97). However, contrary evidence demonstrated that knock-down of BIN1 increases tau aggregation in neurons through an enlargement of Rab5-positive vesicles (100), and reduces lysosomal degradation of BACE1 thereby increasing Aβ production (101). Since BIN1 is largely expressed in mature oligodendrocytes and white matter (102), it is unclear how it could affect AD pathology in neurons.
CD2-associated protein (CD2AP) is an adaptor and scaffolding protein, and its locus is an AD risk factor identified through association of an intronic variant rs10948363 (27, 30, 32). CD2AP affects APP endocytosis in neurons, but shows a mild effect on Aβ levels in vitro and no effect on plaque load in APP/PS1 mice with CD2AP haploinsufficiency (103, 104). CD2AP is expressed in endothelial cells and ablation of CD2AP in mice leads to reduced blood-brain barrier integrity, suggesting its contribution to AD may be APP independent (105).
Phosphatidylinositol-binding clathrin assembly gene (PICALM) is nominally associated with AD through protective variants rs10792832 and rs3851179, which are located 40 kb upstream of PICALM, which is the nearest gene (29, 32). Studies of PICALM in neuronal cells show that it regulates cleaved APP C-terminal fragment degradation via autophagosomes (106) and clathrin-mediated endocytosis of gamma-secretase (107). However, the effect on Aβ levels in vitro and amyloid-β load in vivo is variable and may depend on the level of APP overexpression (106, 108, 109). Interestingly, PICALM is reduced in brain endothelium from AD patients, and PICALM haploinsufficiency in an AD mouse model led to a reduction in Aβ clearance through the blood-brain barrier and concomitant increase in amyloid-β load (110).
These genes suggest a defect in synaptic function, further supported by the association of MEF2C and PTK2B loci with AD progression (32). MEF2C and PTK2B are reported to be involved in regulation of hippocampal synapses and long-term depression, respectively (111, 112), although the functional consequences of variation in these loci awaits validation. One should interpret these data with caution, as validation was mostly performed in cell lines or neurons in relation to APP and secretase trafficking. However, SORL1, BIN1, CD2AP and PICALM gene expression is substantial in microglia (113), and in the context of AD defective internalization of Aβ, APOE, CLU and tau has been reported.
Immune Response
The third group of genes involved in AD pathogenesis based on genetic studies belongs to immune system pathways. Complement receptor 1 (CR1) is a highly replicated AD risk factor associated with an intronic SNP rs6656401 but also in high LD with 2 other genes of the complement family (28, 32). CR1 is expressed on blood cells and microglia, and its function is to inhibit complement activation through C3b and C4b (114). The complement system is activated in AD (115), while in mice challenged with oligomeric Aβ it leads to increased synapse elimination by phagocytic microglia (116). Intragenic copy number variation in CR1 is associated with AD risk, which functionally results in overproduction of the longer CR1 isoform increasing the number of C3b/C4b sites, which might explain the AD risk in this locus (117).
CD33 is a receptor for sialic acid-modified proteins that is found on myeloid and microglia cells and is involved in the anti-inflammatory immune response. Association of CD33 with AD risk was identified by GWAS through rs3865444 located in the promoter region (27, 30). Although CD33 was only suggestively associated with AD risk in the largest meta-analysis of AD GWAS (i.e. not genome-wide significant) (32), there is strong evidence from functional studies that changes in CD33 expression affect Aβ levels in vivo. The exon 2 polymorphism rs12459419 in tight LD with rs3865444 has been proposed to be the causal variant modulating alternative splicing of CD33 (118, 119). Increased inclusion of exon 2 in the presence of the rs3865444 risk allele produces full length CD33 with an IgV domain, which likely mediates sialic acid binding leading to receptor activation. At baseline CD33 inhibited uptake of Aβ42 in mouse primary microglial cells and CD33 ablation in APP/PS1 mice alleviated plaque pathology (120). In accordance, monocytes from rs3865444 risk allele carriers show an increase in CD33 expression and reduced capacity to phagocytose Aβ, which correlates with an increase in brain Aβ load (121).
