Abstract
DNA methylation is an important epigenetic modification that regulates a wide range of biological processes including immune response. However, information on the epigenetics-mediated immune mechanisms in insects is limited. Therefore, in this study, we examined transcriptomes and DNA methylomes in the fat body and midgut tissues of silkworm, Bombyx mori with or without B. mori cytoplasmic polyhedrosis virus (BmCPV) infection. The transcriptional profile and the genomic DNA methylation patterns in the midgut and fat body were tissue-specific and dynamically altered after BmCPV challenge. KEGG pathway analysis revealed that differentially methylated genes (DMGs) could be involved in pathways of RNA transport, RNA degradation, nucleotide excision repair, DNA replication, etc. 27 genes were shown to have both differential expression and differential methylation in the midgut and fat body of infected larvae, respectively, indicating that the BmCPV infection-induced expression changes of these genes could be mediated by variations in DNA methylation. BS-PCR validated the hypomethylation of G2/M phase-specific E3 ubiquitin-protein ligase-like gene in the BmCPV infected midgut. These results demonstrated that epigenetic regulation may play roles in host-virus interaction in silkworm and would be potential value for further studies on mechanism of BmCPV epithelial-specific infection and epigenetic regulation in the silkworm.
Introduction
Pathogens infection, silkworm strain, and climatic condition have drastic effects on the yield and quality of the silk produced. BmCPV is a major virus that specifically infects the epithelial cells of the silkworm midgut1,2. BmCPV is a double-stranded RNA (dsRNA) virus that belongs to the CPV subfamily within the genus Cypovirus, family Reoviridae3. The genome of BmCPV is composed of 10 discrete dsRNA segments among which S1-S4 and S6-S7 encode the viral structural proteins, while S5 and S8-S10 encode nonstructural proteins4. The infected silkworms are characterized by hypogenesis, emaciation, and sluggishness. As the infection advances, white wrinkles can be observed in the posterior part of the midgut, which is the typical symptom of CPV infection-induced disease1.
Studies on BmCPV-host interaction have reported as follows: (1) Gene expression profiles in the silkworm midgut changed after BmCVP infection based on its interactions with the pathogen5–7. For example, some genes related to immune response, such as serpin5, heat shock protein genes, amino peptidases N enzymes, were induced by BmCPV, while those genes related to metabolic pathway were down-regulated7. (2) Differentially expressed microRNAs are observed in BmCPV-infected silkworm midguts8. Cellular microRNAs and BmCPV genome encoded microRNAs could affect BmCPV replication9,10, suggesting that microRNAs may play important roles in BmCPV-host interaction. (3) Autophagy is only slightly induced by BmCPV infection, and could not prevent the invasion and replication of the virus11. (4) Establishing a BmCPV infection depends on other factors interacting with integrin beta and receptor for activated protein kinase C (RACK1) to form a receptor complex for the entry of BmCPV12. Thus far, the molecular mechanism underlying the midgut infection of BmCPV and the antivirus immune response to BmCPV infection is not clearly understood due to the complexity in dsRNA virus infection.
DNA methylation, one of the most important epigenetic modifications in eukaryotes, has been proven to play key roles in a broad range of biological processes including the regulation of gene expression, tumorigenesis, embryonic development, virus infection, and antiviral defenses13–17. Compared to vertebrates and plants, there are some unique features in the DNA methylation of insects. It is mainly mediated by DNA methyltransferases (Dnmts), which vary greatly in different insect species18–21. In B. mori only two have been reported, DNMT1 and DNMT2. BmDNMT-1 retained the function as maintenance DNMT, but its sensitivity to metal ions is different from mammalian DNMT-122. In insect genomes, DNA methylation is predominant in gene bodies23,24 and enhances gene expression25. The overall level of DNA methylation in most insects is lower than 1%24,26–28. In insects, DNA methylation mostly regulates embryonic development, participates in genomic imprinting, regulates caste and wing differentiation, sex determination, and is involved in pesticide resistance24,28–32.
When the host is invaded by pathogens, gene expression is regulated via changes in DNA methylation levels or patterns, leading to the activation or suppression of relevant signaling pathways and triggering a series of immune responses against viral invasion15. Till date, functional analysis of DNA methylation in insect-pathogens interaction is limited. Therefore, in this study, we profiled DNA methylomes and transcriptomes in the fat body and midgut tissues of B. mori with or without BmCPV infection using whole-genome bisulfite sequencing (WGBS) and RNA sequencing (RNA-Seq) technologies. The correlation between gene expression and DNA methylation patterns was analyzed. This is the first report on exploring the epigenetic regulation mechanisms on the immune response of B. mori and our findings provide insight into the transcriptional and epigenetic mechanisms underlying the interactive responses between silkworm and pathogens.
Results and Discussion
BmCPV infection confirmation
BmCPV exclusively infected the epithelial cells of silkworm midguts. However, the fat body, which is the immune organ of silkworm larvae, was hardly infected by BmCPV. Infection of the midgut was confirmed by observing white wrinkles in posterior midguts and multiple polyhedrons through the microscope, while in fat bodies of infected silkworms, no polyhedron was observed. Further, quantitative real-time PCR (qRT-PCR) results revealed that the expression of BmCPV polyhedrin gene in fat bodies of infected silkworms was significantly lower than in midguts, suggesting that fat body was not infected by BmCPV (Supplementary Fig. S1).
Transcriptional profiles of silkworm fat body and midgut are altered upon BmCPV infection
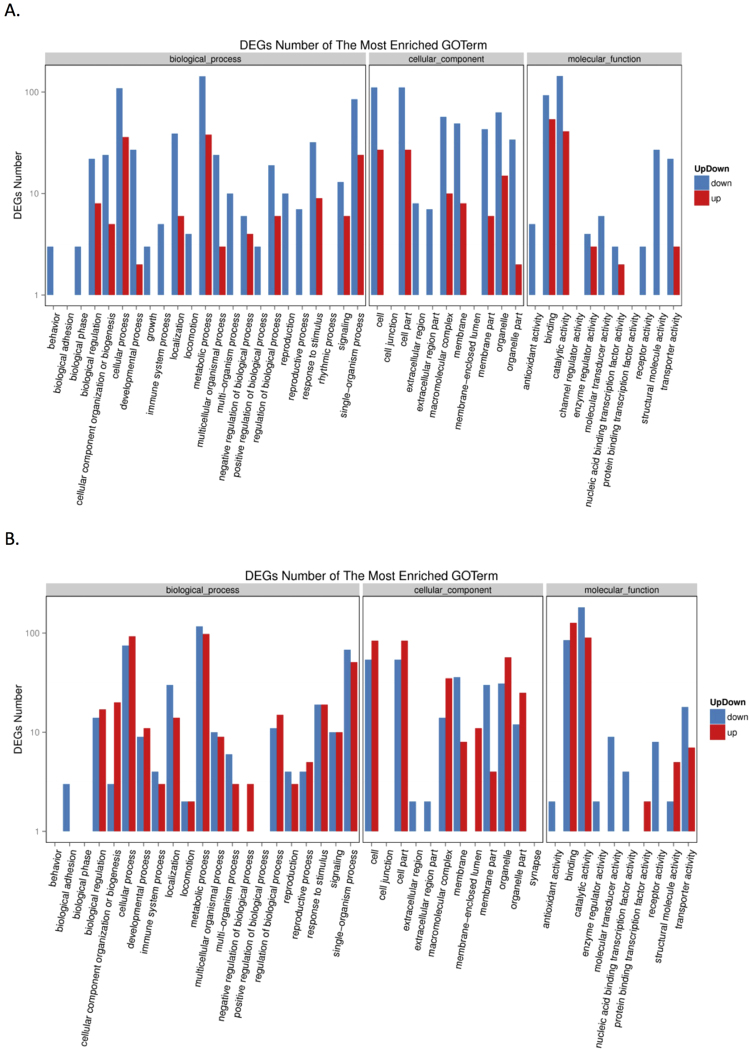
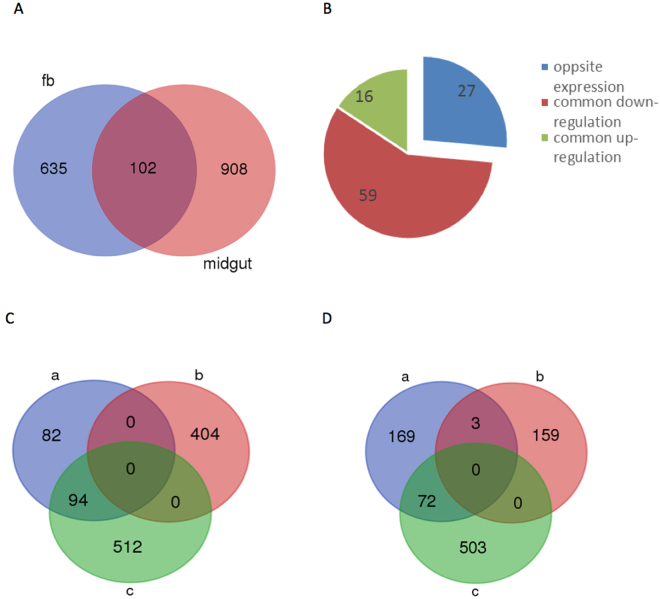
