Abstract
Background
Extensively drug-resistant (XDR) Enterobacteriaceae carrying the blaKPC gene have emerged as a major global therapeutic concern. The purpose of this study was to analyze the complete sequences of plasmids from KPC-2 carbapenemase-producing XDR Escherichia coli sequence type (ST) 1642 isolates.
Methods
We performed antimicrobial susceptibility testing, PCR, multilocus sequence typing (MLST), and whole-genome sequencing to characterize the plasmid-mediated KPC-2-producing E. coli clinical isolates.
Results
The isolates were resistant to most available antibiotics, including meropenem, ampicillin, ceftriaxone, gentamicin, and ciprofloxacin, but susceptible to tigecycline and colistin. The isolates were identified as the rare ST1642 by MLST. The isolates carried four plasmids: the first 69-kb conjugative IncX3 plasmid harbors blaKPC-2 within a truncated Tn4401a transposon and blaSHV-11 with duplicated conjugative elements. The second 142-kb plasmid with a multireplicon consisting of IncQ, IncFIA, and IncIB carries blaTEM-1b and two class 1 integrons. This plasmid also harbors a wide variety of additional antimicrobial resistance genes including aadA5, dfrA17, mph(A), sul1, tet(B), aac(3′)-IId, strA, strB, and sul2.
Conclusions
The complete sequence analysis of plasmids from an XDR E. coli strain related to persistent infection showed the coexistence of a blaKPC-2–carrying IncX3-type plasmid and a class 1 integron-harboring multireplicon, suggesting its potential to cause outbreaks. Of additional clinical significance, the rare ST1642, identified in a cat, could constitute the source of human infection.
Keywords: Escherichia coli, ST1642, blaKPC, Tn4401a, IncX3
INTRODUCTION
Klebsiella pneumoniae carbapenemase (KPC) is the most common class A carbapenemase in the world; the extensive spread of KPC-producing Enterobacteriaceae has become a major therapeutic concern in clinical settings [1]. To date, a total of 21 KPC variants (KPC-2 to KPC-22) have been identified; of these, KPC-2 and KPC-3 are the most prevalent [1]. KPCs are found predominately in K. pneumoniae and less frequently in Escherichia coli [1,2]. Similar to other acquired antimicrobial resistance determinants, the blaKPC gene is disseminated by two means: 1) clonal spread of bacteria carrying the gene, and 2) horizontal transfer of the gene carried on mobile elements such as plasmids and transposons [1,2].
Molecular epidemiological studies on KPC-producing E. coli by multilocus sequence typing (MLST) have revealed that sequence type (ST) 131 strains are the most pervasive, followed by ST410, and less frequently ST69, ST93, ST167, ST354, and ST3948 strains [3,4,5,6]. In E. coli isolates, both blaKPC-2 and blaKPC-3 genes are prevalent, while blaKPC-8 is rarely identified [4]. The blaKPC-2 gene is carried by IncFIA-, IncFIIk-, IncN-, and IncA/C-type plasmids in the USA; IncN-, IncA/C- and IncF-type plasmids in China; an IncFIIA-type plasmid in France; an IncFIIk-type plasmid in Greece; and IncFIIs-, IncN-, and IncHI2-plasmids in Israel; while the blaKPC-3 gene is associated with IncFIA-, IncFIIk-, IncN-, and IncA/C-type plasmids in the USA; an IncFIIk-type plasmid in Italy; IncFII-, IncA/C-, and ColE-type plasmids in Spain; and IncFIIs-, IncN-, and IncHI2-plasmids in Israel [4,6]. Although an IncX3-type plasmid is prevalent in K. pneumoniae carrying the blaKPC gene [7], it has rarely been reported in E. coli.
The highly mobile Tn3-based transposon Tn4401 facilitates dissemination of the blaKPC gene [8]. Tn4401 is comprised of tnpA (transposase), tnpR (resolvase), and two insertion sequences (ISs), ISKpn7 and ISKpn6, as well as the blaKPC gene. The 10-kb transposon has 39-bp imperfect left- and right-inverted repeats and is flanked by 5-bp direct repeats [9]. Seven Tn4401 isoforms, Tn4401a to Tn4401g, have been identified; these are differentiated based on the size (68- to 255-bp) of the deletion between ISKpn7 and blaKPC compared with the prototype Tn4401b [10].
