Abstract
Objective
Patients with Inflammatory Bowel Disease (IBD) display substantial heterogeneity in clinical characteristics. We hypothesize that individual differences in the complex interaction of the host genome and the gut microbiota can explain the onset and the heterogeneous presentation of IBD. Therefore, we performed a case-control analysis of the gut microbiota, the host genome and the clinical phenotypes of IBD.
Design
Stool samples, peripheral blood and extensive phenotype data were collected from 313 IBD patients and 582 truly healthy controls, selected from a population cohort. The gut microbiota composition was assessed by tag-sequencing the 16S rRNA gene. All participants were genotyped. We composed genetic risk scores from 11 functional genetic variants proven to be associated with IBD in genes that are directly involved in the bacterial handling in the gut: NOD2, CARD9, ATG16L1, IRGM and FUT2.
Results
Strikingly, we observed significant alterations of the gut microbiota of healthy individuals with a high genetic risk for IBD: the IBD-genetic risk score was significantly associated with a decrease in the genus Roseburia in healthy controls (FDR 0.017). Moreover, disease location was a major determinant of the gut microbiota: the gut microbiota of colonic CD patients is different from that of ileal CD patients, with a decrease in alpha diversity associated to ileal disease (P = 3.28 × 10−13).
Conclusion
We show for the first time that genetic risk variants associated with IBD influence the gut microbiota in healthy individuals. Roseburia spp are acetate-to-butyrate converters and a decrease has already been observed in IBD patients.
Keywords: Inflammatory bowel disease, Healthy controls, Gut microbiota, Host genetics
BACKGROUND AND AIMS
Inflammatory bowel disease (IBD), comprising Crohn’s Disease (CD) and ulcerative colitis (UC), is a chronic inflammatory disorder of the gastrointestinal tract. In CD, inflammation can occur throughout the gastrointestinal tract whereas, in UC, inflammation is confined to the mucosal layer of the colon. The clinical characteristics of IBD vary greatly between individuals with respect to disease location, disease activity and disease behaviour. The origin of this heterogeneous clinical presentation remains poorly understood.[1,2]
The pathogenesis of IBD consists of an exaggerated immune response in a genetically susceptible host to the luminal microbial content of the gut. Driven by rapidly evolving genotyping and next generation sequencing technologies, tremendous progress has been made in deciphering the host genomic landscape of IBD.[3,4] Systems biology approaches to genomic and biological data clearly show the importance of the interaction between the host genome and the microbial exposure in the gut.[5] Moreover, known and presumed epidemiological risk factors for developing IBD such as mode of birth (vaginal vs. caesarean section), breastfeeding, smoking, hygiene, infections, antibiotics, diet, stress and sleep pattern are all known to cause microbial perturbations, suggesting a key role for the gut microbiota in the pathogenesis of IBD.[6–9]
Previous studies have shown a reduced biodiversity in the gut microbial composition of IBD patients, characterized by a reduction of known beneficial bacteria, such as Faecalibacterium praunitzii, Roseburia intestinalis and other butyrate-producers, and an increase of pathogens or pathobionts, e.g. adherent-invasive Escherichia coli and Shigella species of the Enterobacteriaceae family. However, these studies used a relatively small number of controls, who were usually selected from the patient population of the gastroenterology department after excluding IBD.[10] Because recent gut microbiome research has shown significant effects of stool consistency and functional complaints on the gut microbiota [11–13], previous results could have been influenced by their method of selection of controls.
While the main composition of the gut microbiota in CD has been studied extensively, the composition of the gut microbiota in UC patients has received less attention.[10,14,15] Furthermore, the relationship between the gut microbiota and the clinical characteristics of IBD, including disease activity, disease duration and disease behaviour has only been studied in an exploratory manner.
Recent studies have begun to unravel the complex interaction of host genetics and the gut microbiota. These links between specific genetic variants and the abundance of specific bacteria are called microbiota quantitative trait loci (microbiotaQTLs). Twin studies show that the abundances of bacterial families Ruminococcaceae and Lachnospiraceae containing butyrate-producers and acetate-to-butyrate converters are, to a certain degree, heritable.[16–18] Animal studies in mice specifically designed to discover microbiotaQTLs show the influence of genomic loci on several microbial genera.[19] Moreover, gut microbiota similarities in twins both concordant and discordant for IBD have been shown in several studies, further suggesting host genetics can influence the gut microbiota.[20–22] Furthermore, preliminary data show that specific variants of the NOD2 gene are associated with changes in the abundance of the Enterobacteriaceae family in IBD patients.[23]
We hypothesize that the large heterogeneity between IBD patients is likely to result from individual differences in the complex interaction between the host genome and the gut microbiota. Therefore, improving our knowledge of this interaction is crucial for our understanding of the pathogenesis of IBD.[14] So far, very few studies have been able to elucidate this interaction in an integrated manner. Here, we present a large single-centre case-control analysis of the luminal gut microbiota, the host genetics and clinical phenotypes of both CD and UC. To ensure optimal data quality, we adopted a rigorously standardized approach to collect and process fresh frozen faecal samples of 313 IBD patients from a single hospital in the North of the Netherlands and 582 truly healthy controls from the same geographical area. For all individuals, extensive clinical data, laboratory and endoscopic findings were collected. In addition, host genomic risk variants and risk scores were obtained in both the IBD patient and the healthy controls to analyse host genomic influences on the gut microbial composition.
METHODS
Cohorts
In total 357 IBD patients were recruited from the specialized IBD outpatient clinic at the Department of Gastroenterology and Hepatology of the University Medical Center Groningen (UMCG) in Groningen, the Netherlands. All IBD patients were diagnosed based on accepted radiological, endoscopic and histopathological evaluation. We excluded 44 IBD patients who had a stoma, pouch or short bowel syndrome from further analyses. Healthy controls were selected from the 1174 participants of LifeLines-DEEP, a cross-sectional general population cohort in the Northern provinces of the Netherlands.[24] Data about medical history, medication use and gut complaints were meticulously reviewed by a medical doctor to ensure controls did not have any severe gut complaints or diseases, and did not use any medication that could confound our analysis of the gut microbiota. The selection process is described in detail in the Supplementary Appendix. Pseudonymized data from IBD patients and healthy controls were provided to the researchers. This study was approved by the Institutional Review Board of the UMCG (IRB number 2008.338). All participants signed an informed consent form.
Clinical characteristics and medication use of IBD patients
Extensive data on clinical characteristics and medication use was available for all IBD patients at the time of stool sampling. Pseudonymized data was retrieved from the IBD-specific electronic patient records of the IBD Center at the department of Gastroenterology and Hepatology of the UMCG. Disease activity at the time of sampling was determined by standardized and accepted clinical activity scores: the Harvey Bradshaw Index (HBI) for CD patients and the Simple Clinical Colitis Activity Index (SCCAI) score for UC patients. C-reactive protein (CRP) and faecal calprotectin measurements were also available as indicators of disease activity. Disease localization and behaviour were described according to the Montreal Classification. Disease duration was determined as date of stool sampling in the study minus the date of diagnosis. IBD treatment at the time of sampling was scored (mesalazines, steroids, thiopurines, methotrexate, tumour necrosis factor alpha (TNF-α) inhibitors and other biologicals) as well as the use of other medication: proton pump inhibitors (PPIs), anti-diarrheal medication (loperamide), bile salts, iron, minerals and vitamins at the time of sampling, and antibiotics use within the previous three months. Extra-intestinal manifestations and complications of IBD were scored in several categories: 1. eye; 2. mouth; 3. skin; 4. joints; 5. Other (details in Supplementary Appendix).
Serological measurements for Anti-neutrophil cytoplasmic antibodies (ANCA) and Anti-Saccharomyces cerevisiae antibodies (ASCA) were determined by immunofluorescence. Information on mode of birth, breastfeeding during infancy and self-reported diets (Supplementary Appendix) were collected through questionnaires.
The association between a phenotype and the gut microbiota was only analyzed if there were five or more IBD patients with that phenotype. A list of all phenotypes can be found in the Supplementary Appendix.