Inhibition of CD33 signaling decreases surface expression of triggering receptor 2 on myeloid cells (TREM2) (122). Several studies have reported that rs75932628, which results in the TREM2 missense variant (R47H), increases AD risk by about two-fold (61, 62). TREM2 was first identified in Nasu-Hakola disease patients, a rare recessive disorder associated with an frontotemporal dementia-like syndrome (123), making TREM2 a candidate gene for targeted sequencing in AD patients. TREM2 expression is increased in response to brain injury in AD (124, 125) and is found on both resident microglia and infiltrating monocytes and macrophages (126). Variants in TREM2 reduce transport and cell surface expression of the full length protein, thereby decreasing cell surface shedding and activity, which functionally results in a decrease of phagocytosis (127). TREM2 is a pattern recognition receptor that binds phospholipids, such as phosphatidylserine exposed on cells undergoing apoptosis (128), as well as APOE- and Clusterin-containing lipoprotein particles (129, 130), promoting phagocytosis of Aβ complexed in these lipoprotein particles (131). Deficiency of TREM2 in AD mouse models affects amyloid-β deposition in a temporal manner resolving pathology at early stages, but showing aggravated plaque load and impairment in microglia viability, proliferation and migration in aged mice (128, 132).
There are other loci supporting the role of microglial/myeloid cells in AD, however, more work is required to establish their functional significance in AD risk and disease progression. For example, the MS4A6A locus (rs983392 (32)) including 5 other members of the MS4A gene family, which are specifically expressed in microglia and regulate cell activation (133). The HLA-DRB1/HLA-DRB5 locus containing 9 genes (rs9271192 (32)) of the major histocompatibility complex II family are involved in immunity. The ZCWPW1 locus contains 7 genes (rs1476679 (32)) including PILRA and PILRB immune receptors involved in monocyte and neutrophil infiltration and response during inflammation (134, 135). A recent study that used the fine-mapping approach discussed above to dissect the causal gene in the CELF1 locus, which includes 13 genes (rs10838725 (32)), reports identification of SPI1 as a master regulator of endophenotypes and genes associated with AD (136). A large meta-analysis of exome chip data has also identified novel microglia-expressed genes associated with AD risk, ABI3 and PLCG2 (137).
Future Directions
Although some progress has been made in understanding the underlying pathogenic mechanisms associated with GWAS loci, this needs to be a major focus of future research using methods outlined in this review (Figure 1). If GWAS samples are enlarged, it is clear from other phenotypes that additional loci will be identified. Two recent large meta-analyses of AD samples have indeed identified additional risk loci (138, 139). A lesson from GWAS is that we need to be bold – large datasets will be needed to find strong evidence for individual genes/variants, identified in whole genome/exome sequencing projects. Although novel large effect size associations with AD provide valuable mechanistic insight into disease pathogenesis, association signals should be carefully assessed for the frequency, directionality and effect size that may change dependent on the methods used for patient stratification and variant identification by sequencing or genotyping.
Definitive verification of functional variants will come from in vivo and in vitro functional studies using mouse models and hiPSc-derived neurons, astrocytes and microglia cells that enable us to model sporadic AD in relevant cell types and predict therapeutic interventions based on mechanism. Functional non-coding AD variants can be tested using genome editing tools, such as CRISPR/Cas9 (140), that offer controlled genetic background to dissect the effect size. Evaluation of GWAS variants with protective effects may provide additional information about the pathways that can counteract disease. For example, a protective APP-A673T mutation identified in the Icelandic population leads to decreased Aβ production in vitro compared to APP-A673V, a LOAD mutant at the same amino acid that increases Aβ production (60). It remains to be shown if AD protective variants act by reversing detrimental phenotypes or boosting cell activity to counteract the pathology, which may have implications for designing AD therapies.