The mechanism of the midgut epithelia-specific infection by BmCPV is not reported yet. To explore the potential mechanism, we compared the transcriptional profiles of the midgut and fat body in control and infected silkworm larvae by RNA-Seq. More than 13,000,000 clean reads were obtained from each sample and the summary of the sequenced data is shown in Supplementary Table S1. The sequence data are deposited in the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/sra/) under the accession number PRJNA381877. Hierarchical clustering analysis suggested that midgut and fat body had distinct gene expression profiles and were clearly separated (Supplementary Fig. S2). The number of significantly differentially expressed genes in infected midgut was 1010, with 404 genes up-regulated and 606 genes down-regulated (Supplementary Table S2). In the fat body, we detected 737 differentially expressed genes, among which, the transcript level of 162 genes was increased while 575 genes was decreased in response to BmCPV infection (Supplementary Table S3). KEGG pathway analysis showed that the differentially expressed genes of both midgut and fat body tissues were involved in cellular processes, environmental information processing, genetic information processing, human diseases, metabolism and organismal systems pathways (Supplementary Fig. S3). GO analysis revealed that the down-regulated genes in the fat body were enriched in behavior, biological phase, growth, reproductive process, immune system process, receptor activity, structural molecule activity, etc. (Fig. 1A). In the midgut, we found that the genes classified to the negative regulation of biological process, protein binding transcription factor activity, and membrane-enclosed lumen were induced by BmCPV (Fig. 1B). Further, venn diagram analysis revealed 102 genes, which differentially expressed in both midguts and fat bodies of infected silkworms (Fig. 2A, Supplementary Table S4). Among them, 16 genes were up-regulated and 59 genes were down-regulated in both tissues. Notably, within the 16 up-regulated genes, 7 genes with the GO function of binding, accounted for 78% of the GO annotation, suggesting that these genes may be involved in BmCPV infection and may play similar roles or have similar effects in response to BmCPV infection in different tissues. Interestingly, we found 27 genes to have the opposite expression pattern induced by BmCPV infection (Fig. 2B). For example, the B. mori polyubiquitin-A-like isoform X1 gene (GeneID: 101742698) was up-regulated in fat body of infected larvae while it was significantly down-regulated in infected midgut. Transcript levels of the ribosome protein genes (GeneID: 692424, 692657, and 732854) and several immune-related genes (GeneID: 101738190, 101742015, and 101739771) was up-regulated in the infected midgut but down-regulated in fat body of the infected silkworms. We presume that the opposite expression pattern may indicate that these genes may have different roles or may be involved in different pathways response due to tissue specificity.
Figure 1.
Annotation of differentially expressed genes with WEGO in fat body (A) and midgut (B). GO annotation was performed by mapping genes to GO terms in the GO database (http://www.geneontology.org). GO enrichment analysis was conducted by hypergeometric test with corrected p-value ≤ 0.05 as a threshold. Gene numbers and percentages are listed for each category.
Figure 2.
Transcriptional patterns of silkworm fat body and midgut are altered upon BmCPV infection. (A) Venn diagram shows 737 (left panel) and 1010 (right panel) differentially expressed genes in fat bodies and midguts of infected larvae, respectively compared to uninfected tissues. In total, 102 genes show differential expression in both fat bodies and midguts. (B) Among the common 102 genes, 16 are up-regulated and 59 are down-regulated. The expression pattern of 27 genes in fat bodies is opposite to that in midguts. (C) Venn diagram shows 94 highly or uniquely expressed genes down-regulated in midguts upon BmCPV infection. “a” represents highly or uniquely expressed genes in midguts. “b” represents up-regulated genes in infected midguts. “c” represents down-regulated genes in infected midguts. (D) Venn diagram shows 3 and 72 highly or uniquely expressed genes up-regulated and down-regulated in fat bodies, respectively upon BmCPV infection. “a” represents highly or uniquely expressed genes in fat bodies. “b” represents up-regulated genes in fat bodies of infected larvae. “c” represents down-regulated genes in fat bodies of infected larvae.
We compared the abundance of genes between control fat body and control midgut to determine the tissue-specific expression profile. The results revealed that about 3000 genes displayed differential expression pattern between the two tissues. We selected the genes with log2-ratio value >6.6 (the expression level of these genes in midgut was 100 times greater than in fat body) or <−6.6 (the expression level of these genes in fat body was 100 times greater than in midgut) in control fat body vs. control midgut group as highly or uniquely expressed genes. As a result, a total of 176 and 244 highly or uniquely expressed genes were identified in the midgut and fat body, respectively (Supplementary Table S5). For example, esterase-related genes (GeneID: 101738777, 101745376, 100144572, 101740845, and 692770) were highly expressed in midgut but not expressed in fat body. The genes with unique expression in the midgut or fat body may play key roles in maintaining tissue-specificity. We then explored how the expression level of these genes was changed after the BmCPV challenge. For this, venn diagrams were created with the data of differentially expressed genes and highly expressed genes. The results (Fig. 2C,D) demonstrated that in the midgut, 94 highly or uniquely expressed genes (>50%) were down-regulated after BmCPV infection (Supplementary Table S6), while in the fat body, 30% of the highly or uniquely expressed genes showed differential expression including 73 down-regulated and 3 up-regulated following BmCPV infection (Supplementary Table S6). GO analysis revealed that these down-regulated genes in the fat body were grouped into 25 categories, among which, 12 categories were unique compared to the midgut and included developmental process, enzyme regulator activity, immune system process, etc. (Supplementary Fig. S4).
Changes in transcript levels of host genes appear to be a common consequence of a viral infection7. Comparing the transcriptome of midgut with that of fat body, we can get several features: (1) Although BmCPV hardly infects the fat body, more than 700 genes were observed to have differential expression and 80% accounted for down-regulation (Supplementary Table S3). The proportion of down-regulated genes was higher than that of midgut (60%). (2) Among 102 genes, which differentially expressed in both midguts and fat bodies of infected silkworms, there were 27 genes to have the opposite expression pattern in different tissue induced by BmCPV infection. (3) Most of those DEGs, (>90%), which are highly or uniquely expressed in midgut or fat body were all down-regulated after BmCPV infection and the functions of these DEGs are diverse. These features suggest that the midgut and fat body have distinct expression profiles. The molecular mechanisms against BmCPV may be different between the two tissues. Large quantity of down-regulated genes were detected during the late stage of BmCPV infection (96 h), suggesting that the expression patterns of genes related to BmCPV infection were reorganized for anti-viral responses to diminish the negative effects of viral invasion or to facilitate viral proliferation and avoid host defense triggered by virus.