Here, we investigated the plasmids present in KPC-producing E. coli clinical isolates to elucidate the mechanisms underlying the acquisition of multi-drug resistance, including resistance to carbapenems.
METHODS
1. Patient description
An 85-year-old woman with a history of hypertension was transferred from Hyemin general hospital (Seoul, Korea) to Gangnam Severance hospital (Seoul, Korea) in January 2014 for the management of her cerebral hemorrhage. Brain computed tomography and magnetic resonance imaging revealed a subacute intracerebral hemorrhage in the right thalamus. The patient complained of urinary frequency and retention, and a fever of over 38.6℃ developed on the 8th day post admission. An E. coli isolate (EcU443) was recovered from a urinary specimen on the 8th day, and subsequent E. coli isolates (EcU213 on the 18th day and EcU120 on the 42nd day) were serially recovered from urinary specimens.
2. Bacterial isolates and antimicrobial susceptibility testing
This study involved two E. coli clinical isolates, EcU443 and EcU120, serially recovered from urinary specimens of a patient with a 34-day interval. The isolates were identified as E. coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Bruker Daltonics GmbH, Leipzig, Germany). Anti-microbial susceptibilities were determined using VITEK 2 AST N211 cards (bioMérieux Vitek Inc., Hazelwood, MO, USA) and disk diffusion tests on Mueller-Hinton agar (Oxoid Ltd., Basingstoke, UK) according to the CLSI guidelines [11]. Carbapenemase production was confirmed using the KPC+MBL Confirm ID Kit (Rosco Diagnostica, Taastrup, Denmark) with tablets containing meropenem (10 µg) alone or supplemented with dipico-linic acid (1,000 µg), phenylboronic acid (400 µg), and cloxacillin (750 µg) [12].
3. Genotyping of β-lactamases
The genomic DNA of each isolate was extracted by the boiling lysis method [13]. PCR was performed to detect genes encoding extended-spectrum β-lactamases (CTX-M-1-, CTX-M-9-, TEM-, and SHV-type) and carbapenemases (IMP-1-type, VIM-2-type, NDM, KPC, GES, and OXA-48-like), as previously described [14]. Both strands of the amplicons were sequenced using an automatic sequencer (model 3730xl; Applied Biosystems, Weiterstadt, Germany), and the nucleotide sequences were compared using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast).
4. Multilocus sequence typing
MLST was carried out using partial sequences of seven E. coli housekeeping genes, including adk, fumC, gyrB, icd, mdh, purA, and recA. Nucleotide sequences were compared with those in the MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) to identify allelic types and STs.
5. Bacterial conjugation
E. coli strain EcU443 was used as the donor and sodium azide-resistant E. coli J53 was used as the recipient for the standard agar mating method [15]. Following overnight mating at 37℃ on brain-heart infusion (BHI; MB cell, Los Angeles, CA, USA) agar, transconjugants were selected on BHI supplemented with 100 µg/mL sodium azide and 0.5 µg/mL imipenem.
6. Whole genome sequencing
The complete DNA sequences of the plasmids present in EcU443 were obtained via Single-Molecule Real-Time sequencing on a PacBio RSII instrument (Pacific Biosciences, Menlo Park, CA, USA), according to the manufacturer's instructions. The sequences were annotated using Prokka 1.11 (http://www.vicbioinfor-matics.com/software.prokka.shtml). Resistance genes, IS elements, replication origins, virulent elements, and toxins and anti-toxin systems were identified using ResFinder (https://cge.cbs.dtu.dk//services/ResFinder/), IS-finder (https://www-is.biotoul.fr/), plasmid finder (https://cge.cbs.dtu.dk//services/Pla-smidFinder/), the Virulence Factor Database (http://www.mgc.ac.cn/VFs/), and TA Finder 1.0 (http://202.120.12.133/TAfinder/index.php), respectively.
7. GenBank accession numbers
Nucleotide sequence data for plasmids pECSEV_01 and pECSEV_02 are available under GenBank accession numbers KX683283 and KX683284, respectively.
RESULTS
1. Antimicrobial susceptibilities and molecular typing
The antimicrobial susceptibility profiles of the clinical E. coli strains are presented in Table 1. The isolates exhibited resis-tance to most antibiotics tested, including meropenem, ampicillin, ceftriaxone, gentamicin, and ciprofloxacin, but were susceptible to tigecycline and colistin. Phenotypic carbapenemase differentiation tests showed positive results for KPC production in both isolates (EcU443 and EcU120; Table 2). The MLST assay assigned both isolates as an identical ST1642 (6-4-5-18-11-8-6). PCR and sequencing for β-lactamase genes demonstrated the presence of blaKPC-2, blaSHV-11, and blaTEM-1 in both isolates.
Table 1. Antimicrobial susceptibilities of clinical Escherichia coli isolates*.
| Antibiotics | MIC (mg/L) | Antibiotics | Zone diameter (mm) | Interpretation‡ | |||
|---|---|---|---|---|---|---|---|
| EcU443† | EcU213† | EcU120† | EcU443 | EcU120 | |||
| AMP | ≥ 32 | ≥ 32 | ≥ 32 | AMP | 6 | 10 | R |
| SAM | ≥ 32 | ≥ 32 | ≥ 32 | SAM | 6 | 6 | R |
| TZP | ≥ 128 | ≥ 128 | ≥ 128 | PIP | 10 | 10 | R |
| ATM | ≥ 64 | ≥ 64 | ≥ 64 | TIC | 10 | 10 | R |
| CAZ | 4 | 4 | 16 | CRO | 15 | 16 | R |
| GEN | ≥ 16 | ≥ 16 | ≥ 16 | GEN | 7 | 7 | R |
| LVX | ≥8 | ≥8 | ≥8 | CIP | 6 | 6 | R |
| CFZ | ≥ 64 | ≥ 64 | ≥ 64 | IMP | 20 | 19 | I |
| MEM | ≥ 16 | ≥ 16 | ≥ 16 | MEM | 20 | 20 | I |
| ERM | 4 | 4 | ≥8 | ERM | 17 | 17 | R |
| CTM | ≥ 320 | ≥ 320 | ≥ 320 | CST | 14 | 15 | - |
| TGC | ≤ 0.5 | ≤ 0.5 | ≤ 0.5 | TGC | 24 | 24 | - |
*The breakpoints were applied according to the Clinical and Laboratory Standards Institute (CLSI) guidelines; the resistance values are in bold; †EcU443, EcU213, and EcU120 were isolated on the 8th, 18th, and 42nd day post admission, respectively; ‡The EcU443 disk diffusion tests results were interpreted according to the CLSI guidelines; the results for colistin and tigecycline are not shown because of the lack of suggested breakpoints.
Abbreviations: AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CFZ, cefazolin; CIP, ciprofloxacin; CRO, ceftriaxone; CST, colistin; ERM, ertapenem; GEN, gentamicin; I, intermediate; IMP, imipenem; LVX, levofloxacin; MEM, meropenem; MIC, minimum inhibitory concentration; PIP, piperacillin; R, resistant; SAM, ampicillin-sulbactam; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; TIC, ticarcillin; TZP, piperacillin-tazobactam.
Table 2. Results of carbapenemase differentiation tests and sequence types of Escherichia coli isolates.
| E. coli isolates* | Carbapenemase differentiation test | MLST | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | MEM+DPA | MEM+PBA† | MEM+CLX | ST | adk | fumC | gyrB | icd | mdh | purA | recA | |
| EcU443 | 19 | 19 | 24 | 20 | 1642 | 6 | 4 | 5 | 18 | 11 | 8 | 6 |
| EcU120 | 20 | 20 | 25 | 21 | 1642 | 6 | 4 | 5 | 18 | 11 | 8 | 6 |
*EcU443 and EcU120 were isolated on the 8th and 42nd day post admission, respectively; †A difference between MEM+PBA and MEM >5 mm indicated KPC production.
Abbreviations: CLX, cloxacillin; DPA, dipicolinic acid; MEM, meropenem; MLST, Multilocus sequence typing; PBA, phenylboronic acid.
2. Plasmids pECSEV_01 and pECSEV_02
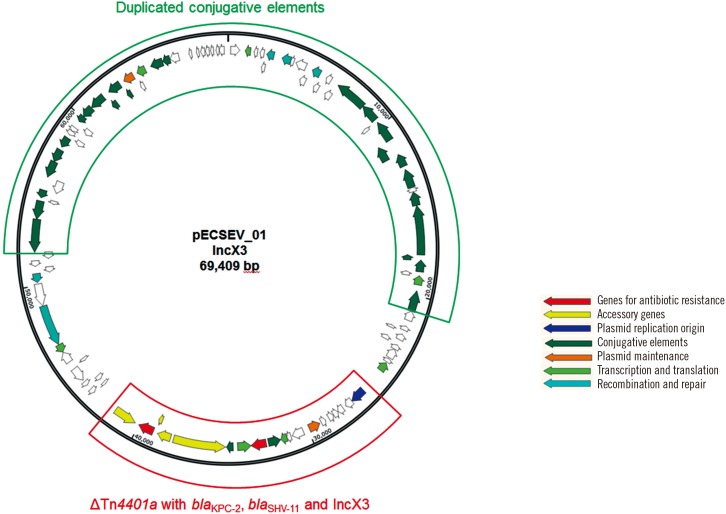
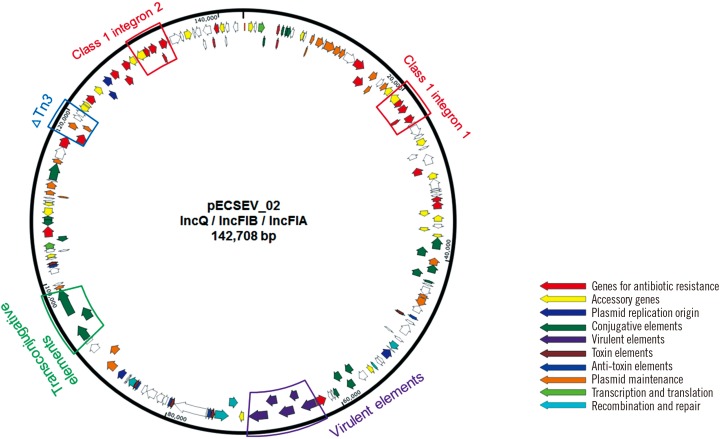
E. coli strain EcU443 had a 4,769,071-bp chromosome and four plasmids. The chromosome did not contain any acquired antimicrobial resistance determinants. The 69,409-bp IncX3 plasmid (pECSEV_01) carried blaKPC-2 and blaSHV-11 for β-lactam resistance; the 142,708-bp multireplicon (IncFIA, IncFIB, and IncQ) plasmid (pECSEV_02) included strA, strB, aadA5, and aac(3)-IId for aminoglycoside resistance, blaTEM-1b for β-lactam resistance, mph(A) for macrolide resistance, sul1 and sul2 for sulfonamide resistance, tet(B) for tetracycline resistance, and dfrA17 for trimethoprim resistance; and the 118,130-bp plasmid (pECSEV_03) harbored blaTEM-1b for β-lactam resistance and qnrS1 for quinolone resistance. Moreover, a cryptic 110,571-bp plasmid was also identified.
Plasmid pECSEV_01 carrying the blaKPC-2 gene belonged to the IncX3 group (Fig. 1). This plasmid could be transferred to recipient E. coli J53 by surface mating. The blaKPC-2 gene was located within a truncated Tn4401 transposon; ΔISKpn7-blaKPC-2-ISKpn6 had a 99-bp deletion between ISKpn7 and blaKPC indicating it was a Tn4401a isoform. The plasmid also harbored the blaSHV-11 gene encoding a broad-spectrum β-lactamase. The replication origin repB gene belonged to the IncX3 incompatibility type. The remaining sections of pECSEV_01 consisted of duplicated type IV secretion systems and conjugative elements.
Fig. 1. Circular map of pECSEV_01 containing blaKPC-2, blaSHV-11, and duplicated conjugative elements.
Plasmid pECSEV_02 possessed three replication origins for the IncFIA, IncFIB, and IncQ groups (Fig. 2). The plasmid carried two identical copies of a class 1 integron, In54, containing the dhfrA17-aadA5-emrE gene cassettes for trimethoprim-, ami-noglycosides-, and multidrug-resistance, as well as a truncated Tn3 transposon harboring blaTEM-1b. The macB, mph(A), aac(3)-IId, strA, strB, and tet(B) drug resistance genes were also identified. In addition to drug-resistance determinants, pECSEV_02 also included a virulence gene cluster containing the iucA, iucB, iucC, iucD, and iutA genes involved in hydroxamate siderophore aerobactin synthesis and two toxin/antitoxin systems, vapBC and ccdAB.
Fig. 2. Circular map of pECSEV_02 containing blaTEM-1, two class 1 integrons, virulence elements, and transconjugative elements.
DISCUSSION
E. coli clinical isolates EcU443 and EcU120 were serially recovered from urinary specimens of a patient with a 34-day interval. Both strains belong to ST1642, exhibit resistance or intermediate resistance to all classes of antimicrobial agents tested, except for tigecycline and colistin, and possess the blaKPC-2, blaSHV-11, and blaTEM-1 β-lactamase genes, indicating that this clone was responsible for the persistent infection in the patient over the 34 days. To the best of our knowledge, E. coli ST1642, first isolated from a pet cat as an extraintestinal pathogenic strain [16], has not been previously reported as a KPC-producer. Although ST131 strains are the most frequently identified in KPC-producer E. coli infections [17], three cases of KPC-2-producer infections due to ST69, ST393, and a new E. coli ST were recently identified in Busan [18]. The three E. coli strains carried an IncX3 plasmid similar to that in our strains, although the E. coli strains were epidemiologically unrelated.
E. coli strain EcU443 possesses four plasmids. The blaKPC-2 gene is on IncX3 plasmid pECSEV_01 containing the blaSHV-11 gene. The plasmid showed 99% identity with IncX3 plasmid pKpS90 from a K. pneumoniae ST258 strain (GenBank accession number JX461340) isolated from a blood culture during a hospital outbreak in France [19]. In pECSEV_01, the truncated Tn4401a carrying the blaKPC-2 gene is integrated in the opposite direction, but at the same location as in pKpS90. The truncated Tn4401a has also been found in IncX3 plasmid pKPC_Kp01 from a clinical K. pneumoniae isolate responsible for a hospital outbreak in Busan [18].
Plasmid pECSEV_02, containing multiple replicons of IncFIA, IncFIB, and IncQ, carries various genes conferring resistance to diverse classes of antimicrobial agents. Interestingly, pECSEV_02 harbors two identical copies of class 1 integron In54. An Enterobacter cloacae strain carrying multiple non-identical class 1 integrons in a plasmid has been identified [20]. Plasmid pECSEV_02 also carries genes for virulence factors and for toxin/antitoxin systems that can enhance bacterial fitness in a human host, thus explaining its long persistence in the patient. The third plasmid, pECSEV_03, also contributed to the extensively drug-resistant (XDR) E. coli via blaTEM-1b and qnrS1.
This study reports a persistent infection case caused by an XDR E. coli ST1642 strain carrying an IncX3 plasmid containing blaKPC-2 associated with a truncated Tn4401a transposon and a plasmid with multireplicons of IncQ, IncFIA, and IncFIB, which contains genes conferring resistance to multiple classes of antimicrobial agents. Sporadic or epidemic infection cases caused by KPC-producers have been increasingly reported in Korea; the blaKPC genes are spreading to K. pneumoniae as well as other species in the family Enterobacteriaceae, including E. coli, via R plasmids. Further studies are needed to investigate the current status of KPC-producers in Korea.
Acknowledgements
This study was supported by the Research Program funded by the Korea Centers for Disease Control and Prevention (2016ER230100#).
References
- 1.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic Context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, et al. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 2014;69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 3.Adler A, Miller-Roll T, Assous MV, Geffen Y, Paikin S, Schwartz D, et al. A multicenter study of the clonal structure and resistance mechanism of KPC-producing Escherichia coli isolates in Israel. Clin Microbiol Infect. 2015;21:230–235. doi: 10.1016/j.cmi.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Piazza A, Caltagirone M, Bitar I, Nucleo E, Spalla M, Fogato E, et al. Emergence of Escherichia coli Sequence Type 131 (ST131) and ST3948 with KPC-2, KPC-3 and KPC-8 carbapenemases from a Long-Term Care and Rehabilitation Facility (LTCRF) in Northern Italy. Adv Exp Med Biol. 2016;901:77–89. doi: 10.1007/5584_2015_5017. [DOI] [PubMed] [Google Scholar]
- 5.Chavda KD, Chen L, Jacobs MR, Bonomo RA, Kreiswirth BN. Molecular diversity and plasmid analysis of KPC-producing Escherichia coli. Antimicrob Agents Chemother. 2016;60:4073–4081. doi: 10.1128/AAC.00452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu G, Jiang Y, An W, Wang H, Zhang X. Emergence of KPC-2-producing Escherichia coli isolates in an urban river in Harbin, China. World J Microbiol Biotechnol. 2015;31:1443–1450. doi: 10.1007/s11274-015-1897-z. [DOI] [PubMed] [Google Scholar]
- 7.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 8.He S, Chandler M, Varani AM, Hickman AB, Dekker JP, Dyda F. Mechanisms of evolution in high-consequence drug resistance plasmids. MBio. 2016;7:e01987–e01916. doi: 10.1128/mBio.01987-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob Agents Chemother. 2008;52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naas T, Cuzon G, Truong HV, Nordmann P. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother. 2012;56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Twenty-sixth informational supplement, M100-S26. Wayne, PA: National Committee for Clinical Laboratory Standards; 2016. [Google Scholar]
- 12.Kim MN, Yong D, An D, Chung HS, Woo JH, Lee K, et al. Nosocomial clustering of NDM-1-producing Klebsiella pneumoniae sequence type 340 strains in four patients at a South Korean tertiary care hospital. J Clin Microbiol. 2012;50:1433–1436. doi: 10.1128/JCM.06855-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, et al. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J Clin Microbiol. 2011;49:579–585. doi: 10.1128/JCM.01588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong S, Kim JO, Jeong SH, Bae IK, Song W. Evaluation of peptide nucleic acid-mediated multiplex real-time PCR kits for rapid detection of carbapenemase genes in gram-negative clinical isolates. J Microbiol Methods. 2015;113:4–9. doi: 10.1016/j.mimet.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 15.Jeong SH, Lee KM, Lee J, Bae IK, Kim JS, Kim HS, et al. Clonal and horizontal spread of the blaOXA-232 gene among Enterobacteriaceae in a Korean hospital. Diagn Microbiol Infect Dis. 2015;82:70–72. doi: 10.1016/j.diagmicrobio.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Harada K, Nakai Y, Kataoka Y. Mechanisms of resistance to cephalosporin and emergence of O25b-ST131 clone harboring CTX-M-27 β-lactamase in extraintestinal pathogenic Escherichia coli from dogs and cats in Japan. Microbiol Immunol. 2012;56:480–485. doi: 10.1111/j.1348-0421.2012.00463.x. [DOI] [PubMed] [Google Scholar]
- 17.O'Hara JA, Hu F, Ahn C, Nelson J, Rivera JI, Pasculle AW, et al. Molecular epidemiology of KPC-producing Escherichia coli: occurrence of ST131-fimH30 subclone harboring pKpQIL-like IncFIIk plasmid. Antimicrob Agents Chemother. 2014;58:4234–4237. doi: 10.1128/AAC.02182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JO, Song SA, Yoon EJ, Shin JH, Lee H, Jeong SH, et al. Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn4401a. Diagn Microbiol Infect Dis. 2017;87:343–348. doi: 10.1016/j.diagmicrobio.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Kassis-Chikhani N, Frangeul L, Drieux L, Sengelin C, Jarlier V, Brisse S, et al. Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013;57:618–620. doi: 10.1128/AAC.01712-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plante I, Centrón D, Roy PH. Direct sequencing and PCR mapping of integrons reveals multiple class 1 integrons in the multiresistant strain Enterobacter cloacae SCH88040794. FEMS Microbiol Lett. 2003;221:59–62. doi: 10.1016/S0378-1097(03)00163-0. [DOI] [PubMed] [Google Scholar]