Stool sample collection and faecal DNA Extraction
Stool samples were collected for 313 IBD cases and 582 controls. Identical protocols were used to collect and process all stool samples. All participants were asked to produce a stool sample at home. These were frozen by the participant within 15 minutes after stool production in the participant’s home freezer. A research nurse visited each participant shortly after stool production to collect the sample on dry ice for transport to the UMCG at −80°C. Samples were subsequently stored at −80°C in the laboratory. All samples remained frozen until DNA-isolation for which aliquots were made and microbial DNA was isolated using the Qiagen AllPrep DNA/RNA Mini Kit cat. # 80204 as previously described.[10]
Host genotyping, variant selection and genetic risk modelling
Host DNA was available for all IBD patients and healthy controls. Host DNA was isolated from peripheral blood as previously described.[25] Genotyping was performed using the Immunochip, an Illumina Infinium microarray comprising 196,524 Single Nucleotide Variants (SNPs) and a small number of insertion/deletion markers, selected based on results from genome-wide association studies of 12 different immune-mediated diseases including IBD. Normalized intensities for all samples were called using the OptiCall clustering program.[26] The genotype prediction was improved via stringent calling with BeagleCall using recommended settings.[27] Marker and sample quality control was performed as previously described.[3] Human leukocyte antigen (HLA) imputation was performed using SNP2HLA. The Type 1 Diabetes Genetics Consortium genotype data was used as a reference panel for imputation. The SNP2HLA imputes the classical HLA alleles and amino acid sequences within the major histocompatibility complex (MHC) region on chromosome 6.[28]
To overcome statistical problems inherent to multiple testing when combining both genome-wide and 16S rRNA microbiota data, we adopted an approach of analysing a set of selected SNPs based on i) their involvement in IBD, ii) their predicted functional consequences and iii) their role in bacterial sensing and signalling in the gut.[23]
Eleven known IBD genetic risk variants were selected for our genome-microbiota interaction analyses. We selected these risk variants ensuring that the selected IBD risk SNPs (as identified in the International IBD Genetics Consortium Immunochip analysis or targeted resequencing studies) are functional variants or are in strong linkage disequilibrium with functional variants that are implicated in the interaction of the host with the gut microbiota.[3,29] We included the following seven genetic variants in NOD2: rs104895431 (S431L), rs2066844 (R702W), rs5743277 (R703C), rs104895467 (N852S), rs2066845 (G908R), rs5743293 (fs1007insC) and rs104895444 (V793M). The variant rs10781499 in CARD9 was selected because Card9 has been shown to mediate intestinal epithelial cell restitution, T-helper 17 responses and control of intestinal bacterial infection in mice.[30] Two variants in FUT2, rs516246 and rs1047781, were selected because these variants have been shown to influence colonic mucosa-associated microbiota in CD.[31] SNPs rs11741861 in IRGM and rs12994997 in ATG16L1 were included because of their role in decreased selective autophagy that results in altered cytokine signalling and decreased anti-bacterial defence.[32,33]
In addition to these 11 genetic variants, we also created risk scores for all 200 known IBD risk variants.[3,5] We also analysed the influence of the HLA-DRB1*01:03 haplotype on the gut microbial composition in colonic disease because this recently identified haplotype is associated with both UC and colonic CD and is suggested to be involved in appropriately controlling the immune response to colonic microbiota.[34]
Determining the gut microbial composition
Illumina MiSeq paired-end sequencing was used to determine the bacterial composition of the stool samples. Forward primer 515F [GTGCCAGCMGCCGCGGTAA] and reverse primer 806R [GGACTACHVGGGTWTCTAAT] of hyper-variable region V4 of the 16S rRNA gene were used. Custom scripts were used to remove the primer sequences and align the paired end reads.[10]
Operational Taxonomic Units: OTU-picking and filtering
The operational taxonomic unit (OTU) selection was performed using the QIIME reference optimal picking, using Usearch (version 7.0.1090) to perform the clustering at 97% of similarity. Greengenes version 13.8 was used as a reference database. In all, 12556 OTUs were identified. Samples with less than 10,000 counts were removed. OTUs that were not present in at least 1% of our samples or with a low abundance (<0.01% of the total counts) were filtered out.
Function prediction
The functional imputation tools PICRUSt and HUMAnN were used to investigate the functional implications of the gut microbiota of IBD patients. More information about the function prediction and the software can be found in the Supplementary Appendix.
Statistical analysis
The richness and the beta-diversity of the microbiota dataset was analysed using QIIME.[35] The Shannon diversity index and the number of observed species per sample were used as alpha diversity metrics. Beta-diversity was calculated using unweighted Unifrac distances and represented in a Principal Coordinate Analyses (PCoA). The Wilcoxon test and Spearman correlations were used to identify differences in Shannon Index and relations between Principal Coordinates. Chi-square tests, Fishers exact tests, Spearman correlations and Wilcoxon-Mann-Whitney tests (WMW tests) were used to determine differences in the clinical characteristics of IBD patients. QIIMETOMAASLIN was used to convert the OTU counts into relative taxonomical abundance. OTUs representing identical taxonomies were aggregated and higher taxon levels were added when multiple OTUs represented that taxon. Due to the limitations of the resolution on taxonomical classification using 16S gene sequencing, we restricted our analysis to genus level and above. The initial 12556 OTUs were classified into 250 taxonomical levels.
We used MaAsLin to identify differentially abundant taxa and pathways: 1) between IBD patients and healthy controls, 2) between different IBD phenotypes and 3) between individuals with diverse amounts of IBD genetic risk variants.[15] MaAsLin performs boosted additive general linear models between metadata and microbial abundance data. The default settings of MaAsLin were used in all analyses. We used the Q-value package implemented in MaAsLin to correct for multiple testing. A false discovery rate (FDR) of 0.05 was used as cut-off value for significance. The effect of the IBD diagnosis (CD or UC) on the gut microbiota composition was analysed by adding the IBD diagnosis versus healthy as a discrete predictor in the MaAsLin general linear mixed model analysis. Unweighted genetic risk scores were calculated for every participant by summing up the risk alleles of the abovementioned SNPs (risk allele = 1; IBD protective allele = 0).[25] Weighted genetic risk scores were calculated for every participant by summing up the log-normalized odds of the genetic variants of the same abovementioned SNPs. Both risk scores were added as a predictor to the additive general linear model in MaAsLin. The analyses of the host genome and the microbiota composition were performed separately in IBD patients and healthy controls.
Correction for factors influencing the gut microbiota
Parameters that potentially influence the gut microbiota were identified by statistical analysis of cohort phenotypes, univariate MaAsLin analyses and literature search, and subsequently added as co-factors to the additive linear model. In every analysis, the parameters age, gender, BMI, read-depth, PPI use, antibiotics use and IBD medication (mesalazines, steroids, thiopurines, methotrexate and TNF-alpha inhibitors) were added as covariates. Stool consistency also affects the gut microbiota. However, since stool consistency, mainly the occurrence of diarrhoea, is a key characteristic of increased IBD disease activity, stool consistency was not used as a covariate in all models. However, stool consistency was incorporated in the analyses, since the clinical disease activity scores used: the Harvey Bradshaw Index (HBI) for Crohn’s Disease and the Simple Clinical Colitis Activity Index (SCCAI) take the number of liquid stools per day (in the HBI) and the number of bowel movements during the day and during the night (in the SCCAI) into account.
RESULTS
The clinical characteristics of IBD patients and the selection of healthy controls
The cohort consists of 313 IBD patients (188 CD, 107 UC and 18 IBDI/IBDU patients) and 582 healthy controls selected from the population cohort LifeLines-DEEP (Selection criteria can be found in the Supplementary Appendix).[24] CD patients were younger than healthy controls (41.3 versus 45.9 years; P = 1 × 10−4, WMW-test) while UC patients were not older than healthy controls (P = 0.32, WMW-test). At the time of sampling, 81 IBD patients (25.8%) had active disease, defined as an HBI of higher than 4 in CD patients or an SCCAI-score higher than 2.5 in UC patients. Of the IBD patients, 23.7% had used antibiotics within the last 3 months. PPI use was more frequent in IBD patients (24.5%) than in healthy controls (4.7%) (P < 0.001, Chi2-test). Extensive information on all clinical characteristics and medication use is presented in Table 1.
Table 1.