Conclusions
The application of functional genomics approaches will finally provide focus for researchers bombarded with the wealth of information from GWAS, transcriptome, proteome and metabolome studies in AD cohorts. Although we may not see an expansion of the number of GWAS common variants associated with AD, whole exome/genome sequencing in specific cohorts will lead the way for discovery of new AD-associated genes. Understanding the mechanisms underlying LOAD genes has shifted our attention from β-amyloid metabolism to other cellular pathways and the contribution of myeloid cell function in AD pathogenesis. Characterization of functional AD-associated variants will broaden our understanding of mechanisms underlying AD progression that is now studied in the context of cell-cell interactions of the brain. In the future it will be important to see how risk variants align to cell specific pathways and predict master regulators of protein hubs that are dysfunctional in AD in order to develop novel therapies.
Acknowledgments
The authors thank the thousands of subjects who have participated in genetic research, advancing our knowledge of the mechanisms underlying disease risk. We thank Ishaan Gupta and Alan Renton for helpful comments. We also acknowledge the following funding sources: the JPB Foundation (AG), NIH (U01AG049508, U01AG052411) (AG), and the Alzheimer’s Association (TR).
Footnotes
Financial Disclosures A.A.P. and T.R. report no biomedical financial interests or potential conflicts of interest. A.M.G. is on the scientific advisory board for Denali Therapeutics and has served as a consultant for AbbVie and Cognition Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Holtzman DM, Morris JC, Goate AM. Alzheimer’s Disease: The Challenge of the Second Century. Sci Transl Med. 2011;3:77. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gómez-Isla T, Price JL, McKeel DW, Morris JC, Growdon JH, Hyman BT. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci. 1996;16:4491–500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray C, Viehman A, Lippa CF. The corpus callosum in Pick’s disease , Alzheimer’s disease , and amyotrophic lateral sclerosis: Gliosis implies possible clinical consequence. Am J Alzheimers Dis Other Demen. 2006;21:37–43. doi: 10.1177/153331750602100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu C, Kivipelto M, Von Strauss E. Epidemiology of Alzheimer’s disease: Occurrence, determinants, and strategies toward intervention. Dialogues Clin Neurosci. 2009;11:111–128. doi: 10.31887/DCNS.2009.11.2/cqiu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 6.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–60. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 7.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Pettingell WH, Yu C, et al. Candidate Gene for the Chromosome 1 Familial Alzheimer’s Disease Locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 8.Rogaev EI, Sherrington R, Rogaeva Ea, Levesque G, Ikeda M, Liang Y, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 9.Guerreiro RJ, Gustafson DR, Hardy J. The genetic architecture of Alzheimer’s disease: Beyond APP, PSENS and APOE. Neurobiol Aging. 2012;33:437–456. doi: 10.1016/j.neurobiolaging.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8:1–14. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 12.Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One. 2009;4:1–6. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, et al. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 14.Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–6. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 15.Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 16.Zhou L, Brouwers N, Benilova I, Vandersteen A, Mercken M, Van Laere K, et al. Amyloid precursor protein mutation E682K at the alternative beta-secretase cleavage beta’site increases Abeta generation. EMBO Mol Med. 2011;3:291–302. doi: 10.1002/emmm.201100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benilova I, Karran E, De Strooper B. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 2012;15:349–57. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 18.Saito T, Suemoto T, Brouwers N, Sleegers K, Funamoto S, Mihira N, et al. Potent amyloidogenicity and pathogenicity of Aβ43. Nat Neurosci. 2011;14:1023–1032. doi: 10.1038/nn.2858. [DOI] [PubMed] [Google Scholar]