We could not however, clearly demonstrate the molecular mechanism for epithelia-specific infection by BmCPV. Supplementary Table S4–S6 provide a lists of candidate genes for future functional characterization. We will particularly focus on the functions of genes involved in membrane binding, trans-membrane transduction, receptor activity, and immune response by using transgenesis and RNAi technologies.
Identifying changes in DNA methylation patterns in the host cell genome after infection with BmCPV
Alternations in global DNA methylation patterns due to pathogen invasion have been widely reported in mammals and plants14,33–35. To evaluate epigenetic changes in insect cells caused by BmCPV infection, methylation of genomic DNA was determined in four samples, namely fat body 96t, midgut 96t (BmCPV infected), fat body 96c, and midgut 96c (BmCPV uninfected) using bisulfite sequencing. We observed that about 0.6% of genomic cytosines are methylcytosines, and that 81% of these were CG dinucleotides (Fig. 3A). Xiang et al. reported that non-CG mCs are either nonexistent or very rare in the silkworm25. Here, we found about 19% mCs at CHG and CHH sites (H = A, C or T), further study need to be carried out for verifying the true or false positive. The average methylation level at CG locations in the whole genome was about 1%, which is significantly higher than 0.67% in the silk glands of silkworm25. In order to understand the relationship between DNA methylation profiles and gene expression, DNA methylation profiles were divided into seven distinct genomic functional regions to study the changes in methylation levels (Fig. 3B). In agreement with previous studies on other insects23–25,27, we found that in silkworm DNA methylation is mostly targeted to gene bodies. DNA methylation levels peaked in the first exon and sharply decreased in intron regions, with 3′ downstream region showing more methylation than the 5′ upstream region. This result is similar to the pattern reported in other insects25,28,33. Fractional methylation levels in gene regions exhibited a different pattern between infected and non-infected samples. Genomic DNA methylation levels of each of the four samples were distinct as shown in Supplementary Fig. S5, suggesting that DNA methylation is tissue-specific and dynamically responds to stimuli.
Figure 3.
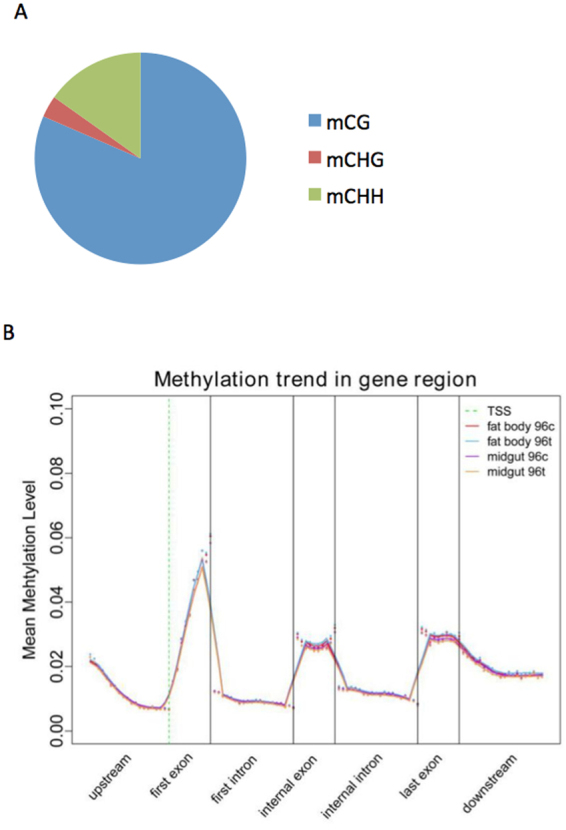
Average DNA methylation level and the distribution in the whole genome in four samples. (A) The average proportion of different types of methylated cytosines in the four samples. (B) Distribution of DNA methylation levels of genes in four samples. The gene structure is defined by seven different features, denoted by the x-axis. The length of each feature was normalized and divided into equal numbers of bins. Each dot denotes the mean methylation level per bin and the respective lines denote the 5-bin moving average. Each feature was analyzed separately for the numbers listed in the table below the figure. The green vertical line indicates the mean location of the transcription start sites.
Differential DNA methylation genes (DMGs) in response to BmCPV infection
DNA methylation is reported to be involved in antiviral responses15,17. To explore the potential function of DNA methylation in insect immune response, we compared the DMR of the whole genome between BmCPV infected midgut or fat body with their corresponding uninfected control tissues. The results revealed that 394 DMRs were present in the midgut 96t vs. midgut 96c group with 271 located in the gene region. Among these, 119 were hypermethylated and 151 genes were hypomethylated (Supplementary Table S7). In the fat body, 591 DMRs were observed and 410 were associated with genes, among which 223 genes were hypermethylated and 187 were hypomethylated (Supplementary Table S8). KEGG pathway analysis of DMGs revealed that DMGs were involved in pathways of RNA transport, RNA degradation, nucleotide excision repair, DNA replication, etc. (Fig. 4), suggesting that variation in DNA methylation could regulate the expression of genes involving in multiple important pathways.
Figure 4.
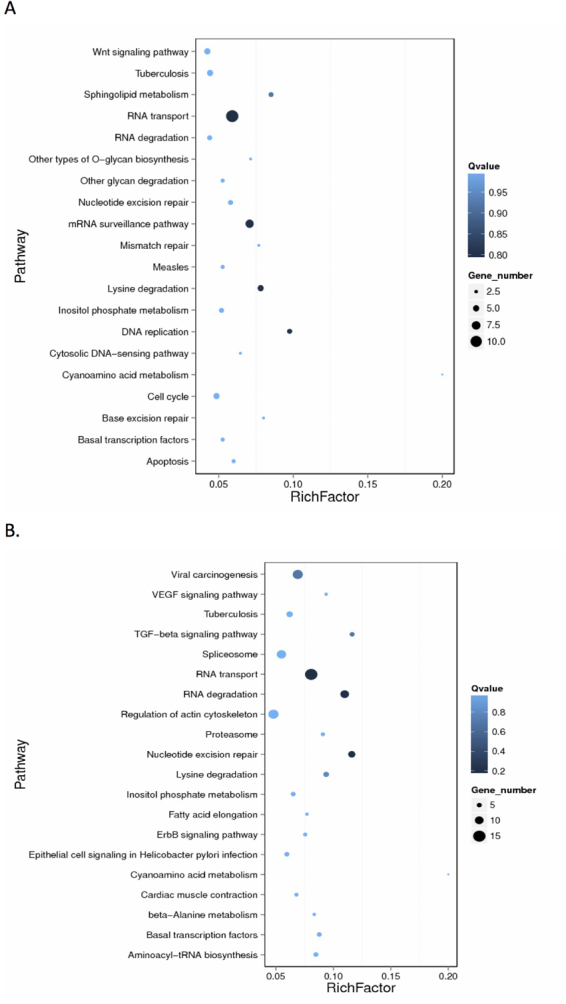
KEGG pathway enrichment of differentially methylated genes following BmCPV infection. (A) Differentially methylated genes in infected midguts. (B) Differentially methylated genes in fat bodies of infected larvae. RichFactor is the ratio of the number of differentially expressed genes in this pathway term to the number of all genes in this pathway term. Greater RichFator means greater intensiveness. Qvalue is corrected p-value ranging from 0–1, and less Qvalue means greater intensiveness. Only the top 20 of the enriched pathway terms are displayed here.
Effects of DMR on gene expression level in silkworm after infection with BmCPV
To determine whether the changes in CG methylation observed in silkworm following BmCPV infection altered gene expression, we synthetically analyzed the data of DEGs and DMRs. As a result, we found 27 DMGs showing differential mRNA levels in the fat body of the infected silkworm (Table 1) and midgut, respectively (Table 2), indicating that these genes related to BmCPV infection may change their expression level via variation in DNA methylation. Analysis of the distribution of DNA methylation on these genes revealed that CG methylation in all 27 genes in the midgut occurred in the gene body, predominantly in the introns of genes (Supplementary Fig. S6). However, in the fat body, DNA methylation was observed in the 5′ or 3′ UTR regions of 6 genes. Both in midgut and fat body, there were parts of genes, whose intron-extron boundaries were also DNA methylated, suggesting that in B. mori DNA methylation is likely associated with alternative splicing36,37.