Clinical characteristics of IBD patients and healthy controls
| Average (SD) or Count (%) | CD only | ileal CD only | colonic CD only | ileocolonic CD only | UC only | IBDU/IBDI only | IBD | Healthy controls |
|---|---|---|---|---|---|---|---|---|
| number of samples | 188 | 68 | 36 | 78 | 107 | 18 | 313 | 582 |
| sequence read depth (SD) | 47730 (37278) | 44610 (36541) | 49870 (39818) | 48060 (37803) | 49090 (37050) | 66653 (43157) | 48820 (37539) | 48740 (29705) |
| Demographics | ||||||||
| Age (SD) | 41.3 (14.5) | 42.54 (14.15) | 42.39 (14.5) | 39.9 (14.7) | 47.3 (14.6) | 44.1 (16.8) | 43.6 (14.8) | 45.9 (13.7) |
| Gender (M/F) (%) | 62/126 (33/67%) | 23/45 (33/66%) | 12/24 (33/66%) | 25/53 (32/67%) | 52/55 (48/51%) | 7/11 (39/61%) | 122/191 (39/61%) | 302/280 (52/48%) |
| Weight and BMI | ||||||||
| Weight (SD) | 75.7 (16.2) | 77.5 (17.2) | 74.4 (14.3) | 74.9 (16.3) | 81.27 (16.1) | 84.9 (27.1) | 78.2 (17.1) | 77.4 (13.3) |
| BMI (SD) | 24.9 (4.6) | 25.0 (4.9) | 25.1 (4.6) | 24.7 (4.7) | 26.46 (4.4) | 27.9 (8.3) | 25.4 (4.9) | 24.9 (3.7) |
| Disease location | ||||||||
| ileum (%) | 68 (36%) | 68 (100%) | NA | NA | NA | NA | 68(4%) | NA |
| colon (%) | 36 (19%) | NA | 36 (100%) | NA | 106 (99%) | 8 (44%) | 152 (48%) | NA |
| both (%) | 78 (41%) | NA | NA | 78 (100%) | NA | 2 (11%) | 80 (25%) | NA |
| Disease activity | ||||||||
| CRP (SD) | 10.7 (16.63) | 11.1 (21.3) | 15.0 (18.8) | 8.8 (9.4) | 6.2 (7.3) | 7.29 (8.6) | 8.9 (13.7) | NA |
| fecal calprotectin (SD) | 390.1 (535.1) | 296.9 (533.3) | 445.6 (693.2) | 432.4 (437.6) | 776.6 (1986.8) | 870.2 (1166.4) | 531.9 (1220.3) | NA |
| Harvey Bradshaw Index (SD) | 3.45 (3.86) | 3.25 (3.18) | 3.8 (4.7) | 3.6 (4.13) | NA | NA | 3.45 (3.86) | NA |
| SCCAI (SD) | NA | NA | NA | NA | 1.8 (2.2) | 1.4 (2.0) | 1.8 (2.2) | NA |
| Disease duration and age at diagnosis | ||||||||
| Disease duration in years (SD) | 12.36 (9.13) | 12.73 (9.0) | 12.14 (8.6) | 12.14 (9.7) | 11.21 (8.48) | 10.5 (8.93) | 11.8 (8.8) | NA |
| Age at diagnosis [years] (SD) | 28.9 (12.4) | 29.9 (11.3) | 30.2 (15.3) | 27.8 (11.8) | 36.08 (14.4) | 32.5 (17.2) | 31.8 (13.7) | NA |
| Disease behavior Crohn’s Disease | ||||||||
| Montreal Classification B1 | 104 (55%) | 33 (48%) | 28 (77%) | 41 (52%) | NA | NA | 104 (55%) | NA |
| Montreal Classification B2 | 59 (31%) | 26 (38%) | 5 (13%) | 25 (32%) | NA | NA | 59 (31%) | NA |
| Montreal Classification B3 | 25 (13%) | 9 (13%) | 3 (8%) | 12 (15%) | NA | NA | 25 (13%) | NA |
| Disease severity Ulcerative Colitis | ||||||||
| Montreal Classification S1 | NA | NA | NA | NA | 6 (5%) | NA | 6 (5%) | NA |
| Montreal Classification S2 | NA | NA | NA | NA | 39 (36%) | NA | 39 (36%) | NA |
| Montreal Classification S3 | NA | NA | NA | NA | 44 (41%) | NA | 44 (41%) | NA |
| Montreal Classification S4 | NA | NA | NA | NA | 17 (15%) | NA | 17 (15%) | NA |
| Serology | ||||||||
| ANCA pos/neg (%) | 55/127 (30/67%) | 18/49 (26/72%) | 14/19(39/52%) | 18/49(72/26%) | 53/49(50/45%) | 8/10(44/56%) | 116/186(37/69%) | NA |
| ASCA pos/neg (%) | 85/92 (44/49%) | 38/28 (56/41%) | 8/24(22/66%) | 37/38(47/49%) | 16/85(80/14%) | 2/16(11/88%) | 102/194(32/62%) | NA |
| Birth & Breastfeeding | ||||||||
| Vaginal birth | 160 (85%) | 58 (85%) | 31 (86%) | 66 (84%) | 93 (87%) | 18 (100%) | 269 (85%) | NA |
| Caesarian section | 5 (2%) | 1 (1%) | 1 (2%) | 3 (3%) | 2 (1%) | 0 (0%) | 7 (2%) | NA |
| Breastfed | 96 (51%) | 36 (53%) | 22 (61%) | 36 (46%) | 62 (58%) | 11 (61%) | 169 (54%) | NA |
| Smoking | ||||||||
| Current smokers | 58 (31%) | 24 (35%) | 12 (33%) | 24 (35%) | 15 (14%) | 4 (22%) | 77 (25%) | 98 (17%) |
| IBD medication | ||||||||
| mesalazines | 12 (6%) | 6 (9%) | 1 (2%) | 5 (6%) | 87 (81%) | 3 (17%) | 113 (36%) | 0 (0%) |
| steroids | 40 (21%) | 8 (11%) | 9 (25%) | 20 (26%) | 18(17%) | 5 (28%) | 60 (19%) | 0 (0%) |
| thiopurines | 67 (36%) | 27 (40%) | 15 (42%) | 23 (30%) | 32(30%) | 6 (33%) | 104 (33%) | 0 (0%) |
| methotrexate | 22 (11%) | 6 (9%) | 3 (8%) | 12 (15%) | 1(1%) | 0 (0%) | 23 (7%) | 0 (0%) |
| anti-TNF alpha | 79 (42%) | 26 (38%) | 18 (50%) | 34 (44%) | 10(9%) | 3 (17%) | 92 (29%) | 0 (0%) |
| Other medication | ||||||||
| Antibiotics | 41 (21%) | 13 (19%) | 11 (39%) | 17 (21%) | 15 (14%) | 4 (22%) | 59 (19%) | 0 (0%) |
| Proton pump inhibitors | 44 (23%) | 16 (24%) | 6 (17%) | 21 (27%) | 13 (12%) | 2 (11%) | 60 (19%) | 26(4%) |
| Antidiarrheal | 29 (16%) | 14 (3%) | 2 (5%) | 13 (17%) | 4 (3%) | 1 (5%) | 33 (11%) | 0 (0%) |
| Bile Salts | 3 (1%) | 2 (3%) | 0 (0%) | 1 (1%) | 3 (2%) | 1 (5%) | 7 (2%) | 0 (0%) |
| Immunosuppressants | 92 (51%) | 32 (51%) | 19 (55%) | 38 (50%) | 38 (36%) | 5 (30%) | 135 (44%) | 0 (0%) |
| Mineral | 5 (2%) | 1 (1%) | 3 (8%) | 1 (1%) | 2 (1%) | 0 (0%) | 7 (2%) | 0 (0%) |
| Osteoporosis medication | 5 (2%) | 2 (3%) | 1 (2%) | 2 (3%) | 1 (1%) | 0 (0%) | 6 (2%) | NA |
| Vitamins | 74 (41%) | 34 (52%) | 5 (14%) | 32 (42%) | 2 (1%) | 2 (11%) | 78 (25%) | 0 (0%) |
| Self-reported diets | ||||||||
| Diabetes diet | 2 (1%) | 0 (0%) | 1 (2%) | 1 (1%) | 4 (4%) | 0 (0%) | 6 (2%) | 0 (0%) |
| Fat limited diet | 6 (3%) | 2 (3%) | 1 (2%) | 2 (3%) | 4 (4%) | 1 (5%) | 11 (4%) | 9 (2%) |
| Vegetarian diet | 9 (5%) | 1 (1%) | 3 (8%) | 5 (7%) | 6 (6%) | 1 (5%) | 15 (5%) | 39(7%) |
| Other diet | 18 (10%) | 6 (9%) | 4 (11%) | 8 (11%) | 10 (10%) | 0 (0%) | 28 (9%) | 23(4%) |
ANCA, Anti-neutrophil cytoplasmic antibodies; ASCA, Anti-Saccharomyces cerevisiae antibodies; BMI, Body Mass Index; CRP, C-reactive protein; CD, Crohn’s Disease; IBD, Inflammatory Bowel Disease; IBDI, Inflammatory Bowel Disease Intermediate; IBDU, Inflammatory Bowel Disease Undetermined; SD, standard deviation; UC, Ulcerative Colitis.
Overall composition of the gut microbiota in IBD patients and healthy controls
The predominant phyla in both IBD patients and healthy controls were Firmicutes (73% in IBD patients, 75% in healthy controls), Actinobacteria (9% in IBD patients, 13% in healthy controls) and Bacterioidetes (14% in IBD patients, 8% in healthy controls). Clostridia was the most abundant class (64% in IBD patients, 68% in healthy controls). An overview of the abundances at all taxonomic levels can be found in Supplementary Table S1.
Alpha diversity
A statistically significant decrease in the Shannon Index was observed in IBD patients compared to healthy controls as depicted in Supplementary Figure S1 (P = 5.61 × 10−14, Wilcoxon test) and Figure 1.