- 19.Chávez-Gutiérrez L, Bammens L, Benilova I, Vandersteen A, Benurwar M, Borgers M, et al. The mechanism of -Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–74. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretner B, Trambauer J, Fukumori A, Mielke J, Kuhn P-H, Kremmer E, et al. Generation and deposition of Aβ43 by the virtually inactive presenilin-1 L435F mutant contradicts the presenilin loss-of-function hypothesis of Alzheimer’s disease. EMBO Mol Med. 2016;8:1–8. doi: 10.15252/emmm.201505952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, et al. The presenilin-1 dE9 mutation results in reduced gamma-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep. 2013;5:974–985. doi: 10.1016/j.celrep.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun L, Zhou R, Yang G, Shi Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Aβ42 and Aβ40 peptides by -secretase. Proc Natl Acad Sci. 2016;114:E476–E485. doi: 10.1073/pnas.1618657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolaou M, Song YQ, Sato CA, Orlacchio A, Kawarai T, Medeiros H, et al. Mutations in the open reading frame of the beta-site APP cleaving enzyme (BACE) locus are not a common cause of Alzheimer’s disease. Neurogenetics. 2001;3:203–206. doi: 10.1007/s100480100123. [DOI] [PubMed] [Google Scholar]
- 24.Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, et al. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate alpha-secretase activity. Hum Mol Genet. 2009;18:3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suh J, Choi S, Romano DM, Gannon MA, Lesinski AN, Kim DY, Tanzi RE. ADAM10 missense mutations potentiate beta-Amyloid accumulation by impairing prodomain chaperone function. Neuron. 2013;80:385–401. doi: 10.1016/j.neuron.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert J, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 29.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jun G, Naj AC, Beecham GW, Wang L-S, Buros J, Gallins PJ, et al. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karch CM, Cruchaga C, Goate AM. Alzheimer’s disease genetics: from the bench to the clinic. Neuron. 2014;83:11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reitz C, Wang L, Lin C, Larson EB, Graff-radford NR, Evans D, et al. Variants in the ATP-Binding Cassette Transporter (ABCA7), Apolipoprotein E4, and the Risk of Late-Onset Alzheimer Disease in African Americans. J Am Med Assoc. 2013;309:1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spampinato M, Rumboldt Z, Hosker R, Mintzer J Alzheimer’s Disease Neuroimaging Initiative. Apolipoprotein E and gray matter volume loss in patients with mild cognitive impairment and Alzheimer disease. Radiology. 2011;258:843–852. doi: 10.1148/radiol.10100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruchaga C, Kauwe JSK, Harari O, Jin SC, Cai Y, Karch CM, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for alzheimer’s disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schott JM, Crutch SJ, Carrasquillo MM, Uphill J, Shakespeare TJ, Ryan NS, et al. Genetic risk factors for posterior cortical atrophy. Alzheimer’s Dement. 2015;11:P168–P169. doi: 10.1016/j.jalz.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187:367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–28. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett DA, Wilson RS, Arvanitakis Z, Boyle PA, De Toledo-Morrell L, Schneider JA. Selected findings from the religious orders study and rush memory and aging project. J Alzheimer’s Dis. 2013:S397–S403. doi: 10.3233/JAD-2012-129007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–23. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li YI, Van De Geijn B, Raj A, Knowles DA, Petti AA, Golan D, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 48.De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014;17:1164–70. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai L-H, Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–9. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Daniel M. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–1712. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Ge B, Casale FP, Vasquez L, Kwan T, Garrido-Martin D, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398–1414. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaiteri C, Mostafavi S, Honey CJ, De Jager PL, Bennett DA. Genetic variants in Alzheimer disease - molecular and brain network approaches. Nat Rev Neurol. 2016;12:413–427. doi: 10.1038/nrneurol.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pottier C, Ravenscroft TA, Brown PH, Finch NA, Baker M, Parsons M, et al. TYROBP genetic variants in early-onset Alzheimer’s disease. Neurobiol Aging. 2016;48:222.e9–222.e15. doi: 10.1016/j.neurobiolaging.2016.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, et al. A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s Disease. Cell Syst. 2016;4:1–13. doi: 10.1016/j.cels.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–9. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 61.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2012;368:107–16. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinberg S, Stefansson H, Jonsson T, Johannsdottir H, Ingason A, Helgason H, et al. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat Genet. 2015;47:445–7. doi: 10.1038/ng.3246. [DOI] [PubMed] [Google Scholar]
- 64.Jin SC, Benitez BA, Karch CM, Cooper B, Skorupa T, Carrell D, et al. Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum Mol Genet. 2014;23:5838–5846. doi: 10.1093/hmg/ddu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pottier C, Wallon D, Rousseau S, Rovelet-Lecrux A, Richard AC, Rollin-Sillaire A, et al. TREM2 R47H variant as a risk factor for early-onset Alzheimer’s disease. J Alzheimer’s Dis. 2013;35:45–49. doi: 10.3233/JAD-122311. [DOI] [PubMed] [Google Scholar]
- 66.Vardarajan BN, Ghani M, Kahn A, Sheikh S, Sato C, Barral S, et al. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol. 2015;78:487–498. doi: 10.1002/ana.24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cuyvers E, De Roeck A, Van den Bossche T, Van Cauwenberghe C, Bettens K, Vermeulen S, et al. Mutations in ABCA7 in a Belgian cohort of Alzheimer’s disease patients: A targeted resequencing study. Lancet Neurol. 2015;14:814–822. doi: 10.1016/S1474-4422(15)00133-7. [DOI] [PubMed] [Google Scholar]
- 68.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2015;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wetzel-Smith MK, Hunkapiller J, Bhangale TR, Srinivasan K, Maloney JA, Atwal JK, et al. A rare mutation in UNC5C predisposes to late-onset Alzheimer’s disease and increases neuronal cell death. Nat Med. 2014;20:1452–1457. doi: 10.1038/nm.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Logue MW, Schu M, Vardarajan BN, Farrell J, Bennett DA, Buxbaum JD, et al. Two rare AKAP9 variants are associated with Alzheimer’s disease in African Americans. Alzheimers Dement. 2014;10:609–618. doi: 10.1016/j.jalz.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones L, Lambert JC, Wang LS, Choi SH, Harold D, Vedernikov A, et al. Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimer’s Dement. 2015;11:658–671. doi: 10.1016/j.jalz.2014.05.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 73.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–81. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fryer JD, Simmons K, Parsadanian M, Bales K, Paul SM, Sullivan PM, Holtzman DM. Human apolipoprotein E4 alters the Amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, et al. Human apoE isoforms differentially regulate brain Amyloid-beta peptide clearance. Sci Transl Med. 2011;3:89ra57-89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chung W-S, Verghese PB, Chakraborty C, Joung J, Hyman BT, Ulrich JD, et al. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc Natl Acad Sci. 2016;113:10186–10191. doi: 10.1073/pnas.1609896113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guerreiro RJ, Beck J, Gibbs JR, Santana I, Rossor MN, Schott JM, et al. Genetic variability in CLU and its association with Alzheimer’s disease. PLoS One. 2010;5:e9510. doi: 10.1371/journal.pone.0009510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, et al. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- 81.Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, et al. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011;31:6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Erk S, Meyer-Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, Kirsch P, et al. Hippocampal function in healthy carriers of the CLU Alzheimer’s disease risk variant. J Neurosci. 2011;31:18180–18184. doi: 10.1523/JNEUROSCI.4960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bettens K, Brouwers N, Engelborghs S, Lambert J-C, Rogaeva E, Vandenberghe R, et al. Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk. Mol Neurodegener. 2012;7:3. doi: 10.1186/1750-1326-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bettens K, Vermeulen S, Van Cauwenberghe C, Heeman B, Asselbergh B, Robberecht C, et al. Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations. Mol Neurodegener. 2015;10:30. doi: 10.1186/s13024-015-0024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cukier HN, Kunkle BW, Vardarajan BN, Rolati S, Hamilton-Nelson KL, Kohli MA, et al. ABCA7 Frameshift Deletion Associated with Alzheimer’s Disease in African Americans. Neurol Genet. 2016;2:e79. doi: 10.1212/NXG.0000000000000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Guennec K, Quenez O, Charbonnier C, Wallon D, Grenier-Boley B, Richard A, et al. ABCA7 rare variants and Alzheimer disease risk. Neurology. 2016;86:2134–7. doi: 10.1212/WNL.0000000000002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jehle AW, Gardai SJ, Li S, Linsel-Nitschke P, Morimoto K, Janssen WJ, et al. ATP-binding cassette transporter A7 enhances phagocytosis of apoptotic cells and associated ERK signaling in macrophages. J Cell Biol. 2006;174:547–556. doi: 10.1083/jcb.200601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan SL, Kim WS, Kwok JB, Hill AF, Cappai R, Rye KA, Garner B. ATP-binding cassette transporter A7 regulates processing of amyloid precursor protein in vitro. J Neurochem. 2008;106:793–804. doi: 10.1111/j.1471-4159.2008.05433.x. [DOI] [PubMed] [Google Scholar]
- 89.Kim WS, Li H, Ruberu K, Chan S, Elliott DA, Low JK, et al. Deletion of Abca7 increases cerebral amyloid-β accumulation in the J20 mouse model of Alzheimer’s disease. J Neurosci. 2013;33:4387–94. doi: 10.1523/JNEUROSCI.4165-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakae N, Liu C-C, Shinohara M, Frisch-Daiello J, Ma L, Yamazaki Y, et al. ABCA7 deficiency accelerates Amyloid-beta generation and Alzheimer’s neuronal pathology. J Neurosci. 2016;36:3848–3859. doi: 10.1523/JNEUROSCI.3757-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramirez LM, Goukasian N, Porat S, Hwang KS, Eastman JA, Hurtz S, et al. Common variants in ABCA7 and MS4A6A are associated with cortical and hippocampal atrophy. Neurobiol Aging. 2016;39:82–89. doi: 10.1016/j.neurobiolaging.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 92.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–77. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pottier C, Hannequin D, Coutant S, Rovelet-Lecrux A, Wallon D, Rousseau S, et al. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17:875–879. doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- 94.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–6. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Caglayan S, Takagi-Niidome S, Liao F, Carlo A-S, Schmidt V, Burgert T, et al. Lysosomal sorting of amyloid-β by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci Transl Med. 2014;6:223ra20. doi: 10.1126/scitranslmed.3007747. [DOI] [PubMed] [Google Scholar]
- 96.Dodson SE, Andersen OM, Karmali V, Fritz JJ, Cheng D, Peng J, et al. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer’s disease. J Neurosci. 2008;28:12877–86. doi: 10.1523/JNEUROSCI.4582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chapuis J, Hansmannel F, Gistelinck M, Mounier a, Van Cauwenberghe C, Kolen KV, et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–34. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lambert JC, Zelenika D, Hiltunen M, Chouraki V, Combarros O, Bullido MJ, et al. Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol Aging. 2011;32:756.e11–756.e15. doi: 10.1016/j.neurobiolaging.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 99.Wijsman EM, Pankratz ND, Choi Y, Rothstein JH, Faber KM, Cheng R, et al. Genome-wide association of familial late-onset Alzheimer’s disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet. 2011;7:e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Calafate S, Flavin W, Verstreken P, Moechars D. Loss of Bin1 Promotes the Propagation of Tau Pathology. Cell Rep. 2016;17:931–940. doi: 10.1016/j.celrep.2016.09.063. [DOI] [PubMed] [Google Scholar]
- 101.Miyagawa T, Ebinuma I, Morohashi Y, Hori Y, Young Chang M, Hattori H, et al. BIN1 regulates BACE1 intracellular trafficking and amyloid-β production. Hum Mol Genet Genet. 2016;25:2948–2958. doi: 10.1093/hmg/ddw146. [DOI] [PubMed] [Google Scholar]
- 102.De Rossi P, Buggia-Prévot V, Clayton BLL, Vasquez JB, van Sanford C, Andrew RJ, et al. Predominant expression of Alzheimer’s disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol Neurodegener. 2016;11:59. doi: 10.1186/s13024-016-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ubelmann F, Burrinha T, Salavessa L, Gomes R, Ferreira C, Moreno N, Guimas Almeida C. Bin1 and CD2AP polarise the endocytic generation of beta-amyloid. EMBO Rep. 2016;18:e201642738. doi: 10.15252/embr.201642738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liao F, Zhang TJ, Mahan TE, Jiang H, Holtzman DM. Effects of CD2-associated protein deficiency on amyloid-β in neuroblastoma cells and in an APP transgenic mouse model. Brain Behav Immun. 2015;47:163–171. doi: 10.1016/j.bbi.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cochran JN, Rush T, Buckingham SC, Roberson ED. The Alzheimer’s disease risk factor CD2AP maintains blood-brain barrier integrity. Hum Mol Genet. 2015;24:6667–74. doi: 10.1093/hmg/ddv371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer’s APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci. 2013;110:17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kanatsu K, Morohashi Y, Suzuki M, Kuroda H, Watanabe T, Tomita T, Iwatsubo T. Decreased CALM expression reduces Aβ42 to total Aβ ratio through clathrin-mediated endocytosis of -secretase. Nat Commun. 2014;5:3386. doi: 10.1038/ncomms4386. [DOI] [PubMed] [Google Scholar]
- 108.Kanatsu K, Hori Y, Takatori S, Watanabe T, Iwatsubo T, Tomita T. Partial loss of CALM function reduces Aβ42 production and amyloid deposition in vivo. Hum Mol Genet. 2016;25:3988–3997. doi: 10.1093/hmg/ddw239. [DOI] [PubMed] [Google Scholar]
- 109.Xiao Q, Gil SC, Yan P, Wang Y, Han S, Gonzales E, et al. Role of Phosphatidylinositol Clathrin Assembly Lymphoid-Myeloid Leukemia (PICALM) in intracellular Amyloid Precursor Protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem. 2012;287:21279–21289. doi: 10.1074/jbc.M111.338376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao Z, Sagare AP, Ma Q, Halliday MR, Kong P, Kisler K, et al. Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance. Nat Neurosci. 2015;18:978–87. doi: 10.1038/nn.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barbosa AC, Kim M-S, Ertunc M, Adachi M, Nelson ED, McAnally J, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–6. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hsin H, Kim M, Wang C, Sheng M. Proline-rich tyrosine kinase 2 regulates hippocampal long-term depression. J Neurosci. 2010;30:11983–11993. doi: 10.1523/JNEUROSCI.1029-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crehan H, Hardy J, Pocock J. Blockage of CR1 prevents activation of rodent microglia. Neurobiol Dis. 2013;54:139–149. doi: 10.1016/j.nbd.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 115.Yasojima K, Schwab C, McGeer EG, McGeer PL. Up-regulated production and activation of the complement system in Alzheimer’s disease brain. Am J Pathol. 1999;154:927–936. doi: 10.1016/S0002-9440(10)65340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brouwers N, Van Cauwenberghe C, Engelborghs S, Lambert J-C, Bettens K, Le Bastard N, et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol Psychiatry. 2012;17:223–33. doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet. 2014;23:2729–2736. doi: 10.1093/hmg/ddt666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, Estus S. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33:13320–5. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of Amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–50. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chan G, White CC, Winn PA, Cimpean M, Replogle JM, Glick LR, et al. CD33 modulates TREM2: convergence of Alzheimer loci. Nat Neurosci. 2015;18:1556–8. doi: 10.1038/nn.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–62. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Suárez-Calvet M, Kleinberger G, Araque Caballero MÁ, Brendel M, Rominger A, Alcolea D, et al. sTREM2 cerebrospinal fluid levels are a potential biomarker for microglia activity in early-stage Alzheimer’s disease and associate with neuronal injury markers. EMBO Mol Med. 2016;8:e201506123. doi: 10.15252/emmm.201506123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Piccio L, Deming Y, Del-Águila JL, Ghezzi L, Holtzman DM, Fagan AM, et al. Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta Neuropathol. 2016;131:925–933. doi: 10.1007/s00401-016-1533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Colonna M, Wang Y. TREM2 variants: new keys to decipher Alzheimer disease pathogenesis. Nat Rev Neurosci. 2016;17:201–7. doi: 10.1038/nrn.2016.7. [DOI] [PubMed] [Google Scholar]
- 127.Kleinberger G, Yamanishi Y, Suárez-Calvet M, Czirr E, Lohmann E, Cuyvers E, et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci Transl Med. 2014;6:243ra86. doi: 10.1126/scitranslmed.3009093. [DOI] [PubMed] [Google Scholar]
- 128.Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Atagi Y, Liu C-C, Painter MM, Chen X-F, Verbeeck C, Zheng H, et al. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2) J Biol Chem. 2015;290:26043–26050. doi: 10.1074/jbc.M115.679043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bailey CC, Devaux LB, Farzan M. The triggering receptor expressed on myeloid cells 2 binds apolipoprotein E. J Biol Chem. 2015;290:26033–26042. doi: 10.1074/jbc.M115.677286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of Amyloid-beta by microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 132.Jay TR, Hirsch AM, Broihier ML, Miller CM, Neilson LE, Ransohoff RM, et al. Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J Neurosci. 2016;37:637–647. doi: 10.1523/JNEUROSCI.2110-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eon Kuek L, Leffler M, Mackay GA, Hulett MD. The MS4A family: counting past 1, 2 and 3. Immunol Cell Biol. 2016;94:11–23. doi: 10.1038/icb.2015.48. [DOI] [PubMed] [Google Scholar]
- 134.Wang J, Shiratori I, Uehori J, Ikawa M, Arase H. Neutrophil infiltration during inflammation is regulated by PILRa via modulation of integrin activation. Nat Immunol. 2013;14:34–40. doi: 10.1038/ni.2456. [DOI] [PubMed] [Google Scholar]
- 135.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of Novel Alzheimer’s Disease Risk Genes in Control and Alzheimer’s Disease Brains. PLoS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang K, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, et al. A common haplotype lowers SPI1 (PU.1) expression in myeloid cells and delays age at onset for Alzheimer’s disease. Nat Neurosci. 2017 doi: 10.1038/nn.4587. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, et al. Novel rare coding variants in PLCG2, ABI3 and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017 doi: 10.1038/ng.3916. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu JZ, Erlich Y, Pickrell JK. Case–control association mapping by proxy using family history of disease. Nat Genet. 2017;49:325–331. doi: 10.1038/ng.3766. [DOI] [PubMed] [Google Scholar]
- 139.Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimer’s Dement. 2017:1–12. doi: 10.1016/j.jalz.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Karch CM, Ezerskiy LA, Bertelsen S, Goate AM ADGC. Alzheimer’s disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 loci. PLoS One. 2016;11:1–22. doi: 10.1371/journal.pone.0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nicolas G, Charbonnier C, Wallon D, Quenez O, Bellenguez C, Grenier-Boley B, et al. SORL1 rare variants: a major risk factor for familial early-onset Alzheimer’s disease. Mol Psychiatry. 2015;21:1–6. doi: 10.1038/mp.2015.121. [DOI] [PubMed] [Google Scholar]
- 143.Ruiz A, Heilmann S, Becker T, Hernández I, Wagner H, Thelen M, et al. Follow-up of loci from the International Genomics of Alzheimer’s Disease Project identifies TRIP4 as a novel susceptibility gene. Transl Psychiatry. 2014;4:e358. doi: 10.1038/tp.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Benitez BA, Jin SC, Guerreiro R, Graham R, Lord J, Harold D, et al. Missense variant in TREML2 protects against Alzheimer’s disease. Neurobiol Aging. 2014:35. doi: 10.1016/j.neurobiolaging.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]