Table 1.
Gene lists of both DMGs and DEGs in fat bodies with BmCPV infection.
| Gene ID | log2Ratio of DEGs (fat_body_96t/fat_body_96c) | PPEE | log2Ratio of DMGs (fat_body_96t/fat_body_96c) | Annotation |
|---|---|---|---|---|
| 101738991 | 1.4241 | 9.36E-04 | −1.0340 | heterogeneous nuclear ribonucleoprotein 87F-like [Bombyx mori] |
| 732899 | 1.3718 | 1.34E-11 | 1.0000 | nascent polypeptide associated complex protein alpha subunit [Bombyx mori] |
| 101746243 | 1.3248 | 7.69E-03 | 1.2630 | protein KRI1 homolog [Bombyx mori] |
| 101742308 | 1.2914 | 7.67E-04 | 1.0510 | isoleucine–tRNA ligase, cytoplasmic-like [Bombyx mori] |
| 101746886 | 1.1813 | 4.92E-04 | 1.1600 | set1/Ash2 histone methyltransferase complex subunit ASH2-like [Bombyx mori] |
| 692990 | 1.1672 | 4.94E-02 | 1.1220 | DnaJ (Hsp40) homolog 2 [Bombyx mori] |
| 101739290 | 1.1455 | 3.88E-03 | −1.0000 | inhibitor of Bruton tyrosine kinase-like [Bombyx mori] |
| 101744580 | 1.0709 | 5.84E-04 | 1.0000 | hypothetical protein KGM_18067 [Danaus plexippus] |
| 692956 | 1.0531 | 2.27E-06 | −1.3790 | nucleoplasmin isoform 2 [Bombyx mori] |
| 101736883 | 1.0098 | 3.21E-07 | −1.0540 | protein DEK-like isoform X1 [Bombyx mori] |
| 101738436 | 1.0063 | 8.12E-07 | −1.6370 | splicing factor 3B subunit 1-like [Bombyx mori] |
| 101741560 | −1.0214 | 9.57E-06 | −1.6440 | uncharacterized protein LOC101741560 [Bombyx mori] |
| 101737841 | −1.0766 | 6.68E-04 | 1.3540 | acetyl-CoA hydrolase-like [Bombyx mori] |
| 693058 | −1.0958 | 1.20E-13 | 1.4590 | ribosomal protein S16 [Bombyx mori] |
| 101745599 | −1.1041 | 4.56E-03 | −1.2220 | enoyl-CoA hydratase precursor 1 [Bombyx mori] |
| 733116 | −1.1306 | 0.00 | −20.0000 | eukaryotic translation initiation factor 3 subunit F [Bombyx mori] |
| 101743877 | −1.1365 | 0.00 | 1.1520 | hypothetical protein KGM_08776 [Danaus plexippus] |
| 101746182 | −1.2844 | 2.22E-16 | 1.4950 | uncharacterized protein LOC101746182 [Bombyx mori] |
| 101738186 | −1.4037 | 1.48E-05 | 1.2220 | aconitate hydratase, mitochondrial-like [Bombyx mori] |
| 732876 | −1.4106 | 0.00 | −1.0930 | proteasome 25 kDa subunit [Bombyx mori] |
| 100134921 | −1.4995 | 2.73E-02 | 1.2060 | ubiquinol-cytochrome c reductase [Bombyx mori] |
| 101738009 | −1.6180 | 1.41E-05 | 1.1150 | ATP synthase lipid-binding protein, mitochondrial; |
| 101738244 | −1.6892 | 2.21E-02 | −1.0000 | putative Serine/threonine-protein kinase WNK3 [Danaus plexippus] |
| 733056 | −1.7736 | 4.38E-07 | −1.0000 | putative alcohol dehydrogenase [Bombyx mori] |
| 100862843 | −1.7892 | 0.00 | 1.9180 | UDP-glycosyltransferase, UGT41 A1recursor [Bombyx mori] |
| 101744219 | −1.9049 | 0.00 | 1.0700 | glycosyltransferase-like protein LARGE1-like [Bombyx mori] |
| 101735659 | −1.9256 | 0.00 | 2.3220 | glycine-rich cell wall structural protein 1.0-like [Bombyx mori] |
Table 2.
Gene lists of both DMGs and DEGs in midguts with BmCPV infection.
| Gene ID | log2Ratio of DEGs(midgut_96t/midgut_96c) | PPEE | log2Ratio of DMGs (midgut_96t/midgut_96c) | Annotation |
|---|---|---|---|---|
| 101735936 | 1.6258 | 5.30E-08 | −1.2730 | uncharacterized protein LOC101735936 [Bombyx mori] |
| 101735989 | 1.8947 | 1.26E-13 | −1.0750 | RNA-binding protein 28-like [Bombyx mori] |
| 101736907 | 1.7464 | 3.63E-03 | −1.0000 | uncharacterized protein LOC101742081 [Bombyx mori] |
| 101737128 | 3.0148 | 0.00 | 1.0440 | nucleolar protein 56-like [Bombyx mori] |
| 101737417 | 1.9544 | 2.45E-03 | 1.7000 | lisH domain-containing protein C1711.05-like [Bombyx mori] |
| 101739208 | 2.1410 | 9.18E-06 | −1.0700 | G2/M phase-specific E3 ubiquitin-protein ligase-like [Bombyx mori] |
| 101739510 | −1.5049 | 0.00 | −1.3920 | testin-like [Bombyx mori] |
| 101740026 | 1.9136 | 2.87E-04 | −1.5360 | nucleoporin NUP188 homolog [Bombyx mori] |
| 101740154 | 1.8013 | 1.88E-04 | −1.2220 | telomere-associated protein RIF1-like [Bombyx mori] |
| 101740265 | 1.2050 | 2.21E-02 | −1.2020 | structural maintenance of chromosomes protein 1A-like isoform X1 [Bombyx mori] |
| 101740584 | 1.5595 | 3.45E-11 | 1.1070 | cleavage and polyadenylation specificity factor subunit CG7185-like isoform X1 [Bombyx mori] |
| 101741392 | 2.2260 | 2.41E-02 | 1.3920 | glucosylceramidase-like [Bombyx mori] |
| 101741887 | 2.2111 | 1.63E-03 | 1.3440 | exonuclease 1-like [Bombyx mori] |
| 101742216 | −1.1865 | 3.22E-05 | −1.0420 | phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha-like [Bombyx mori] |
| 101742628 | 1.3424 | 0.00 | −1.0000 | splicing factor 3B subunit 2-like [Bombyx mori] |
| 101742901 | 1.0502 | 9.80E-07 | −1.4150 | importin subunit beta-1-like [Bombyx mori] |
| 101744992 | −1.1445 | 1.98E-02 | 1.9330 | apoptosis-stimulating of p53 protein 2-like [Bombyx mori] |
| 101745449 | 1.3282 | 0.00 | 4.3920 | protein lingerer-like [Bombyx mori] |
| 101746062 | −1.5040 | 0.00 | −1.2900 | nucleobindin-2-like isoform X1 [Bombyx mori] |
| 101746099 | 1.2062 | 0.00 | −1.3220 | zinc finger RNA-binding protein-like isoform X1 [Bombyx mori] |
| 101746131 | −1.7917 | 3.33E-16 | −1.1380 | multiple coagulation factor deficiency protein 2 homolog isoform X2 [Bombyx mori] |
| 692502 | 1.2292 | 0.00 | 1.4150 | casein kinase 2 alpha subunit [Bombyx mori] |
| 692744 | 1.2030 | 7.67E-04 | −1.0000 | 50 kDa lectin [Bombyx mori] |
| 692884 | −1.0291 | 3.12E-02 | −1.2020 | 2-hydroxyphytanoyl-CoA lyase [Bombyx mori] |
| 692945 | −2.5874 | 3.44E-11 | −1.2090 | kazal-type proteinase inhibitor precursor [Bombyx mori] |
| 692978 | 3.3555 | 1.33E-05 | 4.8070 | thymidylate synthase isoform 2 [Bombyx mori] |
| 733031 | −1.0130 | 4.80E-05 | 1.1070 | vacuolar ATP synthase subunit H [Bombyx mori] |
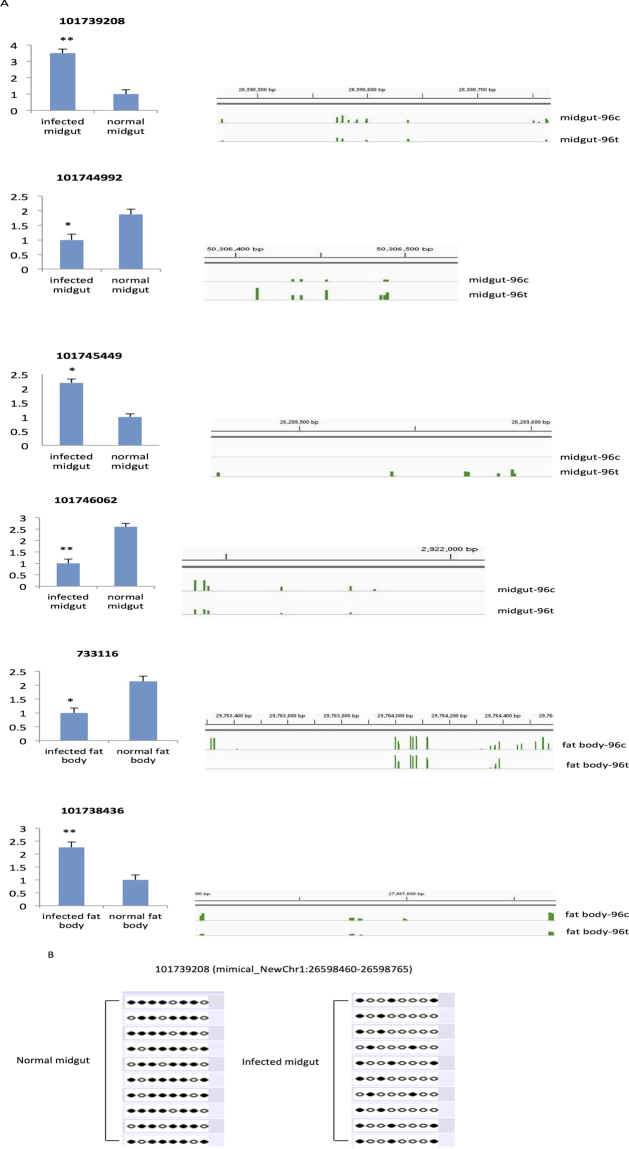