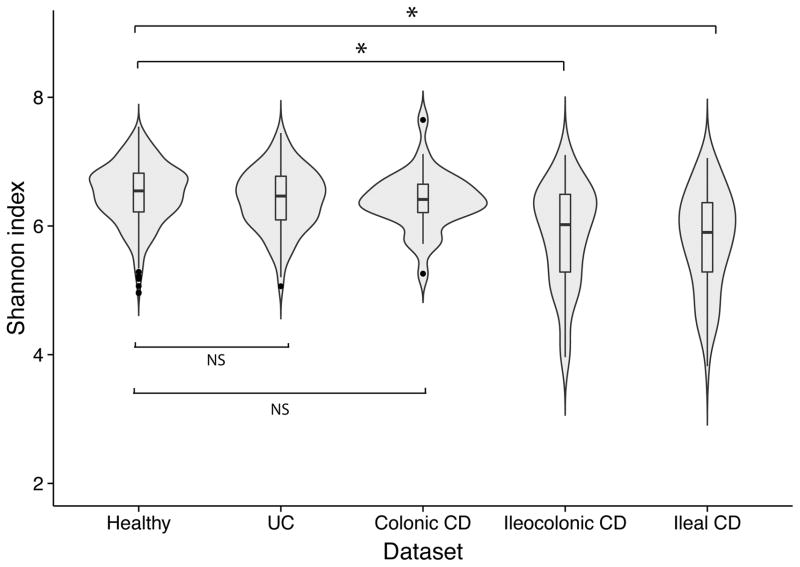
Figure 1.
Alpha diversity (Shannon Index) of the gut microbiota of healthy controls, Ulcerative Colitis (UC) patients, colonic Crohn’s Disease (CD) patients, ileocolonic CD patients and ileal CD patients. Alpha diversity is not decreased in colonic disease (UC and colonic CD) compared to healthy controls. In contrast, in ileal and ileocolonic CD patients, the alpha diversity is statistically significantly decreased (ileal CD patients vs. healthy controls P = 3.28 × 10−13 and ileocolonic CD patients vs. healthy controls P = 3.11 × 10−11, Wilcoxon test).
Principal Coordinate Analysis
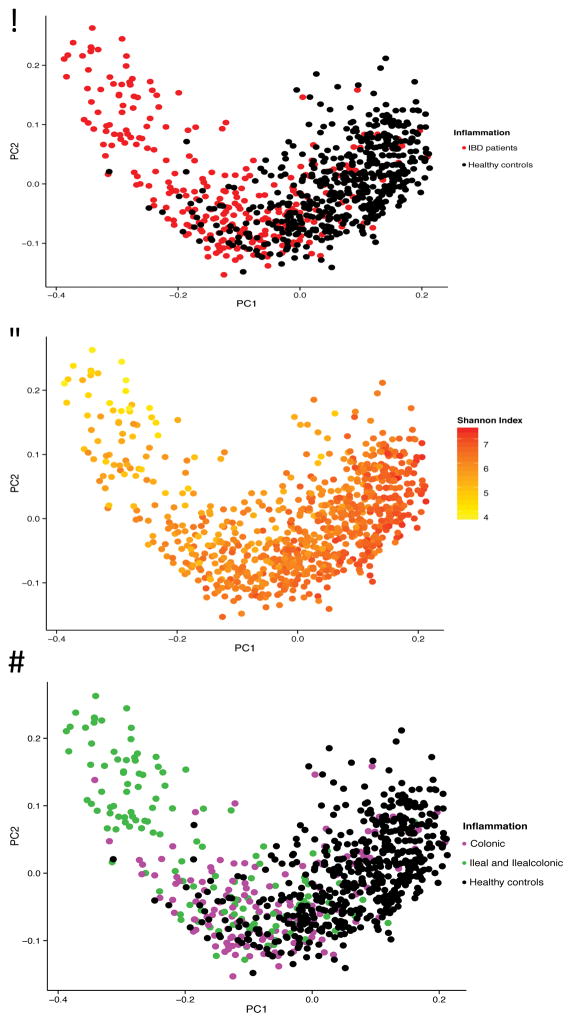
The differences in gut microbial composition between IBD patients and healthy controls were also observed in the PCoA-analysis. Statistically significant differences were found in the first three components (PCoA1 P = 2.62 × 10−68, PCoA2 P = 0.033, PCoA3 P = 1.50 × 10−10, Wilcoxon test). The gut microbiota of healthy controls clustered together, while the gut microbiota of IBD patients were more heterogeneous, partially overlapping the healthy controls. The shape of the PCoA-plot is mainly explained by disease location and the Shannon Index (see results below) as depicted in Figure 2A–2D.
Figure 2.
Principal Coordinate Analysis (PCoA) of stool samples of 313 IBD patients and 582 healthy controls. (A) The gut microbiota of IBD patients is different from the gut microbiota of healthy controls, with only partial overlap. (B) The first component is related to the Shannon Index. (C, D) There is more overlap between colonic disease (Ulcerative Colitis and colonic Crohn’s Disease combined) and healthy controls than between ileal disease (ileal Crohn’s Disease and ileocolonic Crohn’s Disease combined) and healthy controls. The first component is related to disease location (PCoA1 rho=0.63, P = 7.39 × 10−91, Spearman correlation) and colonic CD patients differ from ileal CD patients (P = 5.42 × 10−9).
IBD genetic risk variants are associated to unfavourable gut microbiota changes in healthy controls
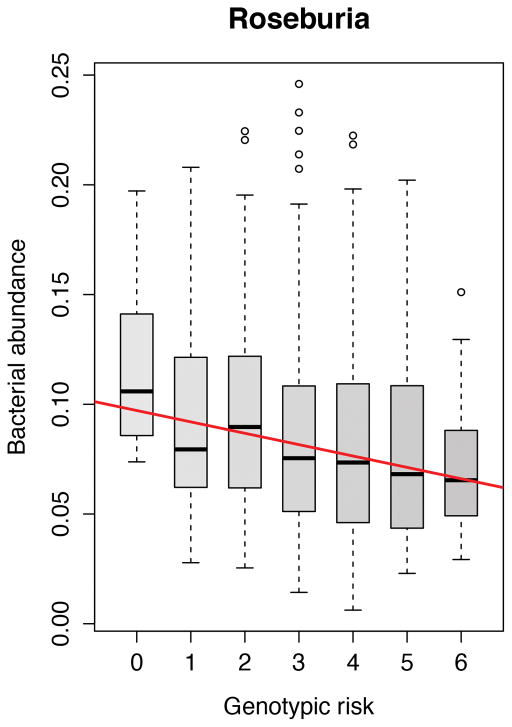
The role of 11 functional genomic variants associated to IBD in the genes NOD2, CARD9, ATG16L1, IRGM and FUT2 was investigated. In the unweighted analysis in healthy controls, a higher number IBD risk alleles was associated with a decrease in the abundance of the genus Roseburia of the phylum Firmicutes (FDR = 0.017) as depicted in Figure 3. In IBD patients as well as subsets of IBD patients (CD patients, UC patients, ileal CD patients, ileocolonic CD patients and colonic CD patients) neither the single genetic risk variants, the HLA-DRB1*01:03 haplotype nor the weighted or unweighted composite scores of genetic risk alleles showed any statistically significant effect on the gut microbiota composition. All results of the analyses with the risk scores of 11 SNPs can be found in Supplementary Table S3. Risk scores including all 200 IBD risk SNPs did not show any significant relations with the gut microbiota composition.
Figure 3.
Increased risk score of 11 IBD related genetic variants in gut bacterial handling genes (NOD2, CARD9, IRGM, ATG16L1 and FUT2) is statistically significantly associated to decreased abundance of Roseburia spp. in healthy controls (FDR = 0.017).
Dysbiosis in CD and UC patients: new associations
Crohn’s disease
Compared to healthy controls, 69 taxa were statistically significantly altered in CD patients (genus and above; 28%; FDR < 0.05). These alterations are presented in Table 2 and depicted in the cladogram in Supplementary Figure S2A. The phyla Bacteroidetes (FDR = 1.12 × 10−14) and Proteobacteria (FDR = 2.71 × 10−22) were increased, while the phyla Actinobacteria (FDR = 7.15 × 10−10) and Tenericutes (FDR = 1.90 × 10−12) were decreased. Within the phylum Bacteroidetes, the order Bacteroidales was increased (FDR = 1.12 × 10−14) as well as the genus Parabacteroides within the family Porphyromonadaceae (FDR = 0.0016). Within the order Clostridiales of the phylum Firmicutes, seven families were decreased: Mogibacteriaceae, Christensenellaceae, Clostridiaceae, Dehalobacteriaceae, Peptococcaceae, Peptostreptococcaceae and Ruminococcaceae (FDR < 0.05). The family Enterobacteriaceae of the phylum Proteobacteria, containing many known gut pathogens, was increased (FDR = 0.0020). The genera Bifidobacterium, Ruminococcus and Faecalibacterium were also decreased in CD patients (FDR = 2.16 × 10−6, FDR = 4.70 × 10−5 and FDR = 7.82 × 10−23, respectively).
Table 2.
Comparison of altered taxa in Crohn’s Disease patients compared to healthy controls; family level and above
| Gut microbiota alterations in Crohn’s Disease patients (current study: FDR < 0.05) | |||||
|---|---|---|---|---|---|
| Taxon (family and above) | Phylum (or kingdom) | Current studya | Gevers et al.b | Morgan et al.c | Willing et al. d |
| f__Methanobacteriaceae | Archea (kingdom) | Down | Not reported | Not reported | Not reported |
| p__Actinobacteria | Down | Down | Not reported | Up in colonic CD | |
| c__Actinobacteria | Actinobacteria | Down | Down | Not reported | Up in colonic CD |
| f__Micrococcaceae | Actinobacteria | Not reported | Up | Not reported | Not reported |
| f__Bifidobacteriaceae | Actinobacteria | Down | Down | Down, in lower taxonomic levels | Up in colonic CD |
| f__Coriobacteriaceae | Actinobacteria | Down | Down | Not reported | Up in colonic CD |
| p__Bacteroidetes | Up | Down | Not reported | Not reported | |
| o__Bacteroidales | Bacteriodetes | Up | Down | Not reported | Not reported |
| f__Porphyromonadaceae | Bacteriodetes | Up | Down | Down, in lower taxonomic levels | Unknown genus in this family: Down in ileal CD |
| p__Firmicutes | Down, in lower taxonomic levels | Down | Down | Up in colonic CD | |
| c__Bacilli | Firmicutes | Up, in lower taxonomic levels | Up | Associated to ileal involvement | Up in ileal CD |
| f__Aerococcaceae | Firmicutes | Up | Not reported | Not reported | Not reported |
| f__Enterococcaceae | Firmicutes | Up | Not reported | Not reported | Not reported |
| o__Gemellales | Firmicutes | Not reported | Up | Not reported | Not reported |
| f__Gemellaceae | Firmicutes | Not reported | Up | Not reported | Not reported |
| f__Streptococcaceae | Firmicutes | Not reported | Up | Not reported | Not reported |
| c__Clostridia | Firmicutes | Down | Down | Down | Down in ileal CD |
| o__Clostridiales | Firmicutes | Down | Down | Down | Down in ileal CD |
| f__Mogibacteriaceae | Firmicutes | Down | Not reported | Not reported | Not reported |
| f__Christensenellaceae | Firmicutes | Down | Down | Not reported | Not reported |
| f__Clostridiaceae | Firmicutes | Down | Down | Not reported | Not reported |
| f__Dehalobacteriaceae | Firmicutes | Down | Not reported | Not reported | Not reported |
| f__Lachnospiraceae | Firmicutes | Up, but genera in lower levels both going up and down | Down | Down, in lower taxonomic levels | Down, in lower taxonomic levels |
| f__Peptococcaceae | Firmicutes | Down | Not reported | Not reported | Down in ileal CD |
| f__Peptostreptococcaceae | Firmicutes | Down | Not reported | Not reported | |
| f__Ruminococcaceae | Firmicutes | Down | Down | Down | Down in ileal CD |
| f__Veillonellaceae | Firmicutes | Not reported | Up | Up | Up in lower taxonomic levels in ileal CD |
| f__Erysipelotrichaceae | Firmicutes | Down | Down | Associated to ileal involvement | Not reported |
| p__Fusobacteria | Not reported | Not reported | Not reported | Up in ileal CD | |
| o__Fusobacteriales | Fusobacteria | Not reported | Up | Not reported | Up in ileal CD |
| f__Fusobacteriaceae | Fusobacteria | Not reported | Up | Not reported | Up in ileal CD |
| p__Proteobacteria | Up | Up | Up | Up in ileal CD | |
| c__Betaproteobacteria | Proteobacteria | Up | Up | Not reported | Not reported |
| o__Burkholderiales | Proteobacteria | Up | Up | Not reported | Not reported |
| f__ Neisseriaceae | Proteobacteria | Not reported | Up | Not reported | Not reported |
| c__Gammaproteobacteria | Proteobacteria | Up | Up | Up | Up in ileal CD |
| f__Aeromonadaceae | Proteobacteria | Not reported | Not reported | Not reported | Up in ileal CD |
| o__Campylobacterales | Proteobacteria | Not reported | Up | Not reported | Not reported |
| f__Enterobacteriaceae | Proteobacteria | Up | Up | Up | Up in ileal CD |
| f__Pasteurellaceae | Proteobacteria | Not reported | Up | Not reported | |
| p__Tenericutes | Down | Not reported | Not reported | ||
| c__Mollicutes | Tenericutes | Down | Not reported | Not reported | Down in ileal CD, up in colonic CD |
| f__Anaeroplasmataceae | Tenericutes | Not reported | Not reported | Not reported | Down in ileal CD, up in colonic CD |
| f__Verrucomicrobiaceae | Verrucomicrobia | Not reported | Down | Not reported | Not reported |
k__, kingdom; p__; phylum; c__, class; o__, order; f__, family.
313 IBD patients including 188 CD patients; 582 healthy controls; stool only.
Cell Host Microbe 2014; 447 CD patients; 221 controls; stool and biopsy.
Genome Biology 2012; 204 IBD patients including 121 CD patients and 27 controls; stool and biopsy.
Gastroenterology 2010; 40 twin pairs concordant or discordant for CD/UC (23 CD pairs, 15 UC pairs, 2 healthy pairs).
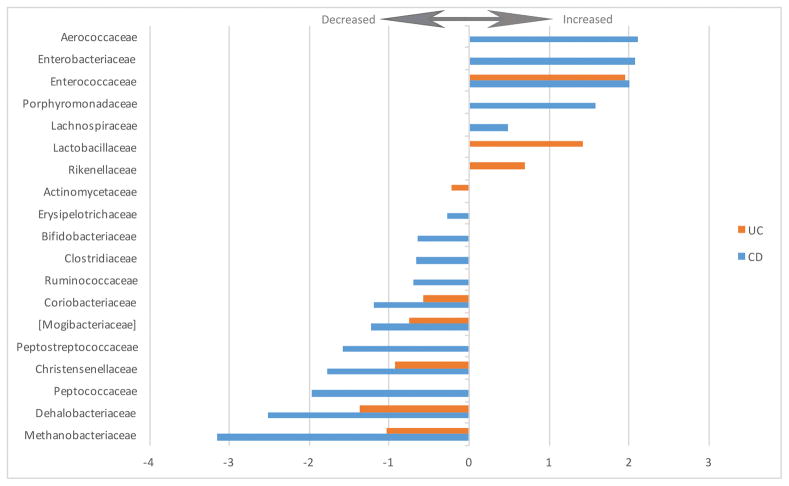
The changes in relative abundance of the statistically significantly altered families are depicted in Figure 4. The complete list of increased and decreased taxa including direction, coefficient and FDR-values is presented in Supplementary Table S2.
Figure 4.
Fold change of increased and decreased bacterial families in UC and CD patients versus healthy controls (FDR < 0.05).
Ulcerative colitis
In UC patients, 38 of the taxa were statistically significantly altered compared to healthy controls (genus and above; 12%; FDR < 0.05). These alterations are presented in Table 3 and depicted in a cladogram in Supplementary Figure S2B. Similar to CD patients, the abundances of the phyla Bacteroidetes (FDR = 8.87 × 10−13) and Proteobacteria (FDR = 4.06 × 10−5) were increased, while the phylum Firmicutes (FDR = 0.0079) was decreased in UC patients. Within the phylum Bacteroidetes, the order Bacteroidales (FDR = 8.87 × 10−13), the family Rikenellaceae (FDR = 0.025) and the genus Bacteroides (FDR = 1.72 × 10−18) are all increased compared to healthy controls. Lachnobacterium and Roseburia, genera in the order Clostridiales of the phylum Firmicutes, were also increased in UC (FDR = 0.023 and FDR = 0.00056, respectively). The changes in relative abundance of the altered families are depicted in Figure 4 (FDR < 0.05). The complete list of increased and decreased taxa, including direction, coefficient and FDR-values, is presented in Supplementary Table S2.
Table 3.
Comparison of significant taxa associations in Ulcerative Colitis patients: family level and above
| Gut microbiota alterations in Ulcerative Colitis patients (current study: FDR < 0.05) | ||||
|---|---|---|---|---|
| Taxon (family and above) | Phylum (or kingdom) | Current studya | Gevers et al.b | Morgan et al.c |
| f__Methanobacteriaceae | Archea | Down | Not reported | Not reported |
| f__Actinomycetaceae | Actinobacteria | Down | Not reported | Not reported |
| f__Coriobacteriaceae | Actinobacteria | Down | Not reported | Not reported |
| p__Bacteroidetes | Up | Not reported | Not reported | |
| o__Bacteroidales | Bacteriodetes | Up | Not reported | Not reported |
| f__Porphyromonadaceae | Bacteriodetes | Not reported | Not reported | Up |
| p__Firmicutes | Down | Down | Not reported | |
| f__Enterococcaceae | Firmicutes | Up | Not reported | Not reported |
| f__Lactobacillaceae | Firmicutes | Up | Not reported | Not reported |
| c__Clostridia | Firmicutes | Down, in lower taxonomic levels | Down | Not reported |
| o__Clostridiales | Firmicutes | Down, in lower taxonomic levels | Down | Not reported |
| f__Mogibacteriaceae | Firmicutes | Down | Not reported | Not reported |
| f__Christensenellaceae | Firmicutes | Down | Not reported | Not reported |
| f__Clostridiaceae | Firmicutes | Down, in lower taxonomic levels | Down, in lower taxonomic levels | Not reported |
| f__Dehalobacteriaceae | Firmicutes | Down | Not reported | Not reported |
| f__Lachnospiraceae | Firmicutes | Within the family genera both going up and down | Down, in lower taxonomic levels | Not reported |
| f__Ruminococcaceae | Firmicutes | Down, in lower taxonomic levels | Down | Not reported |
| f__Veillonellaceae | Firmicutes | Not reported | Up | Not reported |
| f__Erysipelotrichaceae | Firmicutes | Down, in lower taxonomic levels | Not reported | Down, in lower taxonomic levels |
| f__Streptococcaceae | Firmicutes | Not reported | Not reported | Down |
| p__Proteobacteria | Up | Not reported | Not reported | |
| c__Betaproteobacteria | Proteobacteria | Up | Not reported | Not reported |
| o__Burkholderiales | Proteobacteria | Up | Not reported | Not reported |
| p__Tenericutes | Down | Not reported | Down | |
| c__Mollicutes | Tenericutes | Down | Not reported | Down |
| f__Anaeroplasmataceae | Tenericutes | Not reported | Not reported | Down |
| f__Verrucomicrobiaceae | Verrucomicrobia | Down in lower taxonomic levels | Not reported | Not reported |
k__, kingdom; p__; phylum; c__, class; o__, order; f__, family.
313 IBD patients including 188 CD patients; 582 healthy controls; stool only.
Cell Host Microbe 2014; 447 CD patients; 221 controls; stool and biopsy.
Genome Biology 2012; 204 IBD patients including 121 CD patients and 27 controls; stool and biopsy.
Gastroenterology 2010; 40 twin pairs concordant or discordant for CD/UC (23 CD pairs, 15 UC pairs, 2 healthy pairs).
Disease location is a major determinant of the gut microbiota in IBD patients
The principal coordinate analysis depicted in Figure 2C shows the difference between the gut microbiota of patients with colonic disease (colonic CD and UC combined) and patients with ileal disease (ileal CD and ileocolonic CD combined). There is overlap between healthy controls and patients with colonic disease, while in concordance with the alpha-diversity analysis in Figure 1, the gut microbiota of patients with ileal disease deviates more from healthy controls. The statistical analysis of the PCoA supports this result: the first component is related to disease location (PCoA1 rho=0.63, P = 7.39 × 10−91, Spearman correlation) and colonic CD patients differ from ileal CD patients (P = 5.42 × 10−9). The alpha-diversity analysis shows similar results: the gut microbiota of IBD patients with colonic disease is not statistically significantly decreased compared to healthy controls (Shannon index UC patients = 6.41 vs. Shannon index healthy controls = 6.50, P = 0.06; Shannon index colonic CD patients = 6.38 vs. Shannon index healthy controls = 6.50, P = 0.08, Wilcoxon test). On the contrary, IBD patients with ileal disease show a statistically significant decrease in alpha diversity (ileal CD patients vs. healthy controls P = 3.28 × 10−13 and ileocolonic CD patients vs. healthy controls P = 3.11 × 10−11, Wilcoxon test), as depicted in Figure 1.
Whether the IBD genetic risk was associated with disease location was also tested. The genetic risk could not explain the disease location (colonic IBD versus ileal involved IBD; unweighted Genetic Risk Score using 200 SNPs; Spearman correlation; rho 0.045; P = 0.47). The taxonomy analysis of disease location is presented in the Supplementary Appendix.
Effects of IBD disease activity on the gut microbiota
We analysed several readouts for disease activity at the time of sample collection: the clinical HBI scores for CD patients and SCCAI scores for UC patients, as well as CRP and faecal calprotectin level measurements for all IBD patients. A higher HBI was associated with an increase of the family Enterobacteriaceae in CD patients (FDR = 0.036). No significant associations were found between the gut microbiota and the SSCAI in UC patients. Neither CRP nor faecal calprotectin was statistically significantly associated with altered bacterial abundances in the gut. Details of the disease activity analyses can be found in Supplementary Table S5 and S6.
Effects of IBD disease duration on the gut microbiota
The disease duration in IBD patients was measured from date of diagnosis up to the date of sample collection. A longer duration of the disease, corrected for age, was associated with a higher abundance of the phylum Proteobacteria (FDR = 0.045). (Supplementary Table S7)
Analysis of other IBD subphenotypes
Other gut microbial associations with other IBD subphenotypes including medication, smoking behaviour and extra-intestinal manifestations can be found in the Results section of the Supplementary Appendix.
Pathway prediction and gut microbiota function changes in IBD patients
Multiple metabolic pathways including butyrate metabolism, endotoxin metabolism and antibiotics resistance pathways were differentially expressed between IBD patients, UC patients, CD patients, ileal CD, ileocolonic CD and colonic CD as compared to healthy controls. These altered KEGG pathways are presented in Supplementary Figure S3 and Supplementary Table S16. The metabolism of short chain fatty acids (SCFA) was decreased in IBD patients, as indicated by the decrease of the propanoate (also known as propionate) metabolism in CD and UC patients (ko00640; CD: FDR = 2.74 × 10−11 and UC: FDR = 3.59 × 10−5), the decrease of the butanoate (also known as butyrate) metabolism in CD patients (ko00650; FDR = 5.31 × 10−9) and the decreased fatty acid metabolism in CD patients (ko00071; FDR = 4.28 × 10−18). Lipopolysaccharide (LPS) or endotoxin biosynthesis was increased in both CD and UC patients (ko00540; CD: FDR = 4.69 × 10−7 and UC: FDR = 0.027). Beta-lactam resistance metabolism was increased in CD patients (ko00312; FDR = 4.69 × 10−7). There were no significant pathway increases or decreases related to the clinical disease activity score, the HBI, for CD (Supplementary Table S17). More detailed information on the predicted pathways can be found in Results section of the Supplementary Appendix.
CONCLUSIONS
By performing this extensive integrated case-control analysis of the gut microbiota, the host genome and the clinical characteristics of IBD, we have identified new gut microbial associations with IBD and are now able to refine our understanding of the findings of previous studies. We found a relation between host genetic IBD susceptibility variants and the gut microbiota composition in healthy individuals and observed the effect of disease location on the gut microbiota. Moreover, we report microbial associations with multiple IBD subphenotypes.
The onset of IBD: genetic risk factors for IBD associated with pro-inflammatory gut microbiota alterations in healthy individuals
Discovering gene-microbiota interactions is difficult due to the large number of genomic markers as well as microbial taxa, requiring stringent multiple testing correction, thus limiting the possibility of finding statistically significant results. To resolve this issue we created risk scores of known functional IBD risk variants proven to be involved in the bacterial handling in the gut. This hypothesis-based gene-microbiota approach limits the number of tests that need to be done and has proven to be successful.
The gut microbiota interacts with the intestinal epithelium and the host immune system.[18,36–39] Recently, it was hypothesized that the interaction of the immune system with the gut microbiota goes two ways: ‘good’ gut microbiota can ameliorate immune responses, but the gut immune system can also ‘farm’ good bacteria in order to maintain immune-microbe-homeostasis.[36,37] We can show support for this hypothesis: in healthy individuals an increased genetic burden in functional variants in genes involved in bacterial handling (NOD2, IRGM, ATG16L1, CARD9 and FUT2) is associated with a decrease of the acetate-to-butyrate converter Roseburia spp.
The species Roseburia intestinales is one of the 20 most abundant species in the gut microbiota.[40] Importantly, a decrease in Roseburia spp. is already associated to the gut microbiota of IBD patients.[10,15] In an in vitro model, Roseburia spp. specifically colonized the mucins, which govern mucosal butyrate production.[41] Butyrate derived from Clostridium Clusters IV, VIII and XIVa to which Roseburia spp. belong has been shown to induce Treg cells, preventing or ameliorating intestinal inflammation.[38,39] The abundances within the family Lachnospiraceae, to which Roseburia spp. belongs, are significantly more similar in monozygotic twins than in dizygotic twins.[17] Moreover, unaffected siblings of CD patients share a decrease in Roseburia spp.[22]
This finding in healthy individuals carrying IBD genetic risk variants has implications for our understanding of the onset of IBD. We hypothesize that genetic risk factors of the gut immune system lead to ‘farming’ of a more pro-inflammatory gut microbiota and increased susceptibility to IBD. Subsequent unfavourable microbial perturbations due to environmental risk factors could further disturb the immune-microbe-homeostasis in the gut, eventually leading to IBD.
In addition to our genetic risk score based on specific functions, analyses using genetic risk scores of all 200 known IBD susceptibility variants, many of whose function is unknown, did not yield any statistically significant results in either IBD patients or in healthy controls. We could not detect any gene-microbiota interactions in IBD patients, probably due to the already well-established dysbiosis as a consequence of the inflammation in the gut. Another complication is the interrelatedness of the genotype and phenotypes in IBD. For example, NOD2 risk variants are known to be associated with ileal CD and we show that ileal CD has a specific microbial signature. After correction for treatment, disease activity and disease location, we could not find any statistically significant genome-microbiota relations in IBD patients.
Dysbiosis in CD and UC patients: new associations identified, previous associations corrected
The dysbiosis of the gut microbiota in IBD patients is profound: the abundances of 69 taxa in CD patients and 38 taxa in UC patients were altered compared to healthy individuals (FDR < 0.05). We compared our results on the phylum, class, order and family levels to two previous studies looking into the gut microbiota of IBD patients.[10,15,20] This comparison is presented in Table 2 (CD patients) and 3 (UC patients). An important new finding of our study is the increase in the phylum Bacteroidetes in both CD and UC patients. Increased levels of Bacteroidetes have recently been discovered in IBS patients.[13] Since the control groups used in previous IBD studies also had functional gastrointestinal complaints (i.e. IBS), this would have confounded any comparisons between Bacteroidetes levels in IBD patients and controls, masking any meaningful enrichment in IBD.
The genus Bacteroides within the phylum Bacteroidetes is increased in our UC patients. The involvement of Bacteroides spp in the pathogenesis of IBD has been implied in animal studies. In NOD2 knock-out mice the exaggerated inflammatory response in the small intestine was dependent on Bacteroides vulgatus.[42] Bacteroides thetaiotaomicron induced colitis in HLA-B27 transgenic rats.[43] Another study looking into the effects of the vitamin D receptor in mice found increased levels of Bacteroides spp in colitis and increased levels of Bacteroides fragilis in colon biopsies of UC patients.[44]
Increased abundance of the families Streptococcaceae, Micrococcaceae and Veillonellaceae, previously associated with IBD, are now associated to PPI use in our study. PPI use is overrepresented in IBD patients.[45] Since previous studies did not correct for PPI use, we assume that alterations in the abundances of these taxa were wrongly assigned to the effect of IBD.
Our study is the largest gut microbiota study in UC patients to date, and within it we can now begin to resolve the landscape of the UC gut microbiota. We were able to find many new associations, including the association with a decreased abundance of phylum Tenericutes, which we also find to be associated with more extensive UC.
Disease location is a major determinant of the gut microbial composition in IBD
We showed the importance of disease location for the composition of the gut microbiota in IBD patients. In our PCoA, the gut microbiota of colonic CD patients is more similar to the microbiota of UC patients than to that of ileal CD patients. While different clusters of gut microbiota samples are also observed in recent IBD metagenomics research, we have been able to relate these clusters to the disease location phenotype.[46] The importance of disease location also matches recent insights into host genetics, in which, based on genetic risk scores, colonic CD lies between UC and ileal CD.[4] We found that the gut microbiota composition in stool could explain the differences in IBD disease location, while the genetic risk variants in our cohort could not. Moreover, there is important overlap in the clinical presentation of colonic CD and UC, e.g. the risk of developing colorectal carcinoma in colonic CD is similar in UC, but different from ileal CD.[47] Based on both the previous genetic findings and our current microbiota findings, it is becoming more apparent that colonic CD and ileal CD are different diseases within the IBD spectrum.
Through careful selection of healthy controls, meticulous standardization of stool collection, extensive phenotyping and host genotyping, we were able to successfully perform analyses and gain insight into the gut microbiota as key mediator of the IBD pathogenesis. For the first time, we find evidence for a role of the gut microbiota in the onset of IBD: healthy individuals with a high genetic risk load for IBD also have unfavourable changes in their gut microbiota. This relationship warrants further investigation as it might be both a potential target for treatment and a possibility for prevention of IBD in genetically susceptible hosts or their families.
Supplementary Material
Supplementary Figure S1: Alpha diversity (Shannon Index) of IBD patients and healthy controls, depicted in a violin plot.
Supplementary Figure S2: Dysbiosis in Crohn’s Disease (CD) and Ulcerative Colitis (UC) patients. (A) Cladogram of altered gut microbiota taxa in CD patients versus healthy controls (FDR < 0.05) (red = increased; blue = decreased). (B) Cladogram of altered gut microbiota taxa in UC patients versus healthy controls (FDR < 0.05) (red = increased; blue = decreased).
Supplementary Figure S3: Imputed function analysis: increased and decreased KEGG-pathways of the gut microbiota in IBD, CD, UC, ileal CD, colonic CD and ileocolonic CD patients versus healthy controls (FDR < 0.05). Bacterial function was imputed using PICRUSt and HUMAnN.
Supplementary Appendix: includes the supplementary methods.
Supplementary Table S1: Taxa abundances of CD patients, UC patients and healthy controls on phylum, class, order, family and genus levels.
Supplementary Table S10: MaAsLin results on Montreal Classification: stricturing and fistulizing disease in CD patients.
Supplementary Table S11: MaAsLin results on extra-intestinal manifestations and complications in IBD patients.
Supplementary Table S12: MaAsLin results on IBD medication in IBD patients.
Supplementary Table S13: MaAsLin results on mode of birth and breastfeeding of IBD patients.
Supplementary Table S14: MaAsLin results on smoking behavior in IBD patients.
Supplementary Table S15: MaAsLin results on self-reported diets of IBD patients.
Supplementary Table S16: MaAsLin results on imputed function (KEGG-pathways) of the gut microbiota of IBD, CD, ileal CD, ileocolonic CD, colonic CD and UC patients versus healthy controls.
Supplementary Table S17: MaAsLin results on imputed function (KEGG-pathways) of the gut microbiota related to the Harvey Bradshaw Index disease activity metric of Crohn’s Disease patients.
Supplementary Table S2: MaAsLin results on diagnosis and location.
Supplementary Table S3: MaAsLin results on genetic risk scores.
Supplementary Table S4: MaAsLin results on Montreal Classification: extent of UC.
Supplementary Table S5: MaAsLin results on disease activity in IBD patients.
Supplementary Table S6: MaAsLin results on Montreal Classification: severity of UC.
Supplementary Table S7: MaAsLin results on disease duration in IBD patients.
Supplementary Table S8: MaAsLin results on Anti-neutrophil cytoplasmic antibodies.
Supplementary Table S9: MaAsLin results on and Anti-Saccharomyces cerevisiae antibodies.
Summary Box.
What is already known about this subject?
The gut microbiota plays a key role in the pathogenesis of Inflammatory Bowel Diseases.
Known and presumed epidemiological risk factors for developing IBD such as mode of birth, breastfeeding, smoking, hygiene, infections, antibiotics, diet and stress are all known to cause gut microbial perturbations.
The large heterogeneity between IBD patients is likely to result from individual differences in the complex interaction between the host genome and the gut microbiota.
Discovering gene-microbiota interactions is difficult due to the large number of genomic markers as well as microbial taxa, requiring stringent multiple testing correction.
What are the new findings?
Gut microbial changes could precede the onset of IBD. A high IBD-genetic risk score is associated with a decrease in the genus Roseburia in the gut microbiota of healthy controls without gut complaints.
Disease localization is a major determinant of the IBD-associated gut microbiota composition.
The use of a large well-phenotyped healthy control cohort next to an IBD cohort leads to an improved list of IBD-associated gut microbial differences.
How might it impact on clinical practice in the foreseeable future?
Better understanding of gene-microbiota interactions and pro-inflammatory gut microbial changes that precede the onset of IBD can lead to new IBD therapeutics and perhaps even microbial prevention strategies.
Acknowledgments
Funding
RKW, JF and LF are supported by VIDI grants (016.136.308, 864.13.013 and 917.14.374) from the Netherlands Organization for Scientific Research (NWO). EAMF is funded by a career development grant from the Dutch Digestive Foundation (MLDS) (No. CDG-014). Sequencing of the LifeLines-DEEP cohort was funded by a Top Institute Food and Nutrition grant GH001 to CW. CW is further supported by an ERC advanced grant (ERC-671274). AZ holds a Rosalind Franklin fellowship (University of Groningen) and a CardioVasculair Onderzoek Nederland grant (CVON 2012-03).
We thank all the participants of the UR-IBD and Lifelines-DEEP cohorts for contributing stool samples; Dianne Jansen, Jacqueline Mooibroek, Anneke Diekstra, Brecht Wedman, Rina Doorn, Astrid Maatman, Tiffany Poon, Wilma Westerhuis, Daan Wiersum, Debbie van Dussen, Martine Hesselink, Ettje Tigchelaar, Soesma A. Jankipersadsing, Maria Carmen Cenit and Jackie Dekens for logistics support, laboratory support, data collection and data management; the research group of Morris Swertz for providing the high performance computing infrastructure including the Calculon Cluster Computer; the Parelsnoer Institute for supporting the IBD biobank infrastructure; Timothy Tickle, Curtis Huttenhower, Alexandra Sirota, Chengwei Luo and Aleksander Kostic for their help in training the first and second authors; Marten Hofker and Eelke Brandsma for contributing to the scientific discussion and Jackie Senior and Kate Mc Intyre for editing the manuscript.
Abbreviations
- BMI
Body Mass Index
- CRP
C-reactive protein
- CD
Crohn’s Disease
- FDR
False Discovery Rate
- HBI
Harvey Bradshaw Index
- IBD
Inflammatory Bowel Disease
- IBDI
Inflammatory Bowel Disease Intermediate
- IBDU
Inflammatory Bowel Disease Undetermined
- OTU
Operational Taxonomic Unit
- PPI
Proton Pump Inhibitor
- PCoA
Principal Coordinate Analysis
- SCCAI
Simple Clinical Colitis Activity Index
- SD
standard deviation
- UC
Ulcerative Colitis
- WMW tests
Wilcox-Mann-Whitney tests
Footnotes
Competing interests
The authors have no conflicts of interest to declare with this work.
Writing Assistance
This article was edited for language and formatting by Kate McIntyre, Associate Scientific Editor in the Department of Genetics, University Medical Center Groningen.
Author contributions
RKW, DG, AZ, GD, CH and RJX designed the study. FI, MCV, LMS, HMvD, RWFTS, GD and RKW collected the data. FI, AVV, MJB, RA and JF analyzed the data. FI, AVV, EAMF and RKW drafted the manuscript. CW, JF, EAMF, LF, DG, AZ, GD, CH, RJX and RKW critically reviewed the manuscript.
References
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ordás I, Eckmann L, Talamini M, et al. Ulcerative colitis. Lancet. 2012;380:1606–19. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 3.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015 doi: 10.1038/ng.3359. Published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2015;6736:1–12. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–17. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 7.Jost T, Lacroix C, Braegger CP, et al. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 8.Thaiss CA, Zeevi D, Levy M, et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell. 2014;159:514–29. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 9.David La, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevers D, Kugathasan S, Denson LA, et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe. 2014;15:382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tigchelaar EF, Bonder MJ, Jankipersadsing Sa, et al. Gut microbiota composition associated with stool consistency. Gut. 2015 doi: 10.1136/gutjnl-2015-310328. [DOI] [PubMed] [Google Scholar]
- 12.Dupont HL. Review article: Evidence for the role of gut microbiota in irritable bowel syndrome and its potential influence on therapeutic targets. Aliment Pharmacol Ther. 2014;39:1033–42. doi: 10.1111/apt.12728. [DOI] [PubMed] [Google Scholar]
- 13.Chung C-S, Chang P-F, Liao C-H, et al. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand J Gastroenterol. 2015;5521:1–10. doi: 10.3109/00365521.2015.1116107. [DOI] [PubMed] [Google Scholar]
- 14.Kostic AD, Xavier RJ, Gevers D. The Microbiome in Inflammatory Bowel Diseases: Current Status and the Future Ahead. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.02.009. Published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blekhman R, Goodrich JK, Huang K, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich JK, Waters JL, Poole AC, et al. Human Genetics Shape the Gut Microbiome. Cell. 2014;159:789–99. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–84. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leamy LJ, Kelly SA, Nietfeldt J, et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 2014;15:552. doi: 10.1186/s13059-014-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–54. e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 21.Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–7. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 22.Hedin CR, McCarthy NE, Louis P, et al. Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn’s disease and their unaffected siblings. Gut. 2014;63:1578–86. doi: 10.1136/gutjnl-2013-306226. [DOI] [PubMed] [Google Scholar]
- 23.Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tigchelaar EF, Zhernakova A, Dekens JAM. An introduction to LifeLines DEEP: study design and baseline characteristics. 2014:0–21. [Google Scholar]
- 25.Festen EA, Stokkers PC, van Diemen CC, et al. Genetic Analysis in A Dutch Study Sample Identifies More Ulcerative Colitis Susceptibility Loci and Shows Their Additive Role in Disease Risk. Am J Gastroenterol. 2010;105:395–402. doi: 10.1038/ajg.2009.576. [DOI] [PubMed] [Google Scholar]
- 26.Shah TS, Liu JZ, Floyd JAB, et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28:1598–603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browning BL, Yu Z. Simultaneous Genotype Calling and Haplotype Phasing Improves Genotype Accuracy and Reduces False-Positive Associations for Genome-wide Association Studies. Am J Hum Genet. 2009;85:847–61. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing Amino Acid Polymorphisms in Human Leukocyte Antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivas Ma, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokol H, Conway KL, Zhang M, et al. Card9 Mediates Intestinal Epithelial Cell Restitution, T-Helper 17 Responses, and Control of Bacterial Infection in Mice. Gastroenterology. 2013;145:591–601. e3. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rausch P, Rehman A, Künzel S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–5. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mccarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn ‘ s disease. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadaghian Sadabad M, Regeling A, de Goffau MC, et al. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn’s disease patients. Gut. 2014 doi: 10.1136/gutjnl-2014-307289. gutjnl – 2014–307289. [DOI] [PubMed] [Google Scholar]
- 34.Goyette P, Boucher G, Mallon D, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015;47:172–9. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ley RE. The Gene-Microbe Link. Nature. 2015;518:S7. doi: 10.1038/518S7a. [DOI] [PubMed] [Google Scholar]
- 37.Velasquez-Manoff M. Gut Microbiome: The Peacekeepers. Nature. 2015;518:S3–11. doi: 10.1038/518S3a. [DOI] [PubMed] [Google Scholar]
- 38.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 39.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 40.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van den Abbeele P, Belzer C, Goossens M, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7:949–61. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramanan D, Tang MS, Bowcutt R, et al. Bacterial Sensor Nod2 Prevents Inflammation of the Small Intestine by Restricting the Expansion of the Commensal Bacteroides vulgatus. Immunity. 2014;41:311–24. doi: 10.1016/j.immuni.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen JJ, Huang Y, Peterson DA, et al. The colitis-associated transcriptional profile of commensal Bacteroides thetaiotaomicron enhances adaptive immune responses to a bacterial antigen. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0042645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S, Zhang Y, Lu R, et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015;64:1082–94. doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imhann F, Bonder MJ, Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2015 doi: 10.1136/gutjnl-2015-310376. gutjnl – 2015–310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Averboukh F, Ziv Y, Kariv Y, et al. Colorectal carcinoma in inflammatory bowel disease: a comparison between Crohn’s and ulcerative colitis. Color Dis. 2011;13:1230–5. doi: 10.1111/j.1463-1318.2011.02639.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Alpha diversity (Shannon Index) of IBD patients and healthy controls, depicted in a violin plot.
Supplementary Figure S2: Dysbiosis in Crohn’s Disease (CD) and Ulcerative Colitis (UC) patients. (A) Cladogram of altered gut microbiota taxa in CD patients versus healthy controls (FDR < 0.05) (red = increased; blue = decreased). (B) Cladogram of altered gut microbiota taxa in UC patients versus healthy controls (FDR < 0.05) (red = increased; blue = decreased).
Supplementary Figure S3: Imputed function analysis: increased and decreased KEGG-pathways of the gut microbiota in IBD, CD, UC, ileal CD, colonic CD and ileocolonic CD patients versus healthy controls (FDR < 0.05). Bacterial function was imputed using PICRUSt and HUMAnN.
Supplementary Appendix: includes the supplementary methods.
Supplementary Table S1: Taxa abundances of CD patients, UC patients and healthy controls on phylum, class, order, family and genus levels.
Supplementary Table S10: MaAsLin results on Montreal Classification: stricturing and fistulizing disease in CD patients.
Supplementary Table S11: MaAsLin results on extra-intestinal manifestations and complications in IBD patients.
Supplementary Table S12: MaAsLin results on IBD medication in IBD patients.
Supplementary Table S13: MaAsLin results on mode of birth and breastfeeding of IBD patients.
Supplementary Table S14: MaAsLin results on smoking behavior in IBD patients.
Supplementary Table S15: MaAsLin results on self-reported diets of IBD patients.
Supplementary Table S16: MaAsLin results on imputed function (KEGG-pathways) of the gut microbiota of IBD, CD, ileal CD, ileocolonic CD, colonic CD and UC patients versus healthy controls.
Supplementary Table S17: MaAsLin results on imputed function (KEGG-pathways) of the gut microbiota related to the Harvey Bradshaw Index disease activity metric of Crohn’s Disease patients.
Supplementary Table S2: MaAsLin results on diagnosis and location.
Supplementary Table S3: MaAsLin results on genetic risk scores.
Supplementary Table S4: MaAsLin results on Montreal Classification: extent of UC.
Supplementary Table S5: MaAsLin results on disease activity in IBD patients.
Supplementary Table S6: MaAsLin results on Montreal Classification: severity of UC.
Supplementary Table S7: MaAsLin results on disease duration in IBD patients.
Supplementary Table S8: MaAsLin results on Anti-neutrophil cytoplasmic antibodies.
Supplementary Table S9: MaAsLin results on and Anti-Saccharomyces cerevisiae antibodies.