CG methylation in gene body is reported to have positive correlation with gene expression levels25,38. In this study, we found that the effects of DNA methylation in gene body could have a negative or positive effect on gene expression following BmCPV infection. Six genes were selected for mRNA level quantification by using qRT-PCR. The results obtained were consistent with RNA-Seq analysis (Fig. 5A). Moreover, the visual images for DNA methylation level and location in the genome of these six genes were also made by IGV (Version 2.3.83) (Fig. 5A).
Figure 5.
Both differentially expressed and differentially methylated genes in response to BmCPV infection. (A) Six genes were identified to be differentially expressed in midguts or fat bodies upon BmCPV infection by qRT-PCR on left side. RNA was extracted from midguts and fat bodies with or without BmCPV infection. Data represent the relative transcript levels of genes in the infected midguts or fat bodies compared to the control tissues from three independent samples. The error bars indicate standard deviations (**p < 0.01, *p < 0.05). The visualized image of differential methylation genes (right side) was generated by Integrative Genomics Viewer (IGV). The identification of methylated regions was performed by CpG_MPs (v1.1.0) with the required sequencing depth great than 10 and average C methylation ratio great than 0.55, and then converted to wig format by perl script for visualization on IGV. The location of green bars in the images indicates the methylated sites and the height of green bars represents the methylation levels. (B) Bisulfite sequencing validation of differentially methylated 101739208 in normal verse infected midgut. Targeted bisulfite sequencing validation of a region overlapping the DMR is indicated by the rectangle over the genome browser tracks. Dark circles indicate methylated and open circles mean unmethylated cytosines. Each row consists of a single sequenced clone.
In this study, we found that 101739208, a homolog of G2/M phase-specific E3 ubiquitin-protein ligase-like (G2E3), was up-regulated and hypomethylated in the midgut after BmCPV infection (Fig. 5B). G2E3 is an ubiquitin ligase (E3) that is essential for early embryonic development to prevent apoptotic death. It shuttles between the cytoplasm and nucleus, concentrating in the nucleoli and relocalizing to the nucleoplasm in response to DNA damage39,40. Apoptosis is an effective strategy used by insects to defend themselves against pathogen invasion. Loss of G2E3 triggered apoptosis and decreased proliferation of cancer cells41. On the other hand, up-regulation of G2E3 suppressed apoptosis progress and helped effective infection of BmCPV in the midgut. It is suggested that in silkworm, pathogens may successfully establish infection by manipulating the expression of host genes related to the apoptosis pathway via DNA methylation modification.
Moreover, the mRNA levels (p < 0.0001) of SF3B1 (GeneID: 101738436) and SF3B2 (GeneID: 101742628) were increased significantly in BmCPV infected midgut and fat body as detected in this study. In addition, hypomethylation was observed in the introns of SF3B1 and SF3B2 genes. The SF3B1 gene encodes subunit 1 of the splicing factor 3b, which is important for anchoring the spliceosome to precursor mRNA. Numerous studies have indicated that aberrant splicing patterns or mutations in spliceosome components, including SF3B1, SF3B3, and SF3B4, are associated with cancer and tumorigenesis and can act as potential anticancer agents42–44. Depletion of endogenous SF3B1 abrogated the apoptotic effects42. We presume that hypomethylation of SF3B1 and SF3B2 may increase the transcript levels of SF3B1 and SF3B2, which may play important roles in changing the splicing pattern, and attenuating the infectivity by apoptotic cells.
Hypermethylation and down-regulation of apoptosis stimulating protein, p53-2 (ASPP2, GeneID: 101744992), were also observed in BmCPV infected midgut. P53 is a central apoptotic regulator45, and ASPP2 is a damage-inducible p53-binding protein that enhances apoptosis through p53-dependent and p53-independent pathways46. Attenuation of ASPP2 is caused by hypermethylation of the promoter and 5′ UTR regions in native leukemia blasts47. The hypermethylation and down-regulation of ASPP2 in BmCPV infected midgut suggests that BmCPV infection may lead to hypermethylation of ASPP2, which in turn may suppress the expression of ASPP2 and facilitate the proliferation of infected cells.
Significant up-regulation of lingerer gene (Lig, GeneID: 101745449) in infected midgut were also detected (P = 0) in this study. Lig, an UBA domain-containing protein, interacts with the RNA-binding proteins FMR1, rasputin and caprin to restrict tissue growth in Drosophila melanogaster 48. Moreover, Lig functions as a critical growth suppressor to control organ size via bantam (ban) microRNA and Hippo signaling in D. melanogaster 49. Loss of Lig increased organ size and upregulated ban and the expression of Hippo pathway target genes, while overexpression of Lig diminished ban expression and reduced organ size reduction. Previously, we identified the down-regulation of bmo-bantam in BmCPV infected midgut8. Here, the significant up-regulation of Lig gene in infected midgut suggests that suppression of bmo-bantam may retard the growth of infected silkworm by increasing the expression of Lig gene. Notably, we observed hypermethylation of Lig gene in BmCPV infected midgut, indicating that the two epigenetic models, microRNA as well as DNA methylation may co-regulate the expression of Lig gene to regulate maldevelopment and undergrowth of BmCPV infected larvae.
Obvious down-regulation (P = 0) and significant hypomethylation of the subunit F of eukaryotic translation initiation factor 3 (eIF3f, GeneID: 733116) was observed in the fat bodies of infected silkworm larvae. eIF3f plays important roles in cell growth in human, yeast and Arabidopsis50. In addition, eIF3F can stabilize p53 and interact with CLU protein, a secretory heterodimeric glycoprotein thus leading to cancer cell proliferation in humans functioning as a CLU inhibitor51. The overexpression of eIF3f retarded cancer cell growth and induced apoptosis51. Accordingly, the obvious down-regulation (p = 0) and significant hypomethylation of eIF3f in fat bodies of infected silkworm larvae indicates that eIF3f may act as a potential gene that responds to BmCPV infection.
Nucleobindin 2 (GeneID: 101746062) was down-regulated and hypomethylated in BmCPV infected midguts. Nucleobindin 2 has been demonstrated to play critical roles in tumorigenesis and tumor development in breast cancer52. It can enhance cell migration and invasion in breast cancer, prostate cancer, and colon cancer cells53. Reduction in nucleobindin-2 expression inhibited EGF-stimulated MAPK kinase/Erk phosphorylation, cell proliferation54 and abrogate insulin-stimulated GLUT4 translocation55. In this study, the down-regulation and hypomethylation of nucleobindin-2 gene in BmCPV infected midgut suggests that the host may decrease expression level of some genes responsive to BmCPV infection by changing their DNA methylation level to resist the virus infection. Interestingly, another gene, Kazal-type proteinase inhibitor precursor (GeneID: 692945), which plays important roles in physiological processes such as development and immune response56,57 showed the same expression pattern with 101746062, with down-regulated mRNA level and hypomethylation in infected midguts.
In total, 176 and 244 genes were highly or uniquely expressed in the midgut and fat body, respectively. Protein sequences of genes targeted by high levels of methylation are conserved relative to genes lacking methylation. Genes that are methylated in all four invertebrate taxa (honeybee, silkworm, sea squirt, and sea anemone) are enriched for housekeeping functions related to transcription and translation58. The highly or uniquely expressed genes in midgut and fat body detected in this study may be related to tissue-specificity and play distinct roles in tissue function. We analyzed the methylation status of these genes and the results revealed that except four genes (GeneID: 101739540, 101743290, 101737724, and 692564) in fat body, others were all unmethylated or hypomethylated, indicating that DNA methylation is preferentially targeted towards genes with ubiquitous expression, whereas the loss of DNA methylation occurred in genes with tissue-specific functions.
In summary, this is the first report on characterizing genome-wide gene expression and DNA methylation patterns associated with BmCPV infection in a model insect, B. mori. Both the genome-wide transcription profile and DNA methylation patterns of silkworm display tissue-specificity, which is altered upon BmCPV challenge. The rate of genomic CG methylation in the midgut and fat body is much higher than in the silk gland. CG methylation is predominantly in gene bodies, and could either positively or negatively regulate gene expression. Both gene expression level and DNA methylation level can be changed upon BmCPV infection. The mechanism of alteration of gene expression by BmCPV invasion is complicated and not clear until now. It may be regulated probably by non-coding RNA, histone modification and so on. Here, we found 27 DEGs were also DMGs, suggesting that expression level of these genes may be changed via variation in DNA methylation. It also indicated that DNA methylation may be play roles in host-pathogen interaction in insects. Further experiments need to be carried out for demonstrating how the DNA methylation would mediate the expression of important genes involved in response to BmCPV infection. On the other hand, we also found most part of DMRs do not correspond to any DEGs, which may imply that DNA methylation could also be an independent molecular pathway in response to viral infection in B. mori.
Our study is a valuable resource to explore the underlying mechanism of tissue-specific infection of BmCPV, and adds epigenetic modification as a new dimension to host-pathogen interaction in insects. Further experiments need to be carried out for demonstrating how the DNA methylation would mediate the expression of important genes involving in BmCPV infection.
Materials and Methods
Silkworm strain and virus inoculation
Domesticated silkworm strain 4008, European monovoltin and susceptible to BmCPV, was used in this study. They were reared at 25 °C, 80 ± 5% relative humidity under a photoperiod of 12 h of light and 12 h of dark up to fourth molting. Each larva was inoculated about 1 × 106 BmCPV polyhedrons by oral infection. The detailed process about BmCPV inoculation was described in our previous study8.
Confirmation of virus infection
Virus infection was confirmed in the following manners: (1) inoculated silkworm larvae displaying the typical symptom of white wrinkles on the posterior midgut, (2) observation of polyhedra under a microscope, and (3) presence of the BmCPV polyhedrons gene by qRT-PCR.
Sample preparation for RNA-Seq and WGBS
The midgut and fat body of both BmCPV-infected and control larvae (uninfected) were collected at 96 h post-inoculation by dissecting the larvae on ice. Isolated tissues were then quickly rinsed in 0.8% diethylpyrocarbonate (DEPC)-treated physiologicsaline solution and frozen in liquid nitrogen and powdered. Then, the powdered samples in both infected and control groups were individually pooled into three groups for biological replication.
Half of the sample from each group was used to extract the total RNA by using the Trizol reagent (Invitrogen, USA) and for RNA-Seq assay. DNA was isolated from the other half by using DNeasy Blood and Tissue Kit (Qiagen, Germen) and for WGBS assay.
Transcriptome analysis
RNA-Seq experiment was performed by Beijing Genomics Institute (Shenzhen, China). The process is described briefly as follows: firstly, the mRNA was enriched by using the oligo (dT) magnetic beads and fragmented at an elevated temperature. The double strand cDNA was synthesized and purified for each targeted fragments (200–500 bp). Then, the fragments were ligated to sequencing adaptors and enriched by PCR amplification. When the necessary quality control steps were passed, the library products were sequenced via Illumina Hiseq. 4000 platform. Raw reads were filtered into clean reads after removing adaptor sequences, reads consisting more than 10% unknown nucleotides, and low quality reads (more than 20% base Q ≤ 20). Clean reads were mapped to silkworm mRNA (B. mori assembly ASM15162v1) using Bowtie2 (version 2.2.5)59. Differentially expressed genes were identified by EBseq (version 1.4.0)60 with the screened criteria as fold change ≥ 2 and PPEE ≤ 0.05. Gene ontology (GO) annotation was performed by mapping genes to GO terms (http://www.geneontology.org), and GO enrichment analysis of differentially expressed genes (DEGs) was conducted by hypergeometric test based on ‘GO::TermFinder’ (http://www.yeastgenome.org/help/analyze/go-term-finder). The calculated p-value was processed by using Bonferroni Correction and the corrected p-value ≤ 0.05 was used as threshold. KEGG pathway annotation and enrichment analysis were performed similar to GO analysis.
Whole genome bisulfite sequencing
For whole-genome bisulfite sequencing (WGBS), the DNA was fragmented by sonication using a Bioruptor (Diagenode, Belgium) to a mean size of approximately 250 bp, followed by adding dA to 3′ end by bluntend cloning and ligating methylated adaptors. Ligated DNA was bisulfite converted using the EZ DNA Methylation-Gold kit (ZYMO, USA). Different lengths of DNA fragments were excised from a 2% agarose gel and purified and amplified by PCR. Finally, sequencing was performed using the Illumina Hiseq. 4000 platform.
After filtering, the clean data was mapped to the silkworm genome (B. mori assembly ASM15162v1) by using the whole genome bisulfite sequencing mapping program (BSMAP, v2.74)61. Then, the mapping rate and bisulfite conversion rate was calculated for each sample.
The methylation level was determined by dividing the number of reads covering each mC by the total number of reads covering that cytosine, which was also equal to the mC/C ratio at each reference cytosine62.
Identification of differentially methylated regions (DMRs)
Putative DMRs were identified by comparing methylomes from the infected and control tissue using windows that contained at least 5 CpG sites based on the following standards: (1) the whole length of DMRs is ≥ 200 bp; (2) the average methylation rate is >0.2; (3) putative DMRs has 6 × sequencing coverage; and (4) 2-fold change in methylation level and Fisher test p value ≤ 0.05. In addition, two nearby DMRs were considered as one continuous DMR if the genomic region from the start of an upstream DMR to the end of a downstream DMR also had 2-fold methylation level with a p value <= 0.05. The final dataset of DMRs was made with those that were independent from each other. GO and KEGG pathway analysis of genes containing DMRs were performed as described in the “Transcriptome analysis” section.
Methylated regions were identified by CpG_MPs (v1.1.0)63 with the required sequencing depth great than 10 and average C methylation ratio greater than 0.55, and then converted to wig format by using perl script for visualization on IGV (Integrative Genomics Viewer)64.
qRT-PCR Analysis
The total RNA for RNA-Seq analysis was also used for qRT-PCR. qRT-PCR was performed according to the manufacturer’s instructions of the SYBR Premix Ex TaqTM Kit (TaKaRa, China) and run on ABI 7300 machine (Applied Biosystems, USA) with thermal cycling parameters at 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s, 60 °C for 31 s. Following amplification, melting curves were constructed. Three independent experiments with three technical replicates were performed. Data were analyzed and normalized to control gene transcript levels. A relative quantitative method (2−ΔΔCt) and the student’s t test were used to evaluate relative expression differences. All of the primers were listed in Supplementary Table S9.
Bisulfite-PCR validation of DMRs
Genomic DNA from infected and control tissues was prepared for bisulfite conversion as described above. Bisulfite converted DNA was amplified by PCR with Premix Ex Taq™ Hot Start Version (TaKaRa, Japan) according to the manufacturer’s instructions. Nested primers for BS-PCR were designed using the online MethPrimer program (S9 Table). PCR products were purified and cloned into the pMD19-T vector (TaKaRa, Japan). Ten different clones were then sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequencing. DNA methylation of individual CpG sites was analyzed using Quma software (http://quma.cdb.riken.jp/).
Electronic supplementary material
Acknowledgements
The authors acknowledge the financial support of the National Nature Science Foundation of China (Grant Nos 31572463, 31502016); the Natural Science Foundation of Jiangsu Province, China (No. BK20141286) and the National Basic Research Program of China (Grant No. 2012CB114600).
Author Contributions
P.W. and X.G. designed research; P.W., W.J., and T.C. analyzed data; P.W., S.Q. and E.A. performed experiments; J.X. and C.H. prepared the experimental materials; P.W., W.J., and X.G. wrote the manuscript. All authors have read and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Ping Wu and Wencai Jie contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16357-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Magnoler A. Effects of a cytoplasmic polyhedrosis on larval and postlarval stages of the gypsy moth, Porthetria dispar. J Invertebr Pathol. 1974;23:263–274. doi: 10.1016/0022-2011(74)90089-5. [DOI] [PubMed] [Google Scholar]
- 2.Rubinstein R, Harley EH, Losman M, Lutton D. The nucleic acids of viruses infecting Heliothis armigera. Virology. 1976;69:323–326. doi: 10.1016/0042-6822(76)90219-1. [DOI] [PubMed] [Google Scholar]
- 3.Hill CL, et al. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat Struct Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 4.Cao G, et al. Characterization of the complete genome segments from BmCPV-SZ, a novel Bombyx mori cypovirus 1 isolate. Can J Microbiol. 2012;58:872–883. doi: 10.1139/w2012-064. [DOI] [PubMed] [Google Scholar]
- 5.Guo R, et al. The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Appl Microbiol Biotechnol. 2015;99:5175–5187. doi: 10.1007/s00253-015-6634-x. [DOI] [PubMed] [Google Scholar]
- 6.Gao K, et al. Cytoplasmic polyhedrosis virus-induced differential gene expression in two silkworm strains of different susceptibility. Gene. 2014;539:230–237. doi: 10.1016/j.gene.2014.01.073. [DOI] [PubMed] [Google Scholar]
- 7.Wu P, et al. Microarray analysis of the gene expression profile in the midgut of silkworm infected with cytoplasmic polyhedrosis virus. Mol Biol Rep. 2011;38:333–341. doi: 10.1007/s11033-010-0112-4. [DOI] [PubMed] [Google Scholar]
- 8.Wu P, et al. Involvement of microRNAs in infection of silkworm with bombyx mori cytoplasmic polyhedrosis virus (BmCPV) PLoS One. 2013;8:e68209. doi: 10.1371/journal.pone.0068209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu P, Qin G, Qian H, Chen T, Guo X. Roles of miR-278-3p in IBP2 regulation and Bombyx mori cytoplasmic polyhedrosis virus replication. Gene. 2016;575:264–269. doi: 10.1016/j.gene.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Pan ZH, et al. Identification and characterization of two putative microRNAs encoded by Bombyx mori cypovirus. Virus Res. 2017;233:86–94. doi: 10.1016/j.virusres.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Yang, X. et al. Lack of Connection Between Midgut Cell Autophagy Gene Expression and BmCPV Infection in the Midgut of Bombyx mori. J Insect Sci15, 10.1093/jisesa/iev047 (2015). [DOI] [PMC free article] [PubMed]
- 12.Zhang Y, et al. Integrin beta and receptor for activated protein kinase C are involved in the cell entry of Bombyx mori cypovirus. Appl Microbiol Biotechnol. 2017;101:3703–3716. doi: 10.1007/s00253-017-8158-z. [DOI] [PubMed] [Google Scholar]
- 13.Jair KW, et al. De novo CpG island methylation in human cancer cells. Cancer Res. 2006;66:682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- 14.Corley MJ, et al. Comparative DNA Methylation Profiling Reveals an Immunoepigenetic Signature of HIV-related Cognitive Impairment. Sci Rep. 2016;6:33310. doi: 10.1038/srep33310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, Y. H. et al. Changes in methylation of genomic DNA from chicken immune organs in response to H5N1 influenza virus infection. Genet Mol Res1510.4238/gmr.15037382 (2016). [DOI] [PubMed]
- 16.Matsha TE, et al. Genome-Wide DNA Methylation in Mixed Ancestry Individuals with Diabetes and Prediabetes from South Africa. 2016;2016:3172093. doi: 10.1155/2016/3172093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez Sanchez A, Stassen JH, Furci L, Smith LM, Ton J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016;88:361–374. doi: 10.1111/tpj.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Functional CpG methylation system in a social insect. Science. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- 19.Glastad KM, Hunt BG, Yi SV, Goodisman MA. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol. 2011;20:553–565. doi: 10.1111/j.1365-2583.2011.01092.x. [DOI] [PubMed] [Google Scholar]
- 20.Wurm Y, et al. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci USA. 2011;108:5679–5684. doi: 10.1073/pnas.1009690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marhold J, et al. Conservation of DNA methylation in dipteran insects. Insect Mol Biol. 2004;13:117–123. doi: 10.1111/j.0962-1075.2004.00466.x. [DOI] [PubMed] [Google Scholar]
- 22.Mitsudome T, et al. Biochemical characterization of maintenance DNA methyltransferase DNMT-1 from silkworm, Bombyx mori. Insect Biochem Mol Biol. 2015;58:55–65. doi: 10.1016/j.ibmb.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Beeler SM, et al. Whole-genome DNA methylation profile of the jewel wasp (Nasonia vitripennis) G3 (Bethesda) 2014;4:383–388. doi: 10.1534/g3.113.008953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonasio R, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang H, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nature biotechnology. 2010;28:516–520. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- 26.Mandrioli M, Volpi N. The genome of the lepidopteran Mamestra brassicae has a vertebrate-like content of methyl-cytosine. Genetica. 2003;119:187–191. doi: 10.1023/A:1026016021415. [DOI] [PubMed] [Google Scholar]
- 27.Shi YY, et al. Genomewide analysis indicates that queen larvae have lower methylation levels in the honey bee (Apis mellifera) Naturwissenschaften. 2013;100:193–197. doi: 10.1007/s00114-012-1004-3. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, et al. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 2013;9:e1003872. doi: 10.1371/journal.pgen.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patalano S, Hore TA, Reik W, Sumner S. Shifting behaviour: epigenetic reprogramming in eusocial insects. Curr Opin Cell Biol. 2012;24:367–373. doi: 10.1016/j.ceb.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, Chen J, Liang MS, Wang F. Differential DNA methylation between two wing phenotypes adults of Sogatella furcifera. Genesis. 2013;51:819–826. doi: 10.1002/dvg.22722. [DOI] [PubMed] [Google Scholar]
- 31.Bigot Y, Jegot G, Casteret S, Aupinel P, Tasei JN. DNA modifications and genome rearrangements during the development and sex differentiation of the bumble bee Bombus terrestris. Insect Mol Biol. 2011;20:165–175. doi: 10.1111/j.1365-2583.2010.01052.x. [DOI] [PubMed] [Google Scholar]
- 32.Field LM. Methylation and expression of amplified esterase genes in the aphid Myzus persicae (Sulzer) Biochem J. 2000;349(Pt 3):863–868. doi: 10.1042/bj3490863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galbraith DA, Yang X, Nino EL, Yi S, Grozinger C. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera) PLoS Pathog. 2015;11:e1004713. doi: 10.1371/journal.ppat.1004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen SH, et al. DNA methylation alterations in the pathogenesis of lupus. Clin Exp Immunol. 2017;187:185–192. doi: 10.1111/cei.12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li SC, et al. Major methylation alterations on the CpG markers of inflammatory immune associated genes after IVIG treatment in Kawasaki disease. BMC Med Genomics. 2016;9(Suppl 1):37. doi: 10.1186/s12920-016-0197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regulski M, et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013;23:1651–1662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang H, et al. Comparative methylomics between domesticated and wild silkworms implies possible epigenetic influences on silkworm domestication. BMC Genomics. 2013;14:646. doi: 10.1186/1471-2164-14-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks WS, Banerjee S, Crawford DF. G2E3 is a nucleo-cytoplasmic shuttling protein with DNA damage responsive localization. Exp Cell Res. 2007;313:665–676. doi: 10.1016/j.yexcr.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks WS, et al. G2E3 is a dual function ubiquitin ligase required for early embryonic development. J Biol Chem. 2008;283:22304–22315. doi: 10.1074/jbc.M803238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt F, Kunze M, Loock AC, Dobbelstein M. Screening analysis of ubiquitin ligases reveals G2E3 as a potential target for chemosensitizing cancer cells. Oncotarget. 2015;6:617–632. doi: 10.18632/oncotarget.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung FF, et al. Jerantinine A induces tumor-specific cell death through modulation of splicing factor 3b subunit 1 (SF3B1) Sci Rep. 2017;7:42504. doi: 10.1038/srep42504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, W. et al. SF3B4 is decreased in pancreatic cancer and inhibits the growth and migration of cancer cells. Tumour Biol39, 1010428317695913, 10.1177/1010428317695913 (2017). [DOI] [PubMed]
- 44.Gao Y, Trivedi S, Ferris RL, Koide K. Regulation of HPV16 E6 and MCL1 by SF3B1 inhibitor in head and neck cancer cells. Sci Rep. 2014;4:6098. doi: 10.1038/srep06098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D, Padiernos E, Ding F, Lossos IS, Lopez CD. Apoptosis-stimulating protein of p53-2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ. 2005;12:358–368. doi: 10.1038/sj.cdd.4401536. [DOI] [PubMed] [Google Scholar]
- 46.Wang B, et al. [ASPP2 inhibits oxaliplatin-induced autophagy and promotes apoptosis of colon cancer cells] Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2015;31:898–904. [PubMed] [Google Scholar]
- 47.Schittenhelm MM, et al. Attenuated expression of apoptosis stimulating protein of p53-2 (ASPP2) in human acute leukemia is associated with therapy failure. PLoS One. 2013;8:e80193. doi: 10.1371/journal.pone.0080193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baumgartner R, Stocker H, Hafen E. The RNA-binding proteins FMR1, rasputin and caprin act together with the UBA protein lingerer to restrict tissue growth in Drosophila melanogaster. PLoS Genet. 2013;9:e1003598. doi: 10.1371/journal.pgen.1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong L, et al. Growth suppressor lingerer regulates bantam microRNA to restrict organ size. J Mol Cell Biol. 2015;7:415–428. doi: 10.1093/jmcb/mjv045. [DOI] [PubMed] [Google Scholar]
- 50.Xia C, et al. The Arabidopsis eukaryotic translation initiation factor 3, subunit F (AteIF3f), is required for pollen germination and embryogenesis. Plant J. 2010;63:189–202. doi: 10.1111/j.1365-313X.2010.04237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JY, Kim HJ, Rho SB, Lee SH. eIF3f reduces tumor growth by directly interrupting clusterin with anti-apoptotic property in cancer cells. Oncotarget. 2016;7:18541–18557. doi: 10.18632/oncotarget.8105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Qi C, Wang A, Li L, Xu Y. High expression of nucleobindin 2 mRNA: an independent prognostic factor for overall survival of patients with prostate cancer. Tumour Biol. 2014;35:2025–2028. doi: 10.1007/s13277-013-1268-z. [DOI] [PubMed] [Google Scholar]
- 53.Kan JY, et al. Nesfatin-1/Nucleobindin-2 enhances cell migration, invasion, and epithelial-mesenchymal transition via LKB1/AMPK/TORC1/ZEB1 pathways in colon cancer. Oncotarget. 2016;7:31336–31349. doi: 10.18632/oncotarget.9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tagaya Y, Miura A, Okada S, Ohshima K, Mori M. Nucleobindin-2 is a positive modulator of EGF-dependent signals leading to enhancement of cell growth and suppression of adipocyte differentiation. Endocrinology. 2012;153:3308–3319. doi: 10.1210/en.2011-2154. [DOI] [PubMed] [Google Scholar]
- 55.Saito T, et al. Nucleobindin-2 is a positive regulator for insulin-stimulated glucose transporter 4 translocation in fenofibrate treated E11 podocytes. Endocr J. 2014;61:933–939. doi: 10.1507/endocrj.EJ14-0330. [DOI] [PubMed] [Google Scholar]
- 56.Nirmala X, Kodrik D, Zurovec M, Sehnal F. Insect silk contains both a Kunitz-type and a unique Kazal-type proteinase inhibitor. Eur J Biochem. 2001;268:2064–2073. doi: 10.1046/j.1432-1327.2001.02084.x. [DOI] [PubMed] [Google Scholar]
- 57.Zheng QL, et al. Expression, purification and characterization of a three-domain Kazal-type inhibitor from silkworm pupae (Bombyx mori) Comp Biochem Physiol B Biochem Mol Biol. 2007;146:234–240. doi: 10.1016/j.cbpb.2006.10.106. [DOI] [PubMed] [Google Scholar]
- 58.Sarda S, Zeng J, Hunt BG, Yi SV. The evolution of invertebrate gene body methylation. Mol Biol Evol. 2012;29:1907–1916. doi: 10.1093/molbev/mss062. [DOI] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su J, et al. CpG_MPs: identification of CpG methylation patterns of genomic regions from high-throughput bisulfite sequencing data. Nucleic Acids Res. 2013;41:e4. doi: 10.1093/nar/gks